Abstract
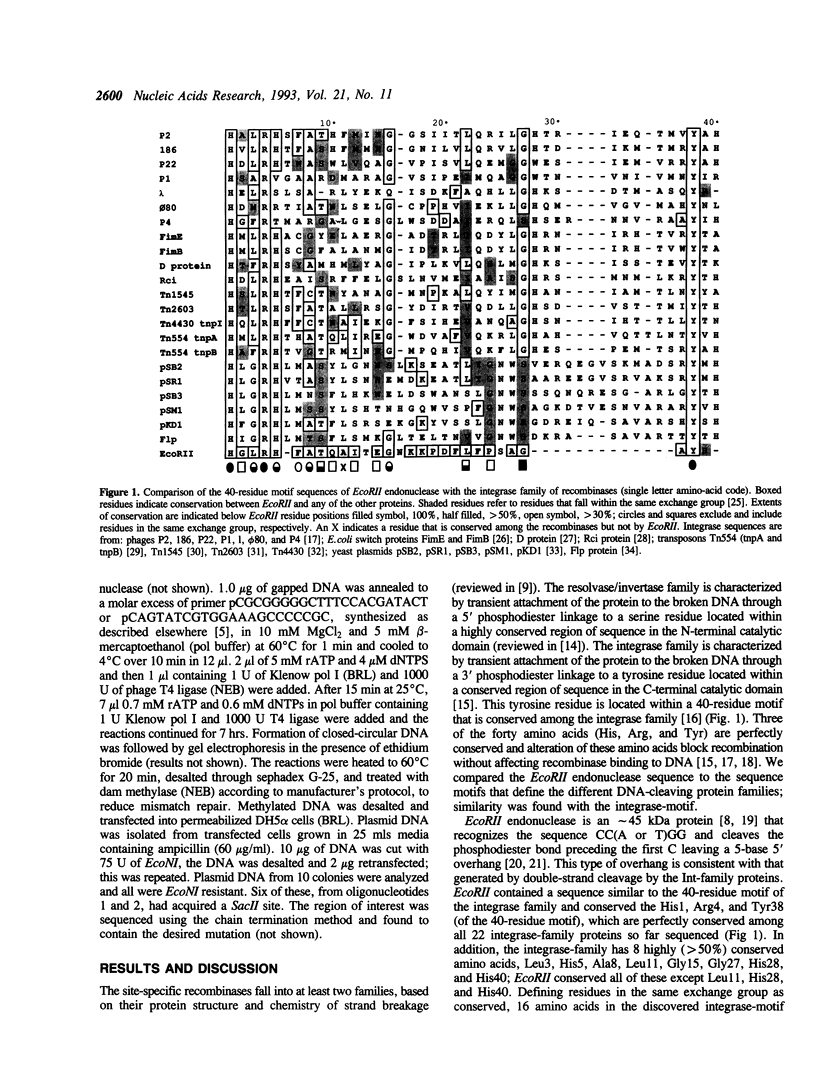
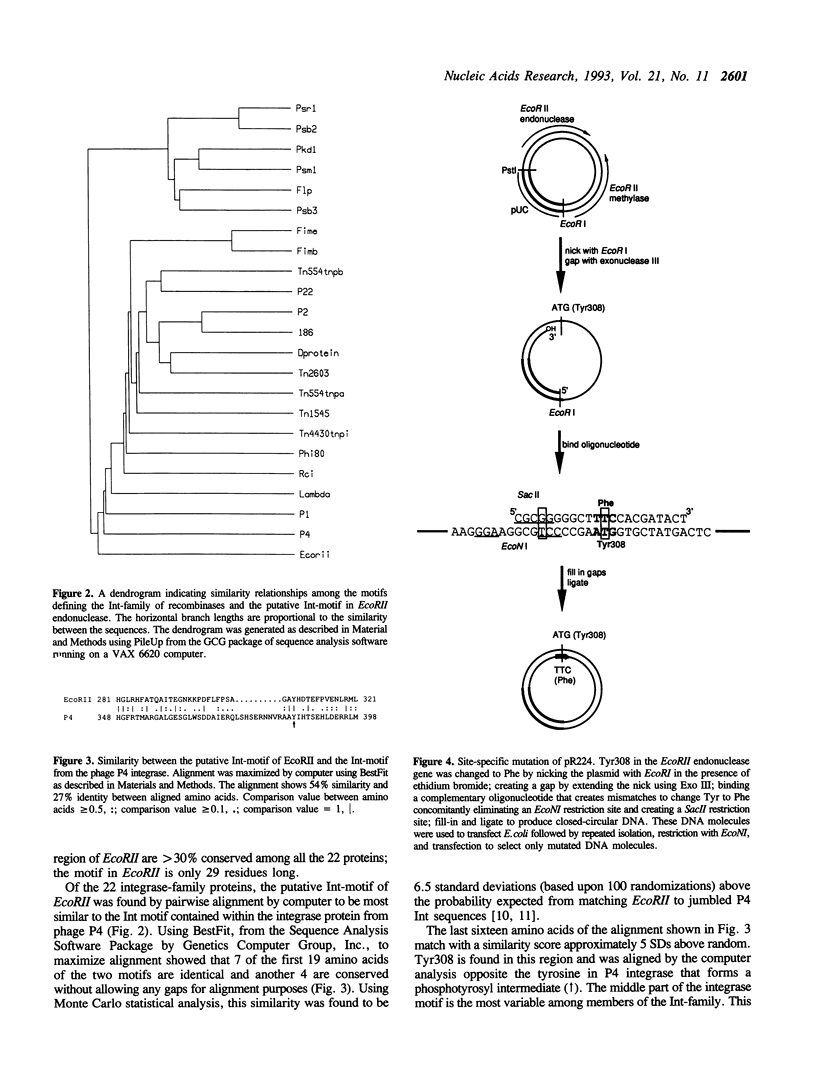
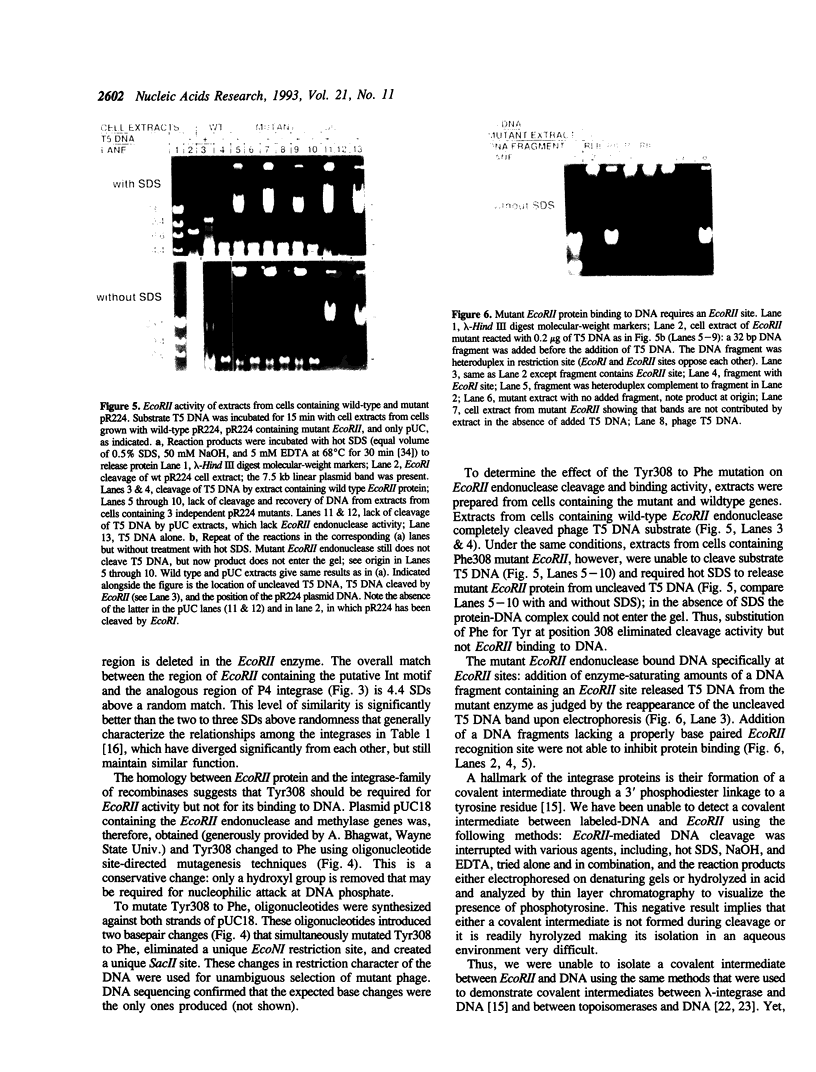
Endonuclease EcoRII is one of a group of type II restriction enzymes, including Nael, Narl, BspMI, HpaII, and SacII, that require binding of an enhancer sequence to cleave DNA. Comparison of the EcoRII amino-acid sequence with the amino-acid consensus motifs that differentiate between recombinase families uncovered similarity between a 29 amino-acid sequence in the carboxyl end of EcoRII and the motif defining the integrase family of recombinases. This similarity implied that EcoRII tyrosine 308 should be involved in catalyzing hydrolysis of the scissile bond. Site-directed mutagenesis was used to mutate Tyr308 to Phe. The phenylalanine-substituted enzyme could not cleave T5 DNA under conditions in which wild-type enzyme completely cleaved this DNA. The Tyr308 to Phe mutation abolished cleavage activity but not specific binding to DNA. No evidence was found for the existence during the cleavage reaction of a covalent linkage between Tyr308 and DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat A. S., Johnson B., Weule K., Roberts R. J. Primary sequence of the EcoRII endonuclease and properties of its fusions with beta-galactosidase. J Biol Chem. 1990 Jan 15;265(2):767–773. [PubMed] [Google Scholar]
- Bigger C. H., Murray K., Murray N. E. Recognition sequence of a restriction enzyme. Nat New Biol. 1973 Jul 4;244(131):7–10. doi: 10.1038/newbio244007a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Conrad M., Topal M. D. DNA and spermidine provide a switch mechanism to regulate the activity of restriction enzyme Nae I. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9707–9711. doi: 10.1073/pnas.86.24.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Gabbara S., Bhagwat A. S. Interaction of EcoRII endonuclease with DNA substrates containing single recognition sites. J Biol Chem. 1992 Sep 15;267(26):18623–18630. [PubMed] [Google Scholar]
- Hall R. M., Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987 Sep 25;15(18):7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986 Jun;5(6):1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Barcak G. J., Reuter M., Smith H. O. EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res. 1988 May 11;16(9):3997–4008. doi: 10.1093/nar/16.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Kusukawa A., Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988 Jul;213(1):30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- Lane D., de Feyter R., Kennedy M., Phua S. H., Semon D. D protein of miniF plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 1986 Dec 22;14(24):9713–9728. [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Wilbur W. J., Smith T. F., Waterman M. S. On the statistical significance of nucleic acid similarities. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):215–226. doi: 10.1093/nar/12.1part1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J., Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988 May;7(5):1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Weber H., Meyer F., Weissmann C. Site-directed mutagenesis in DNA: generation of point mutations in cloned beta globin complementary dna at the positions corresponding to amino acids 121 to 123. J Mol Biol. 1978 Sep 15;124(2):343–358. doi: 10.1016/0022-2836(78)90303-0. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Oller A. R., Vanden Broek W., Conrad M., Topal M. D. Ability of DNA and spermidine to affect the activity of restriction endonucleases from several bacterial species. Biochemistry. 1991 Mar 5;30(9):2543–2549. doi: 10.1021/bi00223a035. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989 Aug;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T. C., Tewey K. M., Liu L. F. Identification of the breakage-reunion subunit of T4 DNA topoisomerase. J Biol Chem. 1984 Jul 25;259(14):9177–9181. [PubMed] [Google Scholar]
- Schiestl R. H., Petes T. D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D., Thresher R. J., Conrad M., Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991 Feb 19;30(7):2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- Tse Y. C., Kirkegaard K., Wang J. C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980 Jun 25;255(12):5560–5565. [PubMed] [Google Scholar]
- Utatsu I., Sakamoto S., Imura T., Toh-e A. Yeast plasmids resembling 2 micron DNA: regional similarities and diversities at the molecular level. J Bacteriol. 1987 Dec;169(12):5537–5545. doi: 10.1128/jb.169.12.5537-5545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A., Kendall M., Abremski K., Hoess R. A mutational analysis of the bacteriophage P1 recombinase Cre. J Mol Biol. 1987 Jun 20;195(4):785–794. doi: 10.1016/0022-2836(87)90484-0. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Topal M. D. Nonidentical DNA-binding sites of endonuclease NaeI recognize different families of sequences flanking the recognition site. Biochemistry. 1992 Oct 13;31(40):9657–9664. doi: 10.1021/bi00155a019. [DOI] [PubMed] [Google Scholar]




