Abstract
Noncoding RNAs have drawn significant attention in biology recently. Whereas the current research is highly inclined to microRNAs, research on other noncoding RNAs has lagged behind. Here, we investigated a novel noncoding RNA that has been known as precursor microRNA miR-886 (pre-miR-886). Pre-miR-886 has been proposed also as a vault RNA, a component of the vault complex implicated in cancer drug resistance. We identified pre-miR-886 as a 102-nucleotide-long, abundant cytoplasmic RNA that is neither a genuine pre-microRNA nor a vault RNA. Pre-miR-886 is physically associated with PKR (Protein Kinase RNA-activated), an interferon-inducible and double-stranded RNA dependent kinase. The suppression of pre-miR-886 activates PKR and its downstream pathways, eIF2α phosphorylation and the NF-κB pathway, leading to impaired cell proliferation. We also found that pre-miR-886 is suppressed in a wide-range of cancer cell lines and in clinical specimens. This study is the first intense characterization of pre-miR-886 as well as the initial report on its function as a PKR regulator, which suggests a critical role in tumorigenesis.
Keywords: cancer, microRNA, noncoding RNA, PKR, vault RNA
INTRODUCTION
It has been noted that most of the mammalian genome is transcribed (Carninci et al. 2005) and most of the transcripts are noncoding, given that protein coding open reading frames comprise only ∼1.2% of the human genome (Lander et al. 2001; Venter et al. 2001). Thus, noncoding RNAs (ncRNAs) have gained significant attention recently. Besides classic ncRNAs such as rRNA, tRNA, small nuclear RNA, small nucleolar RNA, etc., new types of ncRNAs are being discovered through ultra-high-throughput sequencing technology. While small noncoding regulatory RNAs, especially microRNAs (miRNAs), are dominating the current literature, research on other ncRNAs with a size range of 40 nucleotides (nt) or longer has lagged behind. The main reason for this focus is that the role of a miRNA can be inferred from its sequence. Using several target-prediction databases (Min and Yoon 2010), researchers can easily identify miRNA(s) that may regulate their gene of interest. In addition, techniques for miRNA study, such as profiling and overexpression or suppression of miRNAs, have been well established. Exactly the opposite reasons explain why longer ncRNAs have not yet been studied as extensively.
Besides classic ncRNAs and miRNAs, many noncoding transcripts are found to exist in the nucleus (Cheng et al. 2005), and a few have been shown to play nuclear roles such as chromatin modification, regulation of transcription, etc. (for review, see Mercer et al. 2009). Compared to these nuclear roles, the functions of cytoplasmic nonclassical ncRNA have been studied less extensively. Some noncoding transcripts are antisense to known mRNAs and so are suggested to regulate the sense mRNAs at the translational stage; however, this function is still elusive due to lack of solid experimental evidences (for review, see Faghihi and Wahlestedt 2009). Apart from antisense transcripts, human cells express a handful of “well-defined” cytoplasmic ncRNAs, including vault RNAs (vtRNAs), BC RNAs (brain cytoplasmic RNAs) (Tiedge et al. 1993), Y RNA (Lerner et al. 1981), etc. Some of these have been shown to exhibit a specific expression pattern and to be functional. For example, BC RNAs are thought to modulate local protein synthesis in neuronal cells (Wang et al. 2002) and Y RNA is implicated in RNA quality control (Stein et al. 2005).
In this study, we have investigated precursor miR-886 (pre-miR-886). Pre-miR-886 was originally registered in the miRNA database (miRbase; http://www.mirbase.org/) because the mature miR-886-5p and -886-3p were captured in high-throughput sequencing and the two mature miRNAs formed a stem in a predicted stem–loop hairpin structure (Fig. 1A), which is a signature of a miRNA precursor (Landgraf et al. 2007; Yang et al. 2010). It has also recently been proposed that miR-886 is one of the vtRNAs. vtRNAs are components of the vault complex, a hollow, barrel-shaped ribonucleoprotein complex that is thought to play a role in cellular resistance to cancer therapeutic drugs (for review, see Berger et al. 2009). However, the exact role of vtRNAs, as well as the vault complex, is not yet clear. In humans there are three vtRNA genes clustered in chromosome 5 (vtRNA1-1, 1-2, and 1-3) and a pseudogene in chromosome X (van Zon et al. 2001). The human vtRNAs are 88 or 98 nt in length, are RNA polymerase III (Pol III) transcripts, and have a stem–loop secondary structure. All these features are shared by pre-miR-886, and for this reason it was recently proposed to be renamed as vtRNA2-1 (Nandy et al. 2009; Stadler et al. 2009).
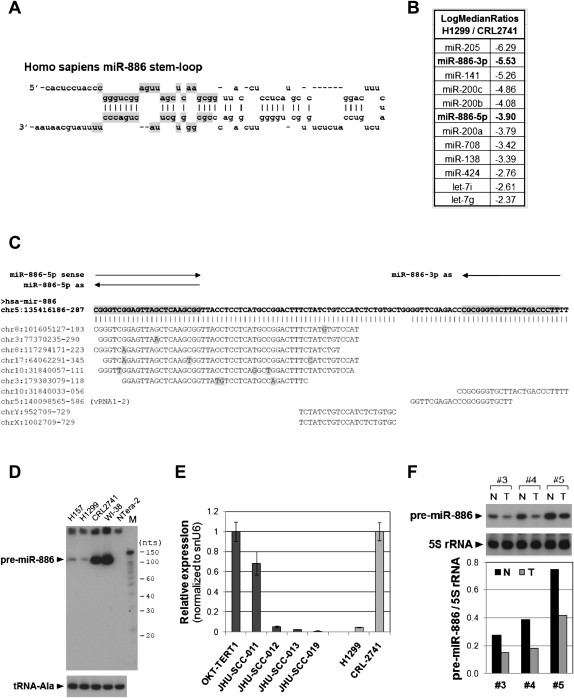
FIGURE 1.
Reduced expression of pre-miR-886 in cancer cell lines and patient specimens. (A) The stem–loop structures of pre-miR-886 (adapted from the miRbase: http://www.mirbase.org/). The mature miR-886-5p and -886-3p portions (top and bottom, respectively), registered in the miRbase from high-throughput sequencing data, are shaded in gray. (B) From the miRNA array data, miRNAs were sorted according to the expression ratio of H1299 to CRL2741, and the 12 miRNAs with the greatest decreases were tabulated as shown. miR-886-5p and -886-3p are highlighted in bold letters. (C) BLAT analysis (using the UCSC genome browser) of pre-miR-886 (with the mature miRNA portions gray highlighted) showing the alignment with genomic loci having identical or highly similar sequences. Mismatched nucleotides are highlighted in gray. The nucleotide coordinates are based on the February 2009 assembly (GRCh37/hg19). Northern probes and qRT-PCR primers are indicated by arrows. (D) Northern hybridization showing pre-miR-886 in indicated cell lines, using “miR-886-3p as” (panel C) as a probe. tRNA–Ala was measured as a loading control. “M” denotes 5′-32P-labeled Decade markers (Applied Biosystems/Ambion); their molecular sizes in nucleotides are shown on the right. (E) qRT-PCR measurement of pre-miR-886 in indicated cell lines (x-axis), using “miR-886-5p sense” and “miR-886-3p as” as primers (panel C). Each value was normalized to that of snU6 (y-axis). For the five oral cell lines, arranged in order of increasing capability of tumorigenesis and metastasis, the value of OKT-TERT1 was set as 1. To validate qRT-PCR assays, H1299 and CRL2741 were included for comparison to panel D. In this pair, the value of CRL2741 was set as 1. (F) Northern hybridization of pre-miR-886 in three pairs of a prostate tumor specimen (T) and the surrounding normal tissue control (N), with 5S rRNA as a loading control. The blot was exposed to the phosphorimager Storm 860 (Molecular Dynamics) and each band was quantified with ImageQuant 5.0 software. The intensity of pre-miR-886 was normalized to that of 5S rRNA (y-axis).
PKR, an interferon-inducible and double-stranded RNA (dsRNA) dependent kinase, is well known for its role in the host defense against viral infection (for review, see Garcia et al. 2007). Upon binding to dsRNA that is a replication intermediate of viruses, PKR undergoes auto-phosphorylation. The phosphorylated PKR, which is an active kinase, phosphorylates its downstream substrates such as eIF2α. Phosphorylation of eIF2α results in shutdown of global protein synthesis, leading to cell death. PKR is also implicated in diverse signaling pathways, for example the NF-κB pathway as the most extensively studied one, and plays critical roles in cell proliferation and apoptosis. Several lines of evidence suggest that PKR is a tumor suppressor (Koromilas et al. 1992; Meurs et al. 1993). However, the molecular mechanism how PKR contributes to the tumorigenesis is not clear yet, although its tumor suppressive role is thought to be attributed to the anti-proliferative activity of PKR.
In this paper, we have identified pre-miR-886 as a new type of ncRNA that is not a canonical miRNA or a vtRNA. We also measured the expression of pre-miR-886 in several cancer cells and revealed that it binds and inhibits PKR activity. This is the first intense characterization of pre-miR-886, a putative tumor suppressive ncRNA.
RESULTS
Reduced expression of pre-miR-886 in cancer
In an attempt to obtain a miRNA expression profile in lung cancer, microarray hybridization was performed in the lung epithelial cell line CRL2741 and the lung cancer cell line H1299 (Fig. 1B). miR-205 and miR-200 family members (miR-200a, -200b, -200c, and -141) were the most decreased in H1299 relative to CRL2741. Our data are consistent with the previous literature that reported the down-regulation of these miRNAs in the epithelial-mesenchymal transition during tumor progression (for review, see Gregory et al. 2008). We also observed lower expression of let-7 miRNAs, whose down-regulation has been well documented in lung cancer (Lee and Dutta 2007). Besides these miRNAs, miR-886-3p and -886-5p were among the most suppressed miRNAs in H1299. We chose miR-886 for further study since it had not been examined in cancer.
In Northern hybridization with a probe specific to miR-886-3p (“miR-886-3p as” in Fig. 1C), we detected a band of ∼100 nt (Fig. 1D), rather than an expected band in the vicinity of a miRNA size (∼22 nt). We wanted to ensure the identity of the band, since our BLAT analysis (http://genome.ucsc.edu/) revealed that sequences identical or very similar to a portion of pre-miR-886 were duplicated in other genomic loci (Fig. 1C). For example, we could not rule out the technical possibility that the 100-nt band originated from the locus at chromosome 10 (nucleotide coordinate 31,840,033–056 in Fig. 1C). We proved that this was not the case: First, a probe specific to miR-886-5p (“miR-886-5p as” in Fig. 1C) yielded the same result as in Figure 1D (data not shown). Second, the ∼100-nt RNA was detected by qRT-PCR (Fig. 1E) using two primers designed for pre-miR-886 (“miR-886-5p sense” and “miR-886-3p as” in Fig. 1C). Third, our RACE (rapid amplification of cDNA ends) data provided unequivocal proof (shown later in Fig. 3C, see below). Thus, the 100-nt band is the pre-miR-886 in chromosome 5.
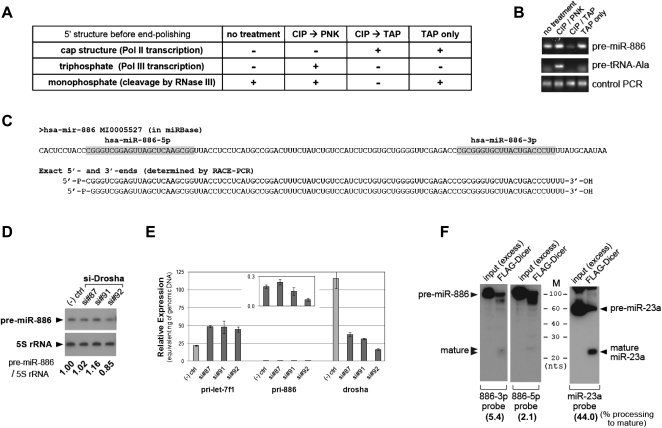
FIGURE 3.
Pre-miR-886 is distinct from a canonical pre-miRNA. (A) Expected 5′-RACE results tabulated for three different 5′-end structures of object RNA after four 5′-end polishing procedures. Before ligation to the 5′-linker oligoribonucleotide (see also Supplemental Fig. S3A for the 5′-RACE scheme), total RNA from CRL2741 (the object RNA) was treated with combinations of PNK (polynucleotide kinase), CIP (calf intestinal phosphatase), or TAP (tobacco acid pyrophosphatase). During 5′-RACE, only 5′-monophosphate of the object RNA can be ligated to the 3′-end of the 5′-linker, and this ligation is a prerequisite for subsequent PCR amplifications. mRNAs have a 5′-cap structure that is a substrate of TAP, which removes a cap and leaves 5′-monophosphate. To be converted to 5′-monophosphate, 5′-triphosphate of Pol III transcripts must first be dephosphorylated by CIP and then treated with PNK, which transfers a monophosphate group to a 5′-OH group (Rezaian 1999). (B) 5′-RACE was performed after the treatments described in panel A. After the second round of PCR, the products were resolved in a 2% agarose gel and visualized by EtBr staining. In comparison to the anticipated results in panel A, it is evident that pre-miR-886 has a 5′-monophosphate. Since the 5′-end of tRNA precursors are known to be triphosphates (Lee et al. 1987), we used the precursor tRNA–Ala (#66 tRNA at chromosome 6 in the tRNA database; http://gtrnadb.ucsc.edu/) as a control, to confirm polishing procedures. As expected from panel A, pre-tRNA–Ala was RACE-amplified only in lane 2 (CIP → PNK). Equal amounts of cDNA among the four samples were confirmed by PCR with two specific primers to pre-miR-886 (bottom panel “Control PCR,” see also Supplemental Fig. S3A). PCR products were cloned and sequenced to determine the precise 5′-end of pre-miR-886 (shown in panel C). (C) miR-886 in the miRbase (top) was aligned to experimentally determined pre-miR-886 sequences (bottom). Our 3′-RACE procedure depended on the 3′-hydroxyl group of object RNA (see Supplemental Fig. S3B and legend for details). Therefore, pre-miR-886 has 5′-monophosphate (5′-P-) and 3′-hydroxyl groups (-3′-OH). The mature miRNAs (in the miRbase) are in gray. Sequencing multiple RACE products revealed two different 5′-ends offset by 1 nt. (D) Northern hybridization after suppression of Drosha. Three siRNAs against Drosha (si#87, #91, #92) and a negative control siRNA were transfected into CRL2741 cells. Transfections were performed twice (at day 0 and 3), before harvesting cells at day 6. Each band was quantified with AlphaView software 2.0.1.1, and the intensity of pre-miR-886 relative to 5S rRNA was calculated (the numbers under each lane). The value of the negative control siRNA was set as 1. (E) qRT-PCR measurement of primary transcripts for let-7f and pre-miR-886 as well as Drosha mRNA. Titrating amounts of genomic DNA from CRL2741 cells were included in qRT-PCR, and Ct (cycle threshold) values from each amount were obtained. A regression line was drawn and used to convert a Ct value from cDNA into a nanogram (ng) amount. The expression levels of each cDNA were thus expressed as “equivalent ng of genomic DNA” (y-axis) and can be directly compared among different primer pairs. To detect primary transcripts for pre-miR-886 (pri-886), we used a reverse primer “miR-886-3p as” (Fig. 1C; Supplemental Table 2) combined with a forward primer (“miR-886-22-1” in Supplemental Table 2) corresponding to a sequence immediately upstream of pre-miR-886. The signal for pri-886 was very low and so was re-plotted against the magnified y-axis in a box within the main graph. (F) In vitro processing of pre-miR-886 (left panels) and pre-miR-23a (right panel) by recombinant Dicer. Cleaved products were detected by Northern hybridization with indicated probes. An excess amount of input was included to ensure the integrity of the synthetic RNA substrates prior to cleavage by Dicer. In each FLAG-Dicer lane, all bands (including the mature miRNA, the unprocessed substrate, and other degradation products) were quantified as described in panel D. Percent processing of mature miRNA (the intensity of mature miRNA relative to that of the sum of all the bands) was calculated and indicated at the bottom of each blot.
The level of pre-miR-886 was significantly lower in two lung cancer cell lines (H1299 and H157) than in CRL-2741 or WI-38 immortalized lung cells. This decrease was not only seen in lung cancer cells; pre-miR-886 was also barely detectable in an embryonic carcinoma cell line, NTera-2 (Fig. 1D), and in other malignant cell lines (Supplemental Fig. S1) such as T47D (breast), MCF-7 (breast), MDA-MB435 (breast or melanoma), and C33A (cervix). The decrease in pre-miR-886 was also evident in a battery of cell lines recapitulating the progression of oral cancer (Fig. 1E). The expression of pre-miR-886 was highest in nontumorigenic oral keratinocyte OKT-TERT1, moderately lower in the tumorigenic but nonmetastatic cell line JHU-SCC-011, and barely expressed in the metastatic cell lines JHU-SCC-012, -013, and -019. Pre-miR-886 was also significantly decreased in clinical specimens of prostate cancer (Fig. 1F). However, not all malignancies lacked pre-miR-886; its expression was evident in some cancer cells including MDA-MB-231 (breast), HeLa (cervix), and HCT116 (colon; Supplemental Fig. S1). On the other hand, pre-miR-886 was detectable in tissues from various human organs (Fig. 2A). We conclude that pre-miR-886 is repressed in a wide range of cancer cells, which suggests a tumor-suppressive role for pre-miR-886.
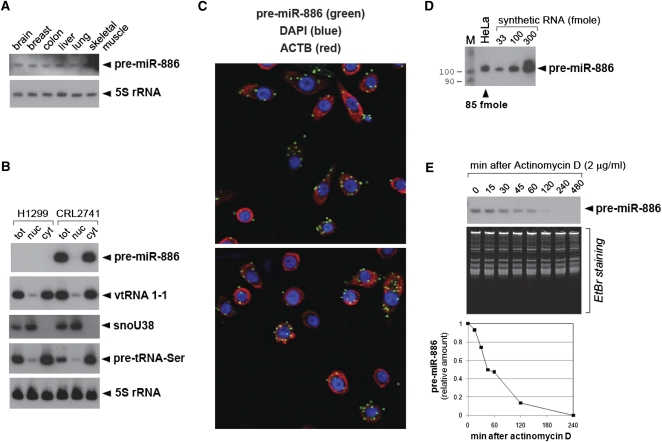
FIGURE 2.
The tissue expression, intracellular localization, copy number, and half-life of pre-miR-886. (A) Northern hybridization of pre-miR-886 in tissue RNAs. All other descriptions are the same as in Figure 1D,F. (B) Northern hybridization of pre-miR-886 and vtRNA1-1 in total (tot), nuclear (nuc), and cytoplasmic RNA (cyt). The quality of fractionation was verified using the nuclear marker snoU38 (Terns et al. 1995) and the cytoplasmic marker precursor tRNA-Ser (Lee et al. 2009). 5S rRNA is a loading control. (C) In situ detection of pre-miR-886 (green), together with DAPI staining (blue) and ACTB mRNA detection (red) for nuclear and cytoplasmic staining, respectively. Two representative fields are shown. (D) Northern hybridization of pre-miR-886 in the total RNA from 5 × 105 HeLa cells and indicated amounts of in vitro transcribed pre-miR-886. The lane “M” is the molecular size marker (as described in Fig. 1D). Each band was quantified with AlphaView software 2.0.1.1 (Alpha Innotech Corp.). A regression line was drawn using the values of the in vitro transcribed pre-miR-886 to estimate its intracellular amount in HeLa. Our calculation yielded 85 fmol of pre-miR-886 per 5 × 105 HeLa cells, which equals ∼105 copies per cell. (E) Northern hybridization of pre-miR-886 after treatment with actinomycin D, an inhibitor of transcription (top panel). EtBr (ethidium bromide) staining of total RNA is shown for equal loading. Each band was quantified as described in panel D, normalized to the intensity of the corresponding EtBr signal, and plotted (bottom panel). The value at time 0 was set as 1. The half-life of pre-miR-886 was estimated to be 45–60 min.
Characterization of pre-miR-886: Its intracellular localization and abundance
Pre-miR-886 is localized in the cytoplasm (Fig. 2B), where it was detected as discrete foci (Fig. 2C). For comparison, we did not observe foci of ACTB mRNA (for β-actin) whose signal was diffused throughout the cytoplasm. The foci formation of pre-miR-886 suggests that its localization is not random but regulated.
Pre-miR-886 is highly abundant; its intracellular quantity was estimated as ∼105 copies per cell in HeLa (see Fig. 2D and legend), CRL2741, and WI-38 (cf. Fig. 1D and Supplemental Fig. S1), which is comparable to those of abundant small nucleolar RNAs and small nuclear RNAs (Reddy and Busch 1988; Lim et al. 2003; Tycowski et al. 2006). For comparison, the copy numbers of the majority of miRNAs range up to ∼103 per cell, and those of the most abundant miRNAs are no more than ∼104 per cell (Lim et al. 2003; Lee et al. 2008; Lu and Tsourkas 2009).
Characterization of pre-miR-886: Is it a pre-miRNA?
Even after longer exposure to obtain a strong signal of pre-miR-886 (Supplemental Fig. S2A), our Northern hybridization did not detect mature miR-886-5p or -886-3p in any of the cell lines tested in this study, including 143B (a human osteosarcoma cell line) and HeLa cell lines where these mature miRNAs were captured in high-throughput sequencing (in the deepBase: http://deepbase.sysu.edu.cn/). This is atypical of most miRNAs (Supplemental Fig. S2B; cf. miR-205 as an example of typical miRNAs). The stem–loop hairpin structure of pre-miR-886 is also atypical in that (1) the stem of the two mature miRNAs harbors many mismatches and bulges, and (2) the loop linking the stem in pre-miR-886 is unusually long (Fig. 1A; also cf. Supplemental Fig. S2C and miR-205). One possible explanation is that pre-miR-886 is not a genuine precursor miRNA. The other possibility is that miR-886-5p and -886-3p are indeed generated from pre-miR-886, but their intracellular levels are too low to be detected by Northern hybridization due to inefficient Dicer processing.
To address this question, we began by characterizing pre-miR-886 and conducting functional assays, rather than simply running a more sensitive qRT-PCR assay to detect miR-886-5p and -886-3p in size-fractionated RNA. Because pre-miR-886 has a high steady-state intracellular level (Fig. 2D) and a short half-life (Fig. 2E; <1 h), it is likely that there are significant amounts of degradation byproducts during the rapid turnover of pre-miR-886. These byproducts could not be distinguished from the genuine mature miRNAs by qRT-PCR assays.
First, we determined the exact sequence of pre-miR-886 by 5′- and 3′-RACE (Fig. 3A–C). The end structure of pre-miR-886 features a Drosha product (Lee et al. 2003): (1) The 5′-end has a monophosphate group (Fig. 3A,B; Supplemental Fig. S3A) and the 3′-end has a hydroxyl group (Supplemental Fig. S3B); and (2) the actual ends correspond to the 5′- and 3′-ends of miR-886-5p and -886-3p (Fig. 3C). Recently, several groups clearly demonstrated that pre-miR-886 is transcribed by Pol III (Canella et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010). Our 3′-end mapping data are in agreement with Pol III transcription, as an oligo-dT stretch is the 3′-termination signal of Pol III (Hagenbuchle et al. 1979).
Notably, we found the 5′-end of pre-miR-886 to be a monophosphate, whereas the 5′-end of a Pol III primary transcript is a triphosphate (see Fig. 3A,B and legend). Relevant to our observation, Canella and colleagues detected an ∼140-nt band besides the 100-nt band (for pre-miR-886) in an in vitro transcription assay (Canella et al. 2010). Thus, it is a possibility that the ∼140-nt band is the primary miRNA (“pri-886”) from which pre-miR-886 is processed by Drosha. If pri-886 exists, its 3′-end will be the 3′-termination site of Pol III transcription and hence coincides with the 3′-end of pre-miR-886. Thus, pri-886 will lack the 3′-strand to form an extended stem (Han et al. 2006) beyond the stem of pre-miR-886. As an extended stem is necessary for efficient processing by the Drosha–DGCR8 complex, the structure of pri-886 is not suitable for Drosha processing. Consistent with our prediction, siRNA-mediated knockdown of Drosha did not cause a significant change in the level of pre-miR-886 (Fig. 3D) or its barely detectable primary transcript (Fig. 3E). For comparison, the primary transcript of let-7f-1 was accumulated (Fig. 3E), as expected with the depletion of Drosha. Thus, generation of pre-miR-886 is independent of Drosha.
Next, we used in vitro transcribed synthetic pre-miR-886 to test if the pre-miR-886 hairpin is processed into mature miRNAs by Dicer. When incubated with partially purified recombinant Dicer, synthetic pre-miR-886 was diced into smaller pieces including a band at the mature miRNA size, while pre-miR-23a was precisely processed into mature miR-23a (Fig. 3F). Quantitatively, the efficiency in production of mature miR-886s (5.4% and 2.1% for miR-886-3p and -886-5p, respectively) was far lower than that of mature miR-23a (44%). In agreement with these in vitro processing data, pre-miR-886 was not efficiently processed into the mature form when 32P-labeled synthetic RNAs were transfected into 293T cells. A band at ∼24 nt was detectable along the smeared signal; however, it was significantly less prominent in quality and ∼10× less intense in quantity than the discrete ∼22-nt band from pre-miR-23a (Supplemental Fig. S4A). Consistent with the cleavage of pre-miR-886 by recombinant Dicer in vitro, siRNA-mediated knockdown of Dicer resulted in an increased intracellular level of pre-miR-886 (Supplemental Fig. S4B,C), indicating that Dicer degrades pre-miR-886 in cells and contributes to its short half-life. In summary, albeit its degradation by Dicer, pre-miR-886 is processed into mature miRNAs inefficiently.
If pre-miR-886 in CRL-2741 cells gave rise to miR-886-5p and -886-3p that were functional as mature miRNAs, we would expect repression of its predicted target genes in CRL-2741 relative to H1299 cells lacking pre-miR-886. However, our bioinformatics analyses of mRNA expression between the two cell lines indicated that there was no significant depletion of predicted targets of miR-886-5p and -886-3p (Supplemental Fig. S5A and legend). Furthermore, we failed to detect the activity of miR-886-5p and -886-3p by luciferase assays employing sensor plasmids harboring an artificial target site at the 3′ UTR of the luciferase open reading frame (Supplemental Fig. S5B and legend). We conclude that pre-miR-886 is neither generated by Drosha nor processed into functional mature miRNAs.
Further characterization of pre-miR-886: Is it a vtRNA?
As described in the Introduction, it has been claimed that pre-miR-886 is misannotated as a miRNA but is instead a vtRNA, a component of the vault complex, primarily because of the sequence similarity between pre-miR-886 and vtRNAs. The sequence alignment of vtRNAs with pre-miR-886 (Fig. 4A) reveals two blocks of identical sequence corresponding to the A and B boxes for Pol III transcription (van Zon et al. 2001; Nandy et al. 2009; Stadler et al. 2009). While the sequence homology between pre-miR-886 and vtRNAs is limited mostly around the A and B boxes, the homology among the three vtRNAs extends well beyond these boxes. Thus, pre-miR-886 is divergent in sequence from vtRNAs that are conserved among them.
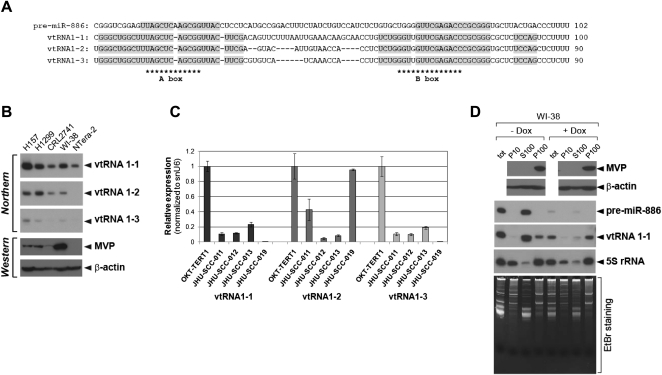
FIGURE 4.
Pre-miR-886 is distinct from vtRNAs. (A) Sequence alignment among pre-miR-886 and vtRNAs. The identical sequence blocks are highlighted in gray. The conserved A and B boxes are indicated by asterisks. (B) Northern hybridization and Western blotting in the five cell lines used in Figure 1D (which contains the loading control for the Northern). (C) qRT-PCR measurement of vtRNAs in the five oral cell lines as described in Figure 1E. (D) Western blotting (top two panels) and Northern hybridization (middle three panels) in subcellular fractions P10 (the nuclear pellet), S100 (the cytoplasmic soluble fraction), and P100 (the cytoplasmic pellet). The same portion from each fraction was loaded and EtBr staining (bottom panel) was included for comparison between fractions. In Northern hybridization, total RNA (tot) was included. In samples in the right half of the panel, WI-38 lung fibroblasts were treated with doxorubicin (4 μg/mL) for 4 h before cell harvest.
Next, we compared the expression levels of vtRNAs among cell lines. Consistent with previous reports (Kickhoefer et al. 1998; van Zon et al. 2001; Stadler et al. 2009), vtRNA1-1 was expressed in a relatively constitutive manner, while vtRNA1-2 and 1-3 were expressed more variably (Fig. 4B,C; Supplemental Fig. S1). The expression of MVP (major vault protein), the primary protein component of the vault complex, was also variable among the cell lines (Fig. 4B). From our expression data on vtRNAs and MVP, we failed to deduce any consensus feature or any correlation. Furthermore, it is clear that the expression pattern of vtRNAs and MVP is not correlated with that of pre-miR-886, which tends to be lower in cancer (Fig. 1D–F).
Both pre-miR-886 and vtRNA1-1 were localized in the cytoplasm (Fig. 2B). However, the more pertinent question was whether pre-miR-886, vtRNA1-1, and MVP were co-localized in the vault complex in the cytoplasm. To test this, we further fractionated the post-nuclear lysate (the cytoplasmic fraction in Fig. 2B) by ultracentrifugation at 100,000g. After ultracentrifugation, high-molecular weight particles such as ribosomes and the vault complex are precipitated (“P100” fraction) and separated from the soluble fraction (“S100” fraction; Kickhoefer et al. 1998). In agreement with previous data (Kickhoefer et al. 1998), MVP was exclusively found in the P100 fraction, where some vtRNA1-1 was also detected (Fig. 4D; Supplemental Fig. S6A). Most importantly, pre-miR-886 was exclusively localized in the S100 fraction, clearly demonstrating that pre-miR-886 is not stably associated with the vault complex.
In the fractionation experiment, we included a set of doxorubicin-treated samples because we knew from our preliminary data that treating cells with this drug decreased pre-miR-886 and vtRNA1-1 (Fig. 4D, cf. lanes 1 and 5). vtRNA1-1 in the S100 fraction was destabilized by doxorubicin, while vtRNA1-1 in the P100 fraction remained stable (Fig. 4D, cf. lanes 3,4 and 7,8). Pre-miR-886, as it was localized mostly in the S100 fraction, was almost completely eliminated after doxorubicin treatment. It should be emphasized that doxorubicin treatment and the subsequent decrease in soluble vtRNA1-1 or pre-miR-886 did not alter the fractionation pattern of MVP and its association with vtRNA1-1, indicating that the integrity of the vault complex was not affected.
In agreement with our biochemical data, in situ hybridization revealed that foci of vtRNA1-1 were enriched in the perinuclear area (Supplemental Fig. S6B), where the vault complex had been shown to be localized (Chugani et al. 1993). For comparison, the pre-miR-886 foci were not confined in the perinuclear area but were detected throughout the cytoplasm (Fig. 2C). In summary, we concluded that pre-miR-886 is not physically or functionally associated with neither MVP nor the vault complex.
Biological roles of pre-miR-886
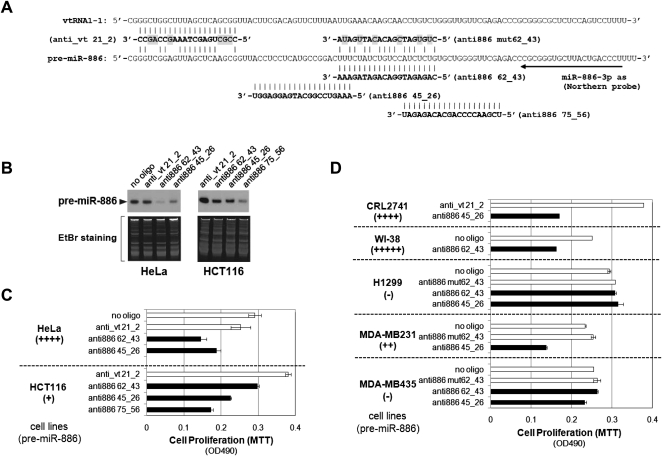
To investigate pre-miR-886's biological roles, we observed cell proliferation upon its depletion by modified antisense oligonucleotides (anti-oligos) (Yoo et al. 2004; Ideue et al. 2009). We designed three anti-oligos (anti886 62_43, anti886 45_26, and anti886 75_56) in the middle portion of pre-miR-886 (Fig. 5A) because that region was the most unique in sequence (see the alignment with vtRNAs in Fig. 4A). The three anti-oligos were off-target from the mature miR-886-5p or -886-3p, if any of them existed.
FIGURE 5.
Suppression of pre-miR-886 inhibits cell proliferation. (A) Three anti-oligos targeting pre-miR-886 (anti886 62_43, anti886 45_26, and anti886 75_56) and two control anti-oligos (anti886 mut62_43 and anti_vt 21_2). Their sequences (in bold letters) are aligned with pre-miR-886 or vtRNA1-1, with mismatched nucleotides highlighted in gray. Arrow indicates Northern probe used in panel B. (B) Northern hybridization of pre-miR-886 after transfection of indicated anti-oligos in HeLa and HCT116 cells. Cells were harvested for RNA preparation 24 h after transfection of each anti-oligo at 40 nM (also for panels C,D). (C,D) Cell proliferation assays (MTT assays) in indicated cell lines after transfecting indicated anti-oligos. Plain bars designate control anti-oligos or no oligo (mock transfection without oligonucleotides), and black bars designate the three targeting anti-oligos. Below each cell line, the relative expression level of pre-miR-886 is indicated (see also Fig. 1D and Supplemental Fig. S1).
Transfection of the anti-oligos into HeLa and HCT116 cells efficiently decreased pre-miR-886 levels (Fig. 5B) and cell proliferation (Fig. 5C). The effectiveness of each oligo was variable among cell lines. For example, anti886 62_43 was better at repressing pre-miR-886 in HeLa cells, while anti886 75_56 was better in HCT116 (Fig. 5B). Nonetheless, at least one anti-oligo effectively decreased pre-miR-886 and suppressed cell proliferation in each cell line that expressed pre-miR-886 (Fig. 5C,D). The degree of growth inhibition was correlated with that of pre-miR-886 decrease, as shown in HeLa and HCT116 cells (Fig. 5, cf. B and C). Importantly, the anti-oligos did not affect the proliferation of two cell lines lacking pre-miR-886 (H1299 and MDA-MB435 in Fig. 5D). Two control anti-oligos (anti_vt 21_2 and anti886 mut 62_43) did not inhibit cell proliferation, relative to “no oligo” control (mock transfection without oligonucleotides). Therefore, any nonspecific toxicity of modified oligonucleotides could be ruled out. Notably, a control anti-oligo targeting vtRNAs suppressed vtRNA1-1 (data not shown) but did not exhibit an anti-proliferative effect (Fig. 5C,D).
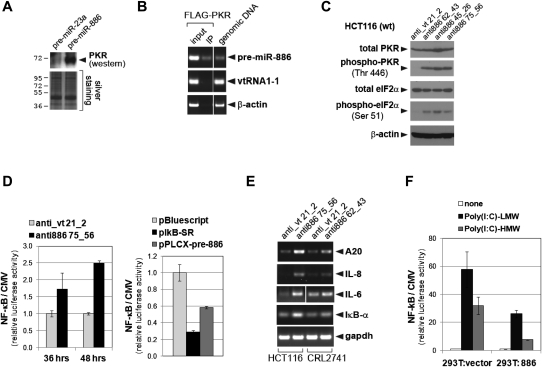
To obtain a clue to pre-miR-886's molecular function underlying its effect on cell proliferation, we searched proteins that physically interact with pre-miR-886 from the HeLa S100 fraction, by using biotinylated synthetic pre-miR-886 as a bait. Our mass spectrometry analysis identified 49 proteins that bound specifically to pre-miR-886, relative to pre-miR-23a as a control (Supplemental Table 1). Among them, we chose PKR for further examination, because it is an RNA binding protein in the cytoplasm and is anti-proliferative upon activation. Our mass spectrometry data were confirmed by immunoblot in which PKR was detected only in the pull-down of pre-miR-886 (Fig. 6A). Pull-down with FLAG-tagged PKR co-precipitated pre-miR-886 specifically (Fig. 6B), reassuring the association between pre-miR-886 and PKR.
FIGURE 6.
Pre-miR-886 is physically associated with PKR and suppresses it from activation. (A) Western blotting of PKR (top panel) in the 0.6 M KCl fraction eluted from the indicated biotinylated RNA–streptavidin bead complex (see Materials and Methods). Silver staining of the gel is shown for equal loading (bottom panel). (B) The FLAG–PKR expressing plasmid was transfected to 293T:886 (a 293T derivative cell line stably expressing pre-miR-886 from integrated pLPCX–pre-886). PKR and associated RNA was immunoprecipitated, according to the protocol of Dicer processing assay (see Materials and Methods) with modifications. Coprecipitated RNA with PKR was eluted by 200 mM KCl, purified by Trizol reagent, and subjected to RT-PCR measurement of pre-miR-886, vtRNA1-1, and β-actin mRNA. (C) Western blotting detection of indicated proteins at 24 h after transfection of indicated anti-oligos. All other descriptions are the same as in Figure 5B. (D) Luciferase assays measuring NF-κB activity at indicated hours (x-axis) after co-transfecting reporter plasmids with indicated anti-oligos (left panel) or plasmids (right panel). pNF-κB-Luc plasmid expresses firefly (Photinus pyralis) luciferase under the control of a NF-κB responsive promoter. As a control, we used pcDNA3.1-Zeo(+)-Pp that expresses firefly luciferase under CMV promoter. Each reporter plasmid was co-transfected with pRL-SV40 (Promega) expressing Renilla luciferase for normalization. Each value of the firefly luciferase (Pp) was first normalized to the Renilla luciferase value (Rr), and then the Pp/Rr value of the NF-κB reporter plasmid was again normalized to that of the CMV control (from pcDNA3.1-Zeo(+)-Pp), yielding the relative luciferase activity of the sensor plasmid (y-axis). pBluescript is a negative control; pLPCX–pre-886 expresses pre-miR-886; pIkB-SR expresses constitutively active IκB-α thus was used as a positive control for NF-κB suppression. (E) RT-PCR measurement of NF-κB target genes after transfecting indicated anti-oligos to HCT116 and CRL2741. All other descriptions are the same as in Figure 5B. (F) Luciferase assays at 24 h after transfecting indicated Poly(I:C) and reporter plasmids into 293T:886 (described in panel B) and 293T:vector (a 293T derivative line harboring pLPCX vector, thus lacking pre-miR-886 expression). All other descriptions are the same as in panel D.
Upon suppression of pre-miR-886 by the anti-oligos, PKR was activated as indicated by induction of phosphorylated PKR (at Thr446, Fig. 6C) and subsequent phosphorylation of its downstream substrate eIF2α (at Ser51, Fig. 6C). It should be noted again that the anti-oligo against vtRNA1-1 (anti_vt 21_2) decreased the intracellular level of vtRNA1-1 (data not shown) but did not activate PKR (Fig. 6C). This effect, specific to pre-miR-886 but not to vtRNA1-1, was in agreement with the specific association of PKR with pre-miR-886, but not with vtRNA1-1 (Fig. 6B). Consistent with our knockdown data, the level of PKR exhibited an anti-correlation with that of pre-miR-886 in the battery of oral cell lines (Supplemental Fig. S7) but showed no correlation with any of the vtRNAs (cf. Supplemental Fig. S7 and Fig. 4C).
It has been known that activated PKR provokes the NF-κB pathway. NF-κB activation was assessed by luciferase reporter assays that measure the activity of a promoter harboring NF-κB responsive elements. The NF-κB activity was induced upon suppression of pre-miR-886 (Fig. 6D, left panel), but attenuated by its ectopic expression (Fig. 6D, right panel). The activation of the NF-κB pathway was also confirmed by our RT-PCR measurement of NF-κB target genes (A20, IL-8, IL-6, IκBα), all of which were induced upon pre-miR-886 suppression (Fig. 6E). Furthermore, induction of NF-κB by a synthetic analog of dsRNA, Poly(I:C), was less efficient in 293T cells stably expressing pre-miR-886 relative to vector control (Fig. 6F). Collectively, our data indicated that pre-miR-886 is a specific inhibitor of PKR (depicted in Fig. 7).
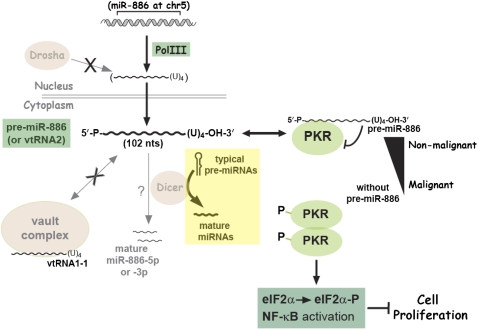
FIGURE 7.
A model depicting the identity, biogenesis, expression, and role of pre-miR-886. See text for details.
DISCUSSION
In this study (summarized in Fig. 7), we scrutinized pre-miR-886 that has been proposed to be a precursor miRNA and/or a vtRNA. Indeed, its revised name “vtRNA2” has been approved by the HUGO Gene Nomenclature Committee (http://www.genenames.org/) and used in databases such as the NCBI (http://www.ncbi.nlm.nih.gov/) and the UCSC genome browser (http://genome.ucsc.edu/). We have provided experimental evidence that pre-miR-886 is neither a vtRNA nor a canonical miRNA, at least in the cell lines examined in this study. Nonetheless, we do not rule out the possibility that pre-miR-886 acts as a pre-miRNA or a vtRNA in certain biological situations. Indeed, mature miR-886-5p and -886-3p were produced by Dicer in vitro, albeit with very low efficiency (Fig. 3F). Also, miRNA-like small RNAs from vtRNAs were reported once previously (Persson et al. 2009). In a recent paper (Pillai et al. 2010), mature miR-886-3p was detected by Northern hybridization in HS5, a human stromal cell line. Also in this case, the signal of pre-miR-886 was much stronger than that of mature miR-886-3p.
The definition of a canonical miRNA is one that is generated by Dicer from its stem–loop structured precursor (Ambros et al. 2003). This definition has become fuzzy, as a growing number of reports have revealed other alternative pathways that generate miRNA-like small RNAs (for review, see Miyoshi et al. 2010). miR-886-5p and -886-3p, at a sub-detectable level by Northern hybridization, may be generated by Dicer or other mechanisms. However, a more plausible scenario is that the seemingly mature miRNAs are only degradation intermediates of pre-miR-886. Our data document that pre-miR-886 is very abundant and unstable (Fig. 2D,E); therefore, a certain level of degradation byproducts is likely to be continuously present in cells expressing pre-miR-886. Close inspection of ultra-high-throughput sequencing data in the deepBase (http://deepbase.sysu.edu.cn/) (Yang et al. 2010) has revealed that other random regions of pre-miR-886 were also captured at significant frequencies, albeit less abundant than those of miR-886-5p and -886-3p. In contrast, the high-throughput sequencing data within a typical pre-miRNA are strongly biased toward three regions—the mature miRNA of one arm of the stem, the miRNA* from the other arm, and the in-between loop. The higher frequency of miR-886-5p and -886-3p than other regions can be explained by our RACE data where their 5′- and 3′-ends respectively match the 5′- and 3′-ends of pre-miR-886, if we assume that the ends are protected from cellular RNases by pre-miR-886's secondary structure or physical association with cellular proteins.
The official name of pre-miR-886 is currently vtRNA2. However, our data clearly show that pre-miR-886 is distinct from genuine vtRNAs; so we suggest that name is inappropriate. In our data (Fig. 4) and others (Kickhoefer et al. 1998), vtRNA1-1 is clearly associated with MVP. Although vtRNA1-2 and 1-3 were not shown to be associated with MVP (Kickhoefer et al. 1998), these two are undoubtedly vtRNAs because their sequences are highly similar to vtRNA1-1. Beyond the transcribed regions, the sequence similarity among the three vtRNAs (vtRNA1-1, 1-2, and 1-3) extends 600–700 nt upstream (>85% sequence identity). Together with their genomic loci being clustered within 15 kb at chromosome 5, this sequence conservation strongly suggests that the three vtRNAs have arisen via gene duplication events during evolution. We found no homology in upstream sequences between pre-miR-886 and the three vtRNAs. As we mentioned earlier, the sequence similarity is restricted to boxes A and B, motifs for Pol III transcription.
So far, the best experimental evidence for pre-miR-886 being a vtRNA is co-fractionation of pre-miR-886 (referred to as CBL-3 therein) with MVP in sucrose density gradient centrifugation (Nandy et al. 2009). It is worth noting in the report (Nandy et al. 2009) that most of pre-miR-886 (97%, according to their description in the text) remained in the S100 fraction. We cannot rule out the possibility that only a miniscule amount of pre-miR-886 is associated with MVP and has a biological function. However, our data (Figs. 4–6) strongly suggest that the role of pre-miR-886 is independent of the vault complex, at least in the cell lines tested in this study. First, the association of pre-miR-886 with MVP was negligible when compared to that of vtRNA1-1. Second, depletion of pre-miR-886 did not affect the fractionation pattern of MVP and vtRNA1-1 at all. Third, the phenotype of pre-miR-886 depletion was not seen with vtRNA1-1 depletion. Fourth, there was no correlation in the expression level among vtRNAs, MVP, and pre-miR-886.
The expression pattern of vtRNAs and pre-miR-886 raises several intriguing questions. All four are Pol III transcripts, but differently expressed (Figs. 1, 4; Supplemental Fig. S1). A significantly different expression pattern is obvious even among the three vtRNAs. At a glance, their transcription rates should not differ, as they possess almost identical sequence in boxes A and B. Given that it is transcribed by Pol III, the decrease of pre-miR-886 in cancer is somewhat paradoxical, as usually Pol III activity is higher in cancerous than in normal tissues (for review, see Marshall and White 2008). This trend was confirmed in our oral and lung cell lines by measuring some tRNA genes (K Lee, N Kunkeaw, and YS Lee, unpubl.).
A very important point is the biological role of pre-miR-886, which we found in this study to be a regulator of PKR. It should be recalled that this role was independent of mature miRNA or vtRNA. Suppression of pre-miR-886 activated PKR; for this we suggest two mechanisms—hypersensitivity or conformational change of PKR. Given that most of the human genome is transcribed, a miniscule amount of dsRNAs is very likely to be present in cells. In our hypersensitivity hypothesis, PKR may be readily activated by sensing a very low level of endogenous dsRNAs, in the absence of pre-miR-886. Alternatively, PKR may undergo conformational change without pre-miR-886 and be activated.
While PKR is activated by long dsRNA (>30 nt), it has been also known to bind to short dsRNAs and be repressed by them (Manche et al. 1992). PKR binds also to hairpin structured single-stranded RNA (ssRNA) (Bevilacqua et al. 1998; Zheng and Bevilacqua 2004). One example is adenoviral Ad2 VAI RNA (virus-associated RNAI of adenovirus type 2) that binds and represses PKR, as one of the viral mechanisms against the host defense system (for review, see Mathews and Shenk 1991). Notably, PKR is activated when a hairpin structured ssRNA is 5′-triphosphorylated, but not when 5′-monophosphorylated (Nallagatla et al. 2007). Our 5′-RACE data demonstrated that pre-miR-886 has a 5′-monophosphate group. As 5′-ends of any Pol III primary transcript (such as tRNA precursors, as shown in Fig. 3) will intrinsically have a 5′-triphosphate group, our data suggest that cells have an active mechanism (for example, by a phosphatase or a ribonuclease) to convert pre-miR-886 into a 5′-monophosphorylated form that is tailored to its role as a PKR repressor.
The impaired cell proliferation upon suppression of pre-miR-886 is explained by the PKR activation and subsequent activation of the eIF2α and NF-κB pathways. It is obvious that cell proliferation is inhibited by eIF2α phosphorylation and the resultant shutdown of protein translation. NF-κB is usually portrayed as pro-proliferative; however, several lines of evidence indicated that NF-κB is pro-apoptotic depending on the cell types, stimuli, etc. (for review, see Prasad et al. 2010). We have shown that the expression of pre-miR-886 was suppressed in some cancers, while previous studies indicated that PKR and NF-κB were activated in some cancers (Fig. 1; Supplemental Figs. S1, S7; for review, see Mounir and Koromilas 2010; Prasad et al. 2010). At a glance, the relationship among pre-miR-886, PKR, and NF-κB looks complicated and even counter-intuitive, in terms of their expression patterns and roles in cell proliferation. However, all of these are explained by our hypothesis that pre-miR-886 plays a tumor surveillance role. We posit two lines of tumorigenic pathways—with or without the loss of pre-miR-886. The latter pathway would explain why not all cancer cell lines lack pre-miR-886 (Supplemental Fig. S1). In the former pathway of tumorigenesis, most tumorigenic cells would be eliminated upon suppression of pre-miR-886 and the resultant activation of PKR. A subset of cancer cells (for example oral cancer cells in Fig. 1 and Supplemental Fig. S7) can apparently develop a bypass pathway to overcome the loss of pre-miR-886 and activation of PKR, explaining our observation that some cancer cell lines do not express pre-miR-886 but nevertheless do proliferate.
In summary, this manuscript is the beginning of the pre-miR-886 story. We have established that pre-miR-886 is a novel ncRNA that is neither a miRNA nor a vtRNA. We also have characterized its definite molecular structure, intracellular abundance, and specific subcellular localization. Most importantly, we revealed that pre-miR-886 plays a putative tumor-suppressive role by modulating PKR activity. To the best of our knowledge, an ∼100-nt cytoplasmic ncRNA with a tumor-suppressive role is unprecedented. As our work is the first earnest investigation on pre-miR-886, much more work will be needed to elucidate its molecular mechanism for PKR regulation, its relevant biological pathways, and the mechanism for its suppression in cancer.
MATERIALS AND METHODS
Cell lines, antibodies, and plasmid DNAs
CRL-2741 (HBE135-E6E7), WI-38, NTera-2 were purchased from the American Type Culture Collection. OKT-TERT1 and JHU-SCC-011, -012, -013, -019 were a kind gift from Dr. Vicente A. Resto (University of Texas Medical Branch). All other cell lines were from our laboratory stock. Antibodies were obtained as follows: MVP and phospho-PKR (Thr446) from Abcam, total eIF2α and phospho-eIF2α (Ser51) from Cell Signaling Technology, total PKR from Santa Cruz Biotechnology, β-actin and FLAG from Sigma-Aldrich. pLPCX–pre-886 was constructed by inserting U6 promoter and pre-miR-886 (102 nt shown in Fig. 3C) into pLPCX vector (Clontech Laboratories). pNF-κB-Luc plasmid was from Stratagene. pIkB-SR (Super-Repressor) expresses constitutively active IκB-α that has mutations at Ser32/Ser36 to Ala/Ala. The plasmid expressing FLAG–PKR was a kind gift from Dr. Yong-Soo Bae (Sungkyunkwan University, Korea).
Tissue RNAs, isolation of cellular RNAs, and measurement/analysis of the small RNAs
Tissue RNAs were purchased from Clontech Laboratories. Isolation of total, nuclear, and cytoplasmic RNA from cell lines was performed as described in Lee and Dutta (2007). Total RNA from prostate cancer specimens was a kind gift from Dr. Anindya Dutta (University of Virginia). miRNA profiling in H1299 and CRL2741 cells was performed by miRCURY LNA arrays (Exiqon Inc.). qRT-PCR measurements were performed with Power SYBR Green PCR master mix and an ABI 7000 real-time PCR system (Applied Biosystems/Ambion) using the primers shown in Supplemental Table 2. For each sample, an average and a standard deviation were calculated from triplicate reactions. 5′- and 3′-RACE were performed with GeneRacer kit (Invitrogen), according to the manufacturer's manual and with modifications described in the figure legend. The sequences of Northern probes are also shown in Supplemental Table 2. In situ detection of pre-miR-886 was performed according to QuantiGene ViewRNA Protocol (Affymetrix), by using mixed probes complementary to nucleotides 28–68 of pre-miR-886 (nucleotide coordinates according to the sequence in Fig. 1C). After in situ hybridization, images were captured on an Applied Precision arrayWorx Multi-format Reader.
siRNAs, antisense oligonucleotides, and synthetic RNAs
siRNAs against Drosha and a negative control duplex were Stealth RNAi siRNA (Invitrogen). The antisense oligonucleotides against pre-miR-886 were from Integrated DNA Technologies. They were 20-nt mixed oligonucleotides containing a backbone phosphorothioate and having 5 nt on each end substituted with 2′-O-methyl ribonucleotides (Yoo et al. 2004; Ideue et al. 2009); the sequences are in Figure 5A. Synthetic pre-miR-886 and pre-miR-23a were made by in vitro transcription using MEGAscript high yield transcription kit (Applied Biosystems/Ambion). DNA templates for in vitro transcription were PCR-amplified (primer sequences are in Supplemental Table 2). Poly(I:C)-HMW(1.5–8 kb) and Poly(I:C)-LMW (0.2–1 kb) were purchased from Invivogen. Transfection of short RNAs was performed with Lipofectamine RNAiMAX reagent or Lipofectamine 2000 reagent (Invitrogen) per the manufacturer's instructions.
Dicer processing assay in vitro
The Dicer expressing plasmid (FLAG-Dicer-pck) was a kind gift from Dr. V. Narry Kim (Seoul National University, Korea). Dicer processing assays were performed as described in Lee et al. (2003). The processed products from the synthetic pre-miR-886 and pre-miR-23a were resolved in a 15% denaturing polyacrylamide gel and subjected to Northern hybridization.
Cell proliferation assays and luciferase assays
Cell proliferation (MTT assay) was measured with CellTiter 96 Aqueous One Solution cell proliferation assay kits (Promega). Luciferase assays were performed by using the Dual-Luciferase Reporter Assay System (Promega). All experiments were performed in triplicate, from which an average and a standard deviation were calculated.
Subcellular fractionation
Subcelluar fractionation was performed as described in Kickhoefer et al. (1998), with modifications. Harvested cells (∼108) were resuspended in 2 mL of buffer A (50 mM Tris-Cl [pH 7.4], 1.5 mM MgCl2, 75 mM NaCl, 0.5% Nonidet P-40) containing 1 mM PMSF (phenylmethylsulfonyl fluoride), 0.1 mL of protease inhibitor cocktail (Sigma-Aldrich), and 10 mM ribonucleoside-vanadyl complex (New England Biolabs). All subsequent steps were performed at 4°C. Resuspended cells were centrifuged at 6000g for 10 min in a TLA-55 rotor using Optima TLX ultracentrifuge (Beckman Coulter). The pellet was the nuclear fraction (designated “P10” in Fig. 4D and Supplemental Fig. S6A). The post-nuclear supernatant was centrifuged at 100,000g for 1 h, separating the resulting supernatant (designated “S100”) from the pellet (designated “P100”). The P10 and P100 fractions were resuspended with Trizol reagent for subsequent RNA preparation or with RIPA buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS) for Western blotting. The S100 fraction was concentrated by adding an equal volume of isopropanol, and the pellet was resuspended with Trizol reagent for subsequent RNA preparation.
Pull-down of pre-miR-886 to identify interacting proteins
HeLa S100 fraction was prepared as described in Dignam et al. (1983). Biotinylated synthetic pre-miR-886 and pre-miR-23a were made by in vitro transcription using biotin-16-UTP (Roche Applied Science). Prior incubation with 6 mg of S100 fraction, 1 nmol of biotinylated RNA was immobilized on 1 mg of streptavidin magnetic beads (New England Biolabs) in 1 mL of buffer B (40 mM Tris-Cl [pH 7.8], 4 mM MgCl2, 150 mM KCl, 0.5 mM dithiothreitol) supplemented with 1 mM PMSF, 10 μL of protease inhibitor cocktail, 10 mM ribonucleoside-vanadyl complex, and 40 units of SUPERase•In (Applied Biosystems/Ambion). After the bead-protein complex was washed with the same buffer containing 300 mM KCl, bound proteins were eluted by increasing salt concentration (0.6 M and 1.2 M KCl in the same buffer) and subjected to mass spectrometry analyses and subsequent identification of proteins (see Supplemental Materials and Methods for details). In comparison to the miR-23a data set, we sifted 49 proteins that specifically bound to pre-miR-886.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Drs. J. Regino Perez-Polo, Satya Prakash, Louise Prakash, and Thomas Wood for sharing equipment; Drs. Vicente Resto, Narry Kim, Yong-Soo Bae, Lee-Jun C. Wong, and Jong In Yook for sharing cell lines and plasmids; Dr. Anindya Dutta for providing RNA from prostate cancer specimens; Dr. David Konkel for critical editing of this manuscript; Drs. Anindya Dutta, Jung-Hoon Yoon, Taeil Kim, Allan Brasier, and Bing Tian for helpful discussion; and Drs. Anindya Dutta and Deog Su Hwang for encouragement. This work was supported by departmental start-up funds from UTMB to Y.S.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2701111.
REFERENCES
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. 2003. A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Steiner E, Grusch M, Elbling L, Micksche M 2009. Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell Mol Life Sci 66: 43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, George CX, Samuel CE, Cech TR 1998. Binding of the protein kinase PKR to RNAs with secondary structure defects: Role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry 37: 6303–6316 [DOI] [PubMed] [Google Scholar]
- Canella D, Praz V, Reina JH, Cousin P, Hernandez N 2010. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res 20: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chugani DC, Rome LH, Kedersha NL 1993. Evidence that vault ribonucleoprotein particles localize to the nuclear pore complex. J Cell Sci 106: 23–29 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Meurs EF, Esteban M 2007. The dsRNA protein kinase PKR: Virus and cell control. Biochimie 89: 799–811 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ 2008. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7: 3112–3118 [DOI] [PubMed] [Google Scholar]
- Hagenbuchle O, Larson D, Hall GI, Sprague KU 1979. The primary transcription product of a silkworm alanine tRNA gene: Identification of in vitro sites of initiation, termination and processing. Cell 18: 1217–1229 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T, Hirose T 2009. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA 15: 1578–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Rajavel KS, Scheffer GL, Dalton WS, Scheper RJ, Rome LH 1998. Vaults are up-regulated in multidrug-resistant cancer cell lines. J Biol Chem 273: 8971–8974 [DOI] [PubMed] [Google Scholar]
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N 1992. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257: 1685–1689 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21: 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, de la Pena P, Tobian JA, Zasloff M, Hatfield D 1987. Unique pathway of expression of an opal suppressor phosphoserine tRNA. Proc Natl Acad Sci 84: 6384–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD 2008. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Hardin JA, Steitz JA 1981. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 211: 400–402 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP 2003. The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tsourkas A 2009. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res 37: e100 doi: 10.1093/nar/gkp482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol 12: 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, White RJ 2008. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer 8: 911–914 [DOI] [PubMed] [Google Scholar]
- Mathews MB, Shenk T 1991. Adenovirus virus-associated RNA and translation control. J Virol 65: 5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Meurs EF, Galabru J, Barber GN, Katze MG, Hovanessian AG 1993. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci 90: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Yoon S 2010. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med 42: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Siomi H 2010. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol Genet Genomics 284: 95–103 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounir Z, Koromilas AE 2010. Uncovering the PKR pathway's potential for treatment of tumors. Future Oncol 6: 643–645 [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC 2007. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 318: 1455–1458 [DOI] [PubMed] [Google Scholar]
- Nandy C, Mrazek J, Stoiber H, Grasser FA, Huttenhofer A, Polacek N 2009. Epstein-barr virus-induced expression of a novel human vault RNA. J Mol Biol 388: 776–784 [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C 2009. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol 11: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Pillai MM, Yang X, Balakrishnan I, Bemis L, Torok-Storb B 2010. MiR-886-3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS ONE 5: e14304 doi: 10.1371/journal.pone.0014304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S, Ravindran J, Aggarwal BB 2010. NF-κB and cancer: How intimate is this relationship. Mol Cell Biochem 336: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Busch H 1988. Small nuclear RNAs: RNA sequences, structure, and modifications. In Structure and function of major and minor small nuclear ribonucleoprotein particles (ed. Birnstiel ML), pp. 1–37 Springer-Verlag, Berlin [Google Scholar]
- Rezaian MA 1999. Synthesis of infectious viroids and other circular RNAs. Curr Issues Mol Biol 1: 13–20 [PubMed] [Google Scholar]
- Stadler PF, Chen JJ, Hackermuller J, Hoffmann S, Horn F, Khaitovich P, Kretzschmar AK, Mosig A, Prohaska SJ, Qi X, et al. 2009. Evolution of vault RNAs. Mol Biol Evol 26: 1975–1991 [DOI] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM 2005. Structural insights into RNA quality control: The Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Grimm C, Lund E, Dahlberg JE 1995. A common maturation pathway for small nucleolar RNAs. EMBO J 14: 4860–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Chen W, Brosius J 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci 13: 2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA 2006. The ever-growing world of small nuclear ribonucleoproteins. In The RNA world (ed. Gesteland RF et al. ), pp. 327–368 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- van Zon A, Mossink MH, Schoester M, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EA 2001. Multiple human vault RNAs. Expression and association with the vault complex. J Biol Chem 276: 37715–37721 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H 2002. Dendritic BC1 RNA: Functional role in regulation of translation initiation. J Neurosci 22: 10232–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Shao P, Zhou H, Chen YQ, Qu LH 2010. deepBase: A database for deeply annotating and mining deep sequencing data. Nucleic Acids Res 38: D123–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BH, Bochkareva E, Bochkarev A, Mou TC, Gray DM 2004. 2′-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res 32: 2008–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Bevilacqua PC 2004. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10: 1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]