Abstract
In Escherichia coli, many small noncoding regulatory RNAs (sRNAs) post-transcriptionally regulate gene expression by base-pairing to mRNAs in a process that is mediated by the RNA chaperone Hfq. Binding of the sRNA to the mRNA can lead to increased or decreased mRNA stability and/or translation. It is not known if proteins other than Hfq are necessary for this process. In order to identify additional genes required for the post-transcriptional regulation of gene expression by Hfq-dependent sRNAs, we developed a novel combined genetic selection and screen for mutants defective in sRNA regulation. In our combined genetic selection and screen, we isolated hfq mutants and mutants in pnp, encoding polynucleotide phosphorylase (PNPase). We show that loss-of-function mutations in pnp result in a decreased stability of several sRNAs including RyhB, SgrS, and CyaR and also decrease both the negative and positive regulation by sRNAs. The defect in stability of CyaR and in negative and positive regulation are suppressed by deletion mutations in RNase E. Altogether, our results suggest that the lack of sRNA-mediated regulation in the absence of an active form of PNPase is due to the rapid turnover of sRNA resulting from an increase in RNase E activity and/or an increase in access of other ribonucleases to sRNAs.
Keywords: Hfq, RNase E, polynucleotide phosphorylase
INTRODUCTION
The post-transcriptional regulation of gene expression by small noncoding RNAs is a critical process that allows bacteria to rapidly adapt to ever-changing internal and environmental conditions. In Escherichia coli, ∼100 sRNAs that typically range in size from 50 to 250 nt have been identified (Argaman et al. 2001; Wassarman et al. 2001; Chen et al. 2002; Vogel et al. 2003; Zhang et al. 2003). Approximately one-third of these sRNAs have been shown to bind to the RNA chaperone Hfq (Wassarman et al. 2001; Zhang et al. 2003), which is homologous to the eukaryotic family of Sm splicing proteins. Hfq was first identified in E. coli as a host factor required for the replication of the phage Qβ (Franze de Fernandez et al. 1972) and later assigned to the gene now called hfq (Kajitani and Ishihama 1991). Its role in sRNA-mediated post-transcriptional regulation was discovered much later when hfq mutants were found to be defective in rpoS translation in both Salmonella and E. coli (Brown and Elliott 1996; Muffler et al. 1996). Hfq catalyzes the pairing of sRNAs to near-complementary sequences, typically in the 5′ untranslated region (5′ UTR) of mRNAs (for reviews, see Valentin-Hansen et al. 2004; Brennan and Link 2007).
In some cases, binding of the sRNA to a sequence in the 5′ UTR of the mRNA can stimulate translation of the mRNA. The best-studied example of this positive regulation is the post-transcriptional regulation of RpoS, the stationary phase sigma factor, by the sRNAs DsrA (Lease et al. 1998; Majdalani et al. 1998), RprA (Majdalani et al. 2001), and ArcZ (Mandin and Gottesman 2010). Base-pairing between an upstream region of the 5′ UTR of the rpoS mRNA and a region encompassing the ribosome binding site normally prevents efficient translation of RpoS. However, binding of DsrA, RprA, or ArcZ to the upstream sequence increases accessibility of the Shine–Dalgarno sequence to ribosomes, resulting in an increased level of translation. Alternatively, binding of an sRNA to an mRNA can inhibit translation by binding to the translation initiation region of the mRNA and, in many cases, can lead to the coupled degradation of the sRNA and mRNA. For example, binding of RyhB sRNA to the translation initiation region of the sodB or sdhCDAB mRNAs leads to an inhibition of translation and the coupled degradation of both RyhB and the targeted mRNAs (Massé et al. 2003).
As mentioned above, the role of Hfq in sRNA regulation was deduced indirectly, as a result of studies of the phenotype of an hfq mutant and the known RNA binding properties of the Hfq protein. While in vitro studies have demonstrated that Hfq alone can promote the association of sRNA and mRNA, no genetic search to determine whether other proteins are essential for in vivo regulation has been reported. We report here the development and application of a combined genetic selection and screen for mutants defective in the post-transcriptional regulation of gene expression by sRNAs. As expected, mutations in hfq were identified. The only other mutations that came out of this search were mutations in pnp, encoding PNPase, a 3′ to 5′ exonuclease, providing insight to an unexpected role of this protein in bacterial sRNA regulation.
RESULTS
Hfq is necessary for both the stability of sRNAs and for sRNA-dependent regulation in E. coli (for review, see Waters and Storz 2008). We sought to identify other genes that are required for the post-transcriptional regulation of gene expression by these Hfq-dependent trans-encoded sRNAs. We developed a combined genetic selection and screen for mutants that are defective in the post-transcriptional regulation of two independent mRNAs, sdhCDAB and cirA, by two different sRNAs, RyhB and OmrB, respectively. sdhCDAB encodes succinate dehydrogenase, which is essential for the ability of E. coli to grow on minimal medium containing succinate as the sole carbon source, and cirA encodes an outer membrane protein that binds an iron-chelating siderophore and some colicins.
The selection portion of this method is based on work of Massé and Gottesman, who found that a fur mutant constitutively expressed the sRNA RyhB, which in turn down-regulated the sdhCDAB mRNA. As a consequence, the fur mutant was unable to grow on minimal medium containing succinate as a sole carbon source. Introduction of an hfq deletion into the fur mutant restored growth on succinate minimal medium (Massé and Gottesman 2002). The genetic screen portion of this method is based on the demonstration that expression of the sRNA OmrB resulted in decreased expression of a cirA-lacZ translation fusion, and that this effect was blocked in an hfq− strain (Guillier and Gottesman 2008).
Our genetic selection/screen assumed that mutations in any gene essential for the post-transcriptional regulation of gene expression by sRNAs would result in a similar phenotype as introduction of an hfq deletion. The strain used was MG1193/pBR-plac-OmrB, a fur− strain harboring a cirA-lacZ fusion and overexpressing OmrB from a Plac promoter on a plasmid. It was unable to grow on succinate and formed white colonies on plates containing isopropyl β-D-1-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal). An hfq deletion derivative of this strain grew on minimal succinate medium (failure of RyhB to repress sdh) and formed blue colonies on minimal medium containing glucose, ampicillin, the β-galactosidase indicator X-Gal, and the inducer IPTG (failure of OmrB to repress cirA-lacZ).
We grew strain MG1193 pBR-plac-OmrB in the presence or absence of the base analog 2-aminopurine, selected for mutants able to grow on minimal medium containing succinate as the sole carbon source, and then screened those mutants for the ability to form blue colonies on minimal glucose plates containing ampicillin, IPTG, and X-Gal. In the absence of mutagen, only one of the six candidates obtained from the selection on minimal succinate plates was able to form blue colonies on the X-Gal-containing plates; in the presence of mutagen, thousands of colonies were obtained from the selection, and 26 of the 29 candidates screened for expression of the cirA′-′lacZ fusion formed blue colonies. The spontaneous mutant and the 26 mutants generated from chemical mutagenesis that showed a defect in the post-transcriptional regulation of both sdhCDAB by RyhB and cirA by OmrB were further characterized by genetic mapping.
Mapping the mutations responsible for defects in sRNA regulation
As noted above, hfq mutants were expected from our selection and screen. The set of 27 mutants was screened for hfq alleles by testing for linkage of the mutant phenotype to a yjfP::cat mutation, located ∼16 kb from hfq, by P1 transduction. P1 grown on an hfq+ yjfP::cat donor was used to transduce the mutants, selecting for chloramphenicol resistance on minimal glucose plates and screening several transductants for loss of the mutant phenotype, that is, for restoration of the negative regulation of sdhCDAB by RyhB (inability to grow on minimal plates containing succinate as the sole carbon source).
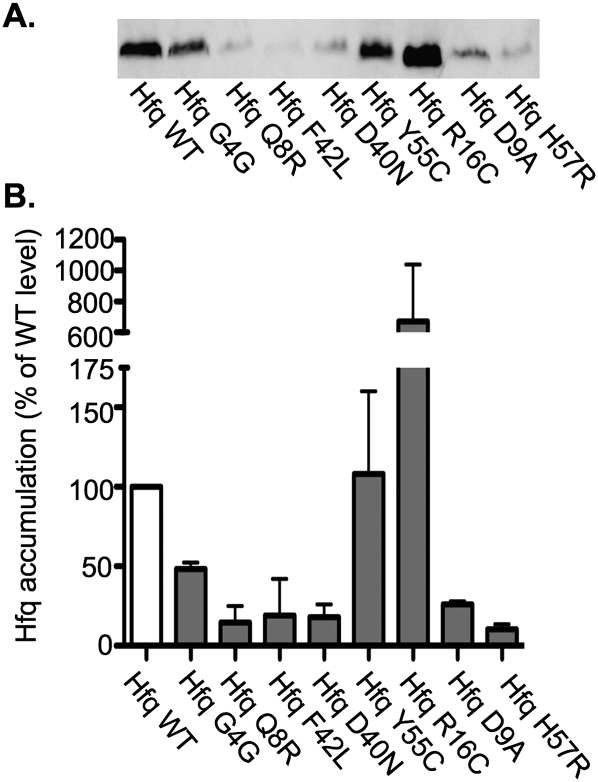
The spontaneous mutant and eight of 26 of the mutants that resulted from the chemical mutagenesis had a mutation in the vicinity of hfq that was necessary for the observed defect in sRNA regulation. Sequencing confirmed that all of these strains had mutations in hfq (Table 1). All but one of these mutations resulted in an amino acid substitution in one of the conserved residues on the proximal face of Hfq that are important for RNA binding (Mikulecky et al. 2004; Sun and Wartell 2006). The remaining mutation in hfq was silent and likely affects the efficiency of translation of this protein. Western blots were performed on crude extract from these mutants and the wild-type strain, and less Hfq was detected in the extracts prepared from all but two mutants, reflecting either unstable Hfq or less efficient detection of the mutant Hfq with the polyclonal antibody. Higher levels of Hfq were detected in the crude extracts from the mutant expressing an Hfq with an R16C substitution (Fig. 1). Since our primary focus was to identify genes other than hfq that are required for the post-transcriptional regulation of gene expression by sRNAs, we did not further characterize these mutants.
TABLE 1.
Mutants obtained from the combined genetic selection and screen
FIGURE 1.
Western blot analysis of Hfq in the mutants obtained from the combined genetic selection/screen. The wild-type strain MG1193 (Hfq WT) and the hfq point mutants obtained from the combined genetic selection and screen, NRD304 (Hfq G4G), NRD307 (Hfq Q8R), NRD309 (Hfq F42L), NRD310 (D40N), NRD314 (Y55C), NRD317 (R16C), NRD323 (D9A), and NRD325 (H57R), were grown overnight in glucose minimal M63 medium. A sample was removed from each culture and processed for Western blotting with anti-Hfq polyclonal antibody as described in Materials and Methods. An equal amount of protein based on the cell density of each culture was applied to the polyacrylamide gel. A representative Western blot is shown in A. The intensity of the bands on the Western blots was quantified using the Multi Gauge software; intensity of Hfq from the wild-type strain was set to 100%, and the signal from all other strains was normalized to this sample. The results shown in B represent the mean of two experiments.
To map the mutation(s) responsible for the defect in sRNA regulation in the remaining 18 mutants, we first identified a transposon insertion linked to the mutants, sequenced the transposon insertion site, and used this information to search candidate genes nearby for mutations. The details are described in Materials and Methods. Using this mapping strategy, three of the mutants that were obtained from chemical mutagenesis, NRD305, NRD311, and NRD324, had a mutation necessary for the Lac+ phenotype that was genetically linked to a transposon insertion between fhuB and hemL, near pcnB, which encodes enzyme poly(A) polymerase. Sequencing revealed that NRD305, NRD311, NRD324, and an additional mutant, NRD308, had missense mutations in pcnB (Table 1). Poly(A) polymerase catalyzes the polyadenylation of the 3′ end of mRNAs, which decreases the stability of mRNAs. Mutations in pcnB have previously been implicated in reducing the copy number of plasmids (Lopilato et al. 1986; Liu and Parkinson 1989; March et al. 1989).
However, for these strains, transduction to replace the mutant pcnB allele with a wild-type gene restored the Lac− phenotype but did not restore the negative regulation of sdhCDAB by RyhB. There was also no difference between a wild-type strain and a pcnB mutant in sodB turnover upon RyhB induction (data not shown). Thus, the linked mutation was necessary for the defect in the negative regulation of cirA by OmrB but not necessary for the defect in the regulation of the sdhCDAB mRNA by RyhB, suggesting that two independent mutations were present in these strains. The simplest explanation is that the pcnB mutations reduced expression of OmrB by lowering the copy number of the plasmid from which it was being expressed, although this has not been confirmed. Since these pcnB mutants were not generally defective for the post-transcriptional regulation of gene expression by sRNAs, we did not characterize them further.
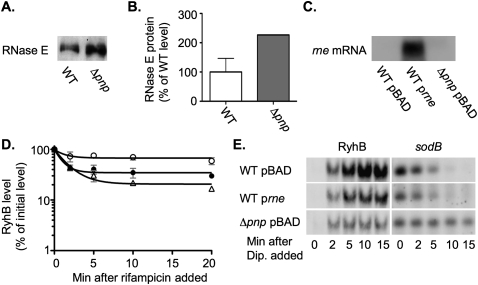
Thirteen of the mutants obtained from chemical mutagenesis had a mutation necessary for both the defect in the regulation of cirA by OmrB and the defect in regulation of sdhCDAB by RyhB in the same region of the E. coli chromosome, far from hfq and genetically linked to transposon insertions in the genes yhbX, yraH, yraN, or yraP. A number of genes implicated in RNA metabolism were in this region of the chromosome, but sequencing of the mutants revealed that the 13 mutants all contained one of five different mutations in pnp, encoding polynucleotide phosphorylase (PNPase) (Table 1). PNPase is a 3′ to 5′ exoribonuclease that degrades single-stranded RNA and binds directly to Hfq (Mohanty et al. 2004), the RNA helicase RhlB (Liou et al. 2002), and the endonuclease RNase E (Carpousis et al. 1994; Py et al. 1994). PNPase binds to a C-terminal unstructured region of RNase E adjacent to the binding site for RhlB (Vanzo et al. 1998). Together, these three proteins, PNPase, RhlB, and RNase E, along with a fourth protein, enolase, form the RNA degradosome (Miczak et al. 1996; Py et al. 1996). In E. coli, polynucleotide phosphorylase is essential at low temperatures (Luttinger et al. 1996) and is up-regulated upon cold shock. All of the substitutions occurred in the N-terminal domain of PNPase in residues that are highly conserved among plants and bacteria (Jarrige et al. 2002).
The genetic analysis described above demonstrated that the pnp alleles were necessary for both growth on succinate and the Lac+ phenotype, since bringing in the wild-type allele reversed both phenotypes. To determine whether the pnp mutations were sufficient to cause a defect in the post-transcriptional regulation of gene expression by sRNAs, three of the mutant alleles, R93C, R315C, and G436D, as well as a deletion of pnp, were introduced into the unmutagenized parent strain, MG1193, harboring pBR-plac-OmrB. Introduction of each of these mutations was sufficient to produce the defect in the post-transcriptional regulation of the cirA-lacZ fusion by OmrB, as judged by the formation of red colonies on MacConkey-lactose plates containing ampicillin and chloramphenicol but was not sufficient to allow growth on minimal plates containing succinate as the sole carbon source. The best explanation for why pnp mutations were found in the selection would be that the original mutants, selected for growth on succinate, harbored a second mutation that improved growth on succinate minimal plates, possibly by increasing expression of sdhCDAB. The second mutation would have arisen in pnp mutant cells, for instance, if these were able to form microcolonies on succinate minimal plates. Experiments described below confirmed that the pnp mutations do compromise RyhB-dependent regulation.
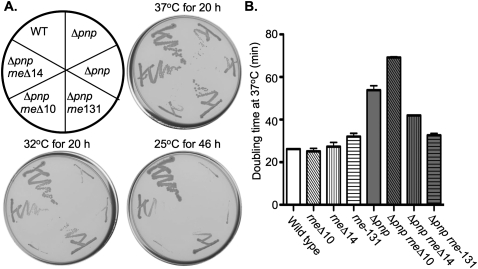
Because a deletion of pnp can abolish OmrB regulation of cirA-lacZ, it seemed likely that the point mutations arising from the screen were loss-of-function, rather than gain-of-function, mutations. This was confirmed for a known phenotype of pnp mutants, cold sensitivity (Luttinger et al. 1996; Zhou and Deutscher 1997). Each of the pnp mutations isolated, as well as a pnp deletion, were transduced into strain DJ624 and tested for growth at 37°C and 25°C on LB plates. The pnp mutants grew significantly slower than the wild type at 37°C and showed no growth after 2 d of incubation at 25°C, whereas the wild-type strain grew well at 25°C (Fig. 2A). The cold sensitivity of the pnp point mutants or pnp deletion mutants was complemented by expression of wild-type pnp from a plasmid (Fig. 2B). These results demonstrated that the pnp point mutants had phenotypes similar to pnp null mutants and were recessive to the wild-type pnp.
FIGURE 2.
The growth of the pnp mutants isolated in the combined genetic selection and screen at 37°C and 25°C. (A) Strains DJ624 (WT), or isogenic derivatives harboring a Δhfq mutation (NRD459), a Δpnp mutation (NRD473), or a point mutation in pnp that encodes a PNPase with an R93C (NRD453), S437F (NRD457), G489D (NRD493), G436D (NRD494), or an R315C (NRD495) substitution were streaked onto two LB plates. One plate was incubated at 37°C for 16 h, and the other was incubated at 25°C for 40 h. (B) The wild-type strain or the pnp mutants from A were transformed with pBAD30 (pBAD) or a plasmid derivative that expresses PNPase from an arabinose-inducible promoter (pNRD412; ppnp), streaked out onto an LB plate containing ampicillin or an LB plate containing ampicillin and arabinose at a final concentration of 0.1%, and then incubated at 25°C for 48 h. The results are representative of three experiments.
pnp mutations have a general effect on sRNA stability
The isolation of multiple loss-of-function mutations in pnp that disrupted sRNA function was unexpected, and the step at which PNPase might act was not immediately apparent. For sRNAs to effectively regulate mRNA targets, they need to accumulate to a level sufficient for activity, interact with Hfq, and, in a process that is not fully understood, interact with their target mRNAs in an Hfq-dependent manner. The interaction between the sRNA and mRNA via base-pairing can lead to changes in translation and/or to changes in mRNA stability.
The first step that was analyzed was sRNA accumulation in pnp mutants. In wild-type cells, sRNAs accumulate rapidly after induction and are stable when new transcription is blocked, for instance, with rifampicin. However, in the absence of Hfq, they are unstable even when transcription is blocked, suggesting that Hfq protects them from ribonucleases (RNase E in the tested cases [Massé et al. 2003]). However, if their half-life is monitored while active cellular transcription proceeds (for instance, when new sRNA synthesis is turned off at the level of the sRNA promoter), they are unstable even in hfq+ cells (Massé et al. 2003). Therefore, Hfq protects sRNAs from degradation, and that protection is apparently abrogated upon pairing. If sRNAs are not properly bound to Hfq, or if Hfq were not able to protect them from degradation, we would expect more rapid degradation of the sRNAs and, therefore, less accumulation, possibly reducing the sRNA levels below that needed for effective regulation.
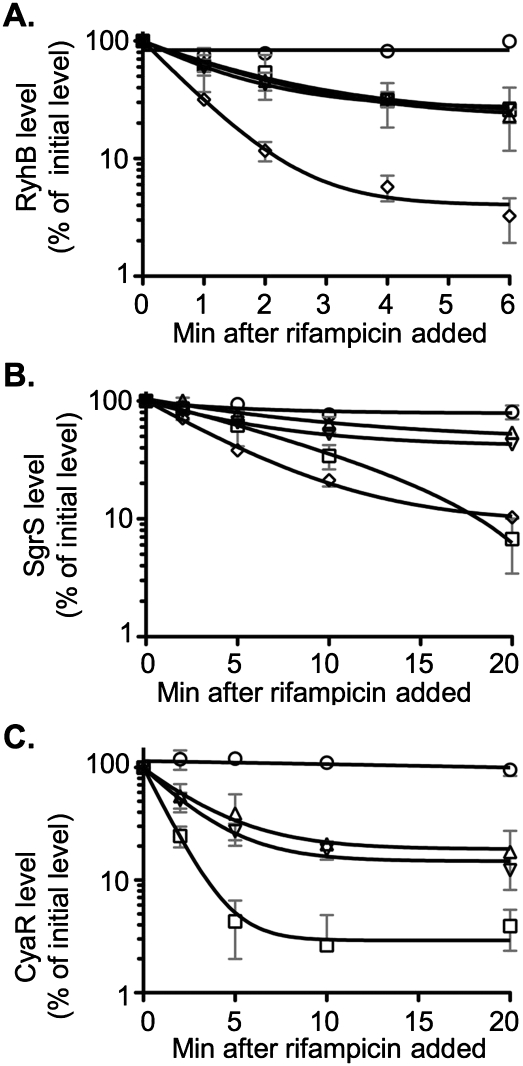
We examined the accumulation and stability of three Hfq-binding sRNAs: RyhB, SgrS, and CyaR. While RyhB accumulation and activity were monitored in the original genetic selection, the other two sRNAs were not. However, we found that each of these sRNAs was more unstable in the pnp mutants than in a wild-type strain (Fig. 3).
FIGURE 3.
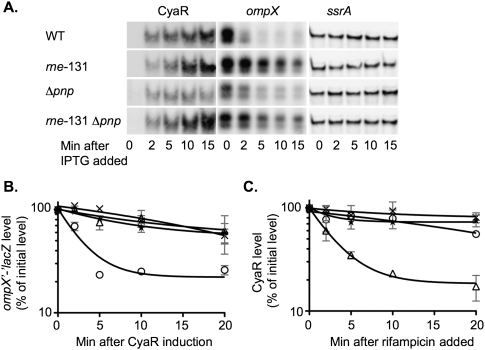
Northern blot analysis of RyhB, SgrS, and CyaR turnover in pnp mutants. (A,B) Overnight cultures of the wild-type strain DJ624 (circles), the hfq deletion mutant NRD459 (squares), the pnp deletion mutant NRD473 (upward pointing triangles), the pnp point mutant NRD494 (downward pointing triangles), or the Δpnp Δhfq double mutant NRD535 (diamonds) were diluted 200-fold into fresh LB. (C) Overnight cultures of a cyaR deletion strain (NRD531; circles), or isogenic derivatives carrying an hfq deletion (NRD532; squares), a pnp deletion (NRD533; upward pointing triangles), or a pnp point mutation (NRD534; downward pointing triangles), all harboring a plasmid expressing CyaR from a lac promoter (pNRD405), were diluted 200-fold in fresh LB containing ampicillin. All strains were grown at 37°C to an OD600 between 0.3 and 0.4, and dipyridyl (A), α-MG (B), or IPTG (C) was then added to each culture to induce sRNA synthesis. A sample was taken from each culture 15 min after induction. Sixteen minutes after induction, rifampicin was added to each culture, and additional samples were taken at the indicated times. RNA was extracted, fractionated on a polyacrylamide gel, and transferred to a nylon membrane. The Northern blot was developed using the 5′-biotinylated RyhB probe (A), SgrS probe (B), or CyaR probe (C) as described in Materials and Methods. The intensity of the bands in the Northern blots was quantified using the Multi Gauge software. The band intensity for the 0-min sample was set to 100%, and other samples were normalized to the 0-min sample. The results represent the mean of two experiments, and the standard deviation is indicated by the light gray bars.
RyhB accumulation and stability were determined by growing cells in LB to mid-log phase and adding dipyridyl to chelate Fe and inactivate the Fur repressor. Samples were taken to monitor RyhB synthesis. Rifampicin was added 16 min after dipyridyl treatment to stop all new synthesis, and samples were taken to monitor decay of the sRNA. As expected, RyhB was rapidly induced to a high level in the wild-type strain and to a much lower level in the hfq mutant (40% of wild type). The level of RyhB was even lower in the pnp mutants (21%–24% of wild-type for two mutants) (Fig. 4A). Turnover of RyhB after rifampicin treatment was rapid in the hfq mutant and slow in the wild-type strain, as previously seen (Massé et al. 2003). In both the pnp deletion and the pnp G463D mutants, RyhB turnover was significantly faster than in the wild-type strain (Fig. 3A).
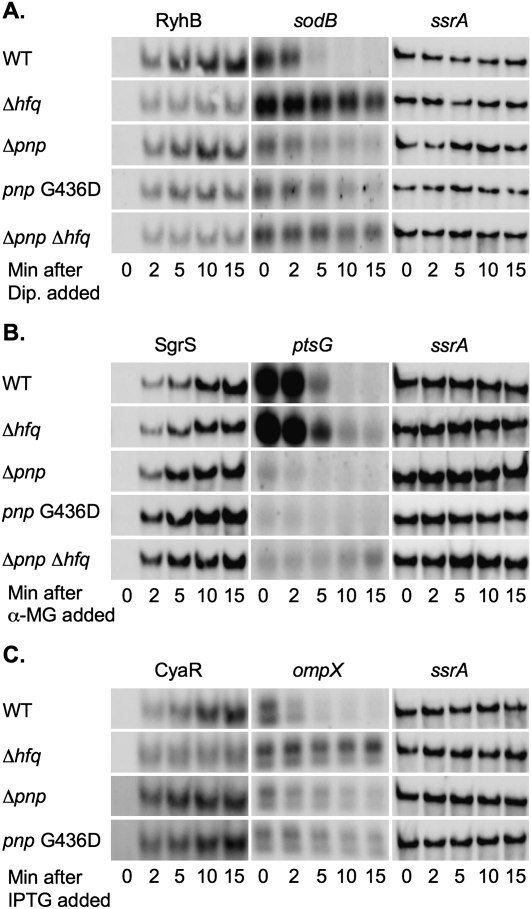
FIGURE 4.
Northern blot analysis of sodB, ptsG, and ompX mRNA fate in pnp mutants. From the same cultures used in Figure 3, samples were removed for RNA isolation from each culture prior to and 2, 5, 10, and 15 min after dipyridyl (A), α-MG (B), or IPTG (C) was added to the culture. An equal amount of RNA from each sample was fractionated on a polyacrylamide or agarose gel, transferred to a nylon membrane, and a Northern blot developed using the 5′-biotinylated RyhB and sodB probe (A), SgrS and ptsG probe (B), or CyaR and ompX probe (C) as described in Materials and Methods. The membranes used for the RyhB, SgrS, or CyaR Northern blots were subsequently stripped by boiling in 0.5% SDS and developed using the ssrA probe as a loading control. The results are representative of two experiments.
The next sRNA tested, SgrS, is induced upon accumulation of glucose-phosphate or after addition of methyl-α-D-glucopyranoside (α-MG) to cells. SgrS induction leads to down-regulation of ptsG, encoding the glucose transporter (Vanderpool and Gottesman 2004). The experiment was similar to that described above, except that methyl-α-D-glucopyranoside (α-MG) was added to induce SgrS expression. Quantification of the Northern blots shown in Figure 4B revealed that SgrS levels after 15 min induction with α-MG were reproducibly 35% lower in the hfq mutant than the wild-type strain. This decrease in SgrS levels in the hfq mutant was less than that observed by Kawamoto et al. (2005); there are differences between their work and ours in the genetic background used, as well as in the growth phase, induction time, and amount of inducer (α-MG). Regardless, the half-life of SgrS was significantly reduced in the pnp point mutant and the pnp deletion mutant as compared to the wild-type strain but was lowest in the hfq mutant (Fig. 3B).
The effect of pnp mutants on the stability of a third sRNA, CyaR, was also determined. While CyaR is regulated by CRP and cAMP, in this case, CyaR was expressed from an inducible lac promoter on a plasmid. As for the experiments above, induction with IPTG was for 15 min, and then the cells were treated with rifampicin. As observed for RyhB and SgrS, CyaR was least stable in the hfq mutant but was less stable in the pnp mutants than in the wild-type strain (Fig. 3C).
In parallel, the activity of each of these sRNAs was monitored by measuring the level of the mRNAs for one of their targets (sodB as a target for RyhB, ptsG as a target for SgrS, and ompX as a target for CyaR). For each of these targets, induction of the appropriate sRNA led to rapid degradation of the mRNA in wild-type cells; in hfq mutants, no, or much slower, degradation of the mRNA took place (Fig. 4A,B,C). In the pnp mutants, two effects were apparent. First, unexpectedly, the levels of the mRNAs were significantly decreased before induction of the sRNA (see 0-min time point, Fig. 4A,B,C). In the case of sodB and ompX, the mRNAs were approximately one-third lower in the pnp deletion or pnp point mutant as compared to the wild-type strain prior to sRNA induction, whereas ptsG mRNA was less than one-tenth the level of the wild-type strain in the pnp mutants before sRNA induction. Second, in the case of sodB and ompX, turnover of the mRNAs was reduced in the pnp mutants (Fig. 4A,C). It was difficult to assess the effect of the pnp mutations on the negative regulation of ptsG mRNA, since all strains showed an initially rapid turnover of the ptsG mRNA after SgrS induction, and the ptsG mRNA level was too low to detect at the time points at which ptsG mRNA stabilized in the hfq mutant (Fig. 4B).
Overall, these experiments pointed to instability of three different sRNAs in the pnp mutants. The decreased stability of the sRNAs may be sufficient to explain the parallel loss in sRNA-dependent regulation. SgrS, which was much more stable in the pnp mutants than RyhB or CyaR and still accumulated to a high level in the pnp mutant, was also still able to carry out some regulation of ptsG. This implied an unexpected protective role for PNPase in allowing sRNA accumulation/stabilization.
The defect in sRNA regulation caused by the pnp mutations was independent of Hfq
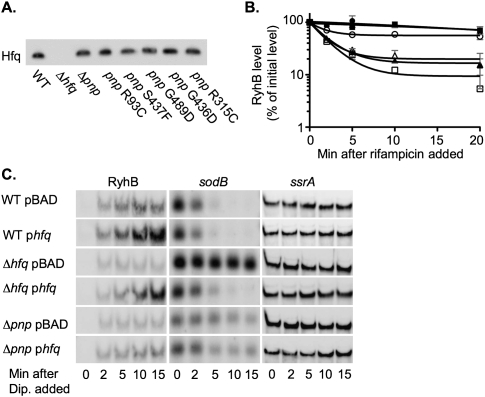
As noted above, and as demonstrated by the isolation of hfq mutations, Hfq is known to be necessary for the sRNAs studied here to act. Thus, our initial model was that loss-of-function mutations in pnp led to reduced expression of Hfq and, as a consequence, reduced sRNA stability and function. To test this model, we examined the level of Hfq by Western blot in isogenic strains that carry the pnp deletion or one of five pnp missense alleles. Hfq levels were similar in a wild-type strain and in the pnp mutants (Fig. 5A). Therefore, pnp does not act by decreasing Hfq levels.
FIGURE 5.
Effect of pnp mutations on Hfq and effects of Hfq overexpression in pnp mutants. (A) Overnight cultures of the wild-type strain DJ624 or derivatives harboring an hfq deletion (NRD459), pnp deletion (NRD473) or pnp allele encoding a PNPase with an R93C (NRD453), S437F (NRD457), G489D (NRD493), G436D (NRD494), or R315C substitution (NRD495) were diluted 200-fold in fresh LB medium. Cells were grown at 37°C to an OD600 of ∼1.0, a sample was taken from each culture, and the protein was TCA-precipitated. An equal amount of protein based on the cell density of each culture at the time of sampling was then processed as in Figure 1. (B,C) An overnight culture of the wild-type strain DJ624 (WT; circles), hfq deletion strain NRD459 (Δhfq; squares), or pnp deletion strain NRD473 (Δpnp; triangles) harboring pBAD30 (pBAD; open symbols) or a plasmid expressing hfq from an arabinose inducible promoter (pNRD414; phfq; solid symbols) was diluted 200-fold in fresh LB medium containing ampicillin. Each culture was grown to an OD600 of 0.3–0.4, and arabinose was added. After 5 min of induction, a sample was taken from each culture (0–min samples), and then dipyridyl added. Additional samples were taken as indicated. Sixteen minutes after dipyridyl addition, rifampicin was added to the cultures and samples taken 2, 5, 10, and 20 min after rifampicin treatment. The RNA was extracted and processed as described in Figure 4, using the 5′-biotinylated RyhB, sodB, and ssrA probe. The results in B represent the mean of two experiments, and the standard deviation is indicated by the gray bars. The blots in A and C are representative of two experiments.
Another way in which Hfq might be unavailable for binding and stabilizing sRNAs would be if the loss of PNPase increased some Hfq-binding RNA(s), making Hfq limiting and therefore unavailable for other RNAs. If so, overexpression of Hfq should lead to an increase in the stabilization of the sRNAs in the pnp mutant and an increase in the RyhB-stimulated turnover of the sodB mRNA. Thus, we grew a wild type strain or isogenic derivatives carrying either an hfq or pnp deletion and harboring either a vector or a plasmid containing hfq under the control of an arabinose promoter, induced expression of Hfq with arabinose, and then induced RyhB expression by the addition of dipyridyl to the culture medium. RyhB synthesis and stability and sodB turnover were assessed as in the experiments described above. Hfq expressed from the plasmid complemented an hfq mutant for sodB turnover and increased the stability of RyhB, confirming the activity of the plasmid (Fig. 5B,C, cf. lines 3 and 4). Overexpression of Hfq in wild-type cells increased the stability of RyhB slightly and increased its accumulation significantly (Fig. 5B,C, cf. lines 1 and 2). However, overexpression of Hfq from a plasmid did not suppress the defects in a pnp deletion mutant; RyhB did not accumulate (Fig. 5C, lines 5 and 6) or increase in stability (Fig. 5B), and sodB did not show any increased degradation (Fig. 5C, cf. lines 5 and 6). Based on these results, we conclude that the defect in the post-transcriptional regulation of gene expression in the pnp null mutants is not due to titration of Hfq.
While Hfq does not appear to be limiting, it seemed possible that lack of PNPase leads to inactivation of Hfq by an unknown mechanism. In this model, the effects of the pnp mutants would be upstream of Hfq, and an hfq mutant would be epistatic to the pnp mutants. However, the half-life of the RyhB and SgrS sRNAs demonstrated additive effects of deleting pnp and hfq. The half-lives of RyhB and SgrS were even shorter in the strain harboring both the hfq and pnp deletion than in a strain harboring either deletion alone (Fig. 3A,B). These results suggested that PNPase was not exerting its effect on the regulation of gene expression by sRNAs solely through Hfq.
Suppression of pnp mutant phenotypes by deletion of the C terminus of RNase E
PNPase is known to bind to RNase E, which is essential for the coupled degradation of several sRNAs and their targeted mRNAs (Massé et al. 2003; Morita et al. 2003; Guillier and Gottesman 2008). Therefore, we considered a model in which the decreased stability of the sRNAs is caused by an increased rate of turnover by RNase E. This increased sRNA degradation may in itself be responsible for the defect in sRNA regulation. In this model, PNPase normally decreases the turnover of sRNAs by modulating RNase E activity, or by blocking the binding of Hfq-bound sRNAs to RNase E, thus protecting them from degradation.
rne is essential, so deleting it entirely was not feasible. Given the known interaction of PNPase with the C terminus of RNase E, we examined the role of the RNase E C terminus in the phenotypes of the pnp mutants. We introduced different alleles of rne, including rne-131, rneΔ10, and rneΔ14, into a wild-type strain or a pnp deletion mutant. rne-131 encodes an RNase E protein lacking the entire unstructured C-terminal domain involved in binding PNPase, enolase, and RhlB. rneΔ14 encodes an RNase E that lacks the region that includes the RhlB and enolase binding sites but retains the PNPase binding site (Leroy et al. 2002). rneΔ10 encodes an RNase E that lacks the region that includes the PNPase binding site but retains the enolase and RhlB site (Leroy et al. 2002). The ability of the resulting strains, an isogenic rne+ pnp deletion mutant and a wild-type strain, to grow at 25°C, 32°C, and 37°C was determined. Unexpectedly, introduction of the rne-131 or rneΔ14 allele into the pnp deletion strain partially suppressed the cold sensitivity of the pnp deletion mutant at 32°C and 25°C and decreased the doubling time of the pnp deletion mutant at 37°C (Fig. 6). On the other hand, introduction of the rneΔ10 allele into the pnp deletion mutant did not suppress the cold sensitivity of the pnp deletion mutant and actually increased the doubling time of the pnp deletion mutant at 37°C (Fig. 6). Introduction of any of these rne alleles into a wild-type strain had little or no effect on growth at 25°C, 32°C, or 37°C (Fig. 6B; data not shown).
FIGURE 6.
The effect of rne truncations on growth of a pnp deletion mutant. (A) A wild-type strain (DJ624; WT) or isogenic derivatives harboring a pnp deletion (NRD473 and NRD579; Δpnp) or a pnp deletion and an rne allele truncating the C-terminal domain of rne were streaked onto LB plates that were incubated at 37°C, 32°C, or 25°C. The rne-131 allele encodes an RNase E that lacks residues 585–1061, the complete unstructured C-terminal domain (NRD578; Δpnp rne-131); rneΔ10 lacks residues 844 to 1045, a region that includes the PNPase binding site (NRD 585; Δpnp rneΔ10); rneΔ14 lacks residues 636 to 845, a region that includes the binding site for RhlB and enolase (NRD589; Δpnp rneΔ14). (B) Overnight cultures of the wild-type (wild type) and pnp deletion strain (Δpnp) harboring the wild-type rne allele or the truncated rne alleles listed in A were diluted 200-fold into fresh LB liquid medium and incubated at 37°C and the doubling time during exponential growth determined. These results represent the mean from two independent experiments.
Suppression of one phenotype of a pnp deletion, cold sensitivity, by rne-131 or rneΔ14 suggested that these rne alleles might also suppress another phenotype, the defect in sRNA-dependent regulation. Testing this is complicated by the involvement of RNase E in degradation of target mRNAs in response to sRNA pairing; because of this, monitoring mRNA levels to evaluate sRNA function would not have been informative (Massé et al. 2003; Morita et al. 2005). To avoid this complication, we took advantage of the observation that translation of mRNAs can be inhibited by sRNAs, even in the absence of mRNA degradation (Maki et al. 2008).
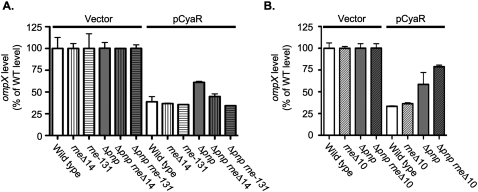
As described above, ompX is negatively regulated by CyaR. While this regulation can be seen at the level of mRNA instability (Johansen et al. 2008; Papenfort et al. 2008; De Lay and Gottesman 2009), mutations such as rne-131 block mRNA degradation (Fig. 7A,B). Therefore, we instead evaluated an ompX-lacZ translational fusion, also negatively regulated by CyaR (Figs. 7B, 8A). In cells expressing CyaR from a plasmid, expression of the fusion was reduced by >60%, and this regulation was not perturbed in an rne-131 mutant cell (Fig. 8A). In parallel with the failure of full regulation of ompX mRNA or the ompX′-′lacZ mRNA in the pnp deletion strain, expression of the fusion was reduced by <40% upon CyaR expression in the Δpnp strain (Fig. 8A). Thus, deletion of pnp partially disrupts the negative regulation of ompX mRNA by CyaR. Introduction of the rne-131 allele into the pnp mutant completely suppressed the defect in sRNA regulation, while introduction of the rneΔ14 allele resulted in partial suppression (Fig. 8A). In contrast, introduction of the rneΔ10 allelle into the pnp mutant did not suppress the defect in the regulation of ompX by CyaR but instead exacerbated the loss of regulation (Fig. 8B).
FIGURE 7.
Analysis of the effect of a pnp deletion and the rne-131 allele on CyaR and ompX turnover. (A,B,C) Overnight cultures of the cyaR deletion strain (NRD377; circles or WT), an rne-131 mutant (NRD674; X's or rne-131), a pnp deletion mutant (NRD677; triangles or Δpnp), and an rne-131 Δpnp double mutant (NRD680; stars or Δpnp rne-131) harboring a plasmid expressing CyaR from a lac promoter (pNRD405) were diluted 200-fold in fresh LB containing ampicillin. Cultures were grown at 37°C to an OD600 between 0.3 and 0.4, a sample was removed, and IPTG was then added to each culture. Additional samples were taken 2, 5, 10, and 15 min after IPTG addition. Sixteen minutes after IPTG was added to each culture, rifampicin was added to each culture, and samples were taken 2, 5, 10, and 20 min after rifampicin treatment. The RNA was extracted and processed as described in Figure 4 using the CyaR, ompX, and ssrA probe (A), lacZ probe (B), or CyaR probe (C). The results in A are representative of two experiments. The intensity of the bands in the Northern blots in B and C was quantified using the Multi Gauge software. The band intensity for the 0-min sample was set to 100%, and each additional sample was normalized to the 0-min sample. The results in B and C represent the mean of two experiments, and the standard deviation is indicated by the gray bars.
FIGURE 8.
Analysis of the effect of a pnp deletion and the rneΔ14 and rne-131 alleles on sRNA regulation. (A) Overnight cultures of the cyaR deletion strain (NRD377; wild type), an rneΔ14 mutant (NRD673; rneΔ14), an rne-131 mutant (NRD674; rne-131), a pnp deletion mutant (NRD677; Δpnp), an rneΔ14 Δpnp double mutant (NRD679; Δpnp rneΔ14), and an rne-131 Δpnp double mutant (NRD680; Δpnp rne-131) harboring either a vector or a plasmid expressing CyaR from a lac promoter (pNRD405) were diluted 200-fold in fresh LB containing ampicillin and IPTG. (B) Overnight cultures of the cyaR deletion strain (NRD377; wild type), an rneΔ10 mutant (NRD672; rneΔ10), a pnp deletion mutant (NRD677; Δpnp), and an rneΔ10 Δpnp double mutant (NRD678; Δpnp rneΔ10) harboring either a vector or a plasmid expressing CyaR from a lac promoter (pNRD405) were diluted 200-fold in fresh LB containing ampicillin and IPTG. Strains were grown to an OD600 between 0.3 and 0.4. Samples were taken from all cultures, and a β-galactosidase assay was performed on those samples as described by Miller (1992).
Consistent with the pnp mutant acting to perturb RNase E-dependent turnover of mRNAs and sRNAs, the rne-131 mutation was epistatic to a deletion of pnp in terms of CyaR accumulation and stability and degradation of either ompX or ompX-lacZ mRNAs. As described above, CyaR was relatively stable in wild-type strains; it was slightly more stable in the rne-131 mutant host (Fig. 7C, cf. X's and circles). As noted above, CyaR was very unstable in a pnp mutant. Introduction of the rne-131 allele into the pnp deletion increased the stability of the CyaR sRNA to a level similar to that seen in the rne-131 mutant alone (Fig. 7C, stars). This increased stability was also reflected in the increased accumulation of CyaR in this double mutant (data not shown).
The rapid turnover of the ompX and ompX′-′lacZ mRNA after CyaR induction seen in a wild-type strain was significantly decreased in the rne-131 host (Fig. 7A,B). In this case, both the pnp mutation and the rne-131 mutation led to slower turnover of ompX RNA, so it is more difficult to judge epistasis. However, the double mutant (Δpnp rne-131) behaved very similarly to the rne-131 mutation, in terms of the turnover of both ompX and ompX′-′lacZ RNA (Fig. 7A,B). Coupled with the suppression of translational inhibition (Fig. 8A), these results suggested that restoring CyaR stability and accumulation was sufficient to restore the function of the sRNA to a pnp mutant. Turnover of the mRNA was a second level of regulation and was abrogated or decreased by the rne-131 allele.
Positive regulation was also defective in a pnp mutant and suppressed by mutations in rne
sRNAs can positively regulate gene expression as well as negatively regulate it. If pnp mutants disrupt sRNA function by reducing sRNA stability, we would expect a role for PNPase in positive regulation as well, and we would predict that the C-terminal deletions in rne should suppress the pnp mutant effect. To determine if PNPase also is required for positive regulation, we examined the effect of the pnp deletion on RpoS expression; RpoS translation is positively regulated by DsrA (Lease et al. 1998; Majdalani et al. 1998) and RprA (Majdalani et al. 2001) and is dependent upon Hfq.
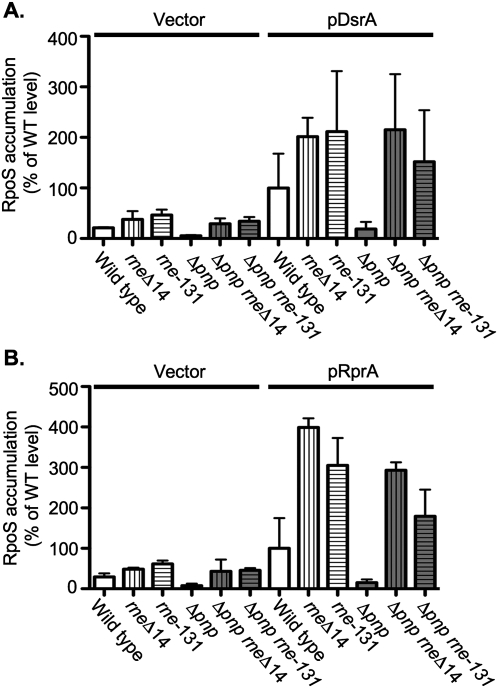
A set of isogenic strains (wild type, pnp, rneΔ14, rne-131, pnp rneΔ14, and pnp rne-131) harboring a vector or a plasmid that expressed either DsrA or RprA from a lac promoter were grown to an OD600 of ∼0.3, DsrA or RprA expression was induced for 20 min, and then samples were taken from each culture. The relative levels of RpoS in each strain, with either a vector or the sRNA-expressing plasmids, were analyzed by Western blot (Fig. 9). When DsrA or RprA were overexpressed in the pnp deletion mutant, RpoS accumulated to a level that was less than one-fifth of the RpoS level observed in the wild-type strain (Fig. 9A,B), indicating that loss of pnp results in the decreased expression of at least one gene positively regulated by sRNAs.
FIGURE 9.
Analysis of the effect of a pnp deletion and the rneΔ14 and rne-131 alleles on positive regulation of RpoS expression by DsrA and RprA. Western blot analysis of RpoS from exponential phase cultures of isogenic pnp+ and Δpnp strains either wild-type for rne (DJ624 and NRD579) or carrying rneΔ14 (NRD475 and NRD589) or rne-131 (NRD476 and NRD578) harboring either a vector or a plasmid that expresses DsrA (A) or RprA (B) from a lac promoter. Overnight cultures were diluted 200-fold in fresh LB medium with ampicillin, growth at 37°C to an OD600 of 0.3–0.4, the sRNA was induced by addition of IPTG for 20 min. Samples were then taken, the protein TCA-precipitated, and an equal amount of protein based on the cell density of each culture processed as described in Materials and Methods and developed using an anti-RpoS polyclonal antibody. The band intensity from each lane was quantified using the Multi Gauge software. The band intensity for the wild-type strain was set to 100%, and other samples were normalized to the wild-type strain. These results represent the mean of two experiments.
Mutations rneΔ14 or rne-131 alone increased expression of RpoS upon DsrA induction (2.5-fold or 2.2-fold) and upon RprA induction (5.6-fold or 3.9-fold), presumably reflecting less rapid turnover of the rpoS mRNA and possibly higher levels of accumulation of the sRNAs (Fig. 9; McCullen et al. 2010). Nonetheless, it was apparent that the rne alleles suppressed the pnp deletion. As for regulation of the ompX::lacZ fusion, the rne mutants were epistatic to the pnp mutants for expression of RpoS. The very low RpoS expression in the pnp mutant was increased in an rne-131 pnp strain to a level 1.5-fold (for DsrA) or 2.1-fold (for RprA) higher, respectively, than the wild-type strain overexpressing DsrA or RprA and only modestly lower than was seen in the pnp+ rne strains (Fig. 9). Similarly, introduction of the rneΔ14 mutation into the pnp mutant increased the expression of RpoS to a level higher than the wild-type strain, i.e., 1.5-fold higher upon DsrA induction and 2.1-fold higher upon RprA induction (Fig. 9). Basal levels of RpoS in the presence of vector were much lower but show a similar pattern to that seen after sRNA expression (low in the pnp mutant, higher in the pnp rne double mutants), presumably reflecting the effects of these mutations on endogenous sRNAs (Fig. 9, vector panels).
Altogether, these results demonstrate that the complete truncation of the C terminus of RNase E or deletion of a region of RNase E that includes the RhlB and enolase binding sites suppresses the cold sensitivity and the defect in sRNA regulation observed in the pnp mutant.
Increased expression of RNase E was not sufficient to cause a defect in sRNA regulation
One possible explanation for our observations was that there was a higher level of RNase E in the pnp mutants and that the higher concentration of RNase E in the cell led to more rapid turnover of sRNAs. In this model, introduction of the rne-131 and rneΔ14 alleles into the pnp mutant counteracts this effect, because the RNase E encoded by these alleles has reduced activity, at least for some substrates (Leroy et al. 2002). In fact, RNase E protein levels were moderately increased in a pnp mutant (Fig. 10A,B). However, if this was the full explanation of the Δpnp phenotype, we would expect overexpression of RNase E to mimic the effect of a pnp null mutation. This was not the case. When RNase E was overexpressed to a level far higher (15–30× greater than wild type) than observed in the pnp deletion mutant (Fig. 10C), the half-life of RyhB decreased but was still significantly higher than what was observed in the pnp mutant (Fig. 10D, cf. Fig. 3A). More importantly, the RyhB-mediated regulation of sodB mRNA was not affected by the overproduction of the RNase E (Fig. 10E). Therefore, we conclude that the pnp mutant has effects beyond the somewhat increased level of RNase E. An alternative model is considered below.
FIGURE 10.
RNase E levels in pnp mutants and the effect of overexpression of RNase E on sRNA function. (A,B) Overnight cultures of the wild-type strain TM338 that expresses a FLAG-tagged RNase E from its native promoter (WT; white bars) or the isogenic pnp deletion mutant NRD558 (Δpnp; gray bars) were diluted 200-fold into fresh LB. The strains were grown at 37°C to an OD600 between 0.3 and 0.4, and a sample was removed from each culture and TCA-precipitated. An equal amount of protein based on the cell density was processed as described in Materials and Methods and developed using an anti-FLAG monoclonal affinity-purified antibody. A representative blot is shown in A. The results shown in B represent the mean of two independent experiments. (C,D,E) Overnight cultures of the wild-type strain (NM534; WT; circles) or a derivative harboring a pnp deletion (NRD571; Δpnp; triangles) and carrying the empty vector (pBAD30; pBAD; open symbols) or a plasmid that expresses RNase E from an araBAD promoter (pNRD416; prne; solid symbols) were diluted 200-fold in LB-ampicillin medium containing arabinose at a final concentration of 0.0009%. The strains were grown at 37°C to an OD600 between 0.3 and 0.4, a sample was taken, and dipyridyl was added to each culture. Additional samples were taken after dipyridyl addition as indicated in E. Sixteen minutes after dipyridyl addition, rifampicin was added to the cultures, and samples were taken as indicated in D. The RNA was extracted and processed as described in Figure 4, using the 5′-biotinylated rne probe, RyhB probe, and sodB probe. The Northern blot in C contains the RNA taken from each sample prior to dipyridyl addition. The blots C and E are representative of two experiments. The results in D represent the mean of two experiments, and the standard deviation is indicated by the gray bars.
DISCUSSION
We have developed and validated a novel genetic approach for identifying genes required for the post-transcriptional regulation of gene expression by sRNAs in E. coli. This combined genetic selection and screen facilitates the isolation of mutants defective in sRNA regulation. The validity of this approach was demonstrated by the isolation of several mutants defective for sRNA regulation that had mutations in hfq, an RNA chaperone that is essential for the post-transcriptional regulation of gene expression by sRNAs. By using chemical mutagenesis rather than transposon insertion mutagenesis, we hoped to isolate point mutations in genes encoding both essential and non-essential proteins involved in sRNA function. While many of the mutants that we obtained had mutations in hfq, most had mutations in pnp, encoding polynucleotide phosphorylase.
Nature of hfq mutations
In all but one case, the hfq mutations led to amino acid substitutions in highly conserved residues on the proximal face of Hfq, including Q8, D9, R16, D40, F42, Y55, and H57. Measurements of Hfq levels suggest lower levels of protein in most of the mutants and unexpectedly higher levels in R16C (Fig. 1). In vitro, alanine substitutions at many of these residues have been previously shown to reduce the affinity of Hfq for poly-A RNAs or DsrA (Mikulecky et al. 2004; Sun and Wartell 2006). A D40A substitution has been shown to destabilize the protein (Mikulecky et al. 2004). Thus, these hfq mutants may be defective in sRNA regulation both because the Hfq protein was destabilized and because the Hfq that was expressed had a low affinity for sRNA binding. Another hfq mutant that we obtained expressed an Hfq with a D9A substitution. Hfq D9A was previously shown to bind a AU5G RNA, DsrA, and rpoS mRNA in vitro with higher affinity than the wild-type Hfq (Mikulecky et al. 2004). The basis for the defect in sRNA regulation in this mutant is not known, and its isolation as defective in sRNA function suggests that it may be interesting to investigate further. Perhaps too tight binding to the sRNA or mRNA is detrimental to pairing. One mutant, NRD304, showed the expected linkage for an hfq mutation, but the only change seen was silent, changing the third codon in hfq from a GGG codon to GGA. We note that the DNA sequence of the first 6 codons of hfq are highly conserved (data not shown). This substitution of the third codon GGG to GGA in hfq reduced the expression of Hfq, although whether that reduction is sufficient to explain the phenotype is not known (Fig. 1). This is the first direct selection of point mutants in hfq that we are aware of, but the overlap with previously studied site-directed mutations suggests that most critical sites have previously been investigated by site-directed mutagenesis.
PNPase mutants
Half of the mutants isolated and characterized had mutations in pnp. PNPase is a 3′–5′ exoribonuclease, which binds directly to the C-terminal scaffold domain of RNase E (Carpousis et al. 1994; Py et al. 1994), along with the RNA helicase RhlB and the glycolytic enzyme enolase, to form the core RNA degradosome. PNPase can also bind to the RNA chaperone Hfq (Mohanty et al. 2004). All of the pnp mutants we obtained had mutations that led to amino acid substitutions in the N-terminal domain of PNPase, and all of the pnp mutants were cold-sensitive, indicating that these were loss-of-function mutations (Fig. 2). This domain contains the two RNase PH domains that are involved in catalysis and in binding to RNase E (Duran-Figueroa et al. 2006).
Unexpectedly for a mutation that inactivates an exonuclease, deletion of pnp decreased the half-life of the RyhB, SgrS, and CyaR sRNAs (Fig. 3) and also led to a decrease in the steady-state levels of the ptsG, sodB, and ompX encoding mRNAs (Fig. 4). These results are inconsistent with a direct effect of PNPase in initiating degradation of these sRNAs and mRNAs, in which case we might have expected increased levels of these RNAs and increased half-lives.
Instead, our data suggest that pnp mutants failed to carry out sRNA regulation because the sRNAs were degraded before they were able to pair with their targets. Prior to this work, Hfq was the only protein known to protect sRNAs from degradation by RNase E. However, a pnp deletion mutant had the same level of Hfq as the wild-type strain (Fig. 5A), ruling out a model in which PNPase regulates Hfq amounts. An alternative model was that PNPase is necessary for Hfq activity, assisting Hfq in binding sRNAs either directly or by displacing already bound sRNAs from Hfq. In this model, the sRNAs were rapidly degraded because they were not binding to Hfq. However, we saw additive effects of pnp and hfq mutants in the degradation of RyhB and SgrS (Fig. 3A,B), strongly suggesting that the effect of PNPase was not fully dependent upon Hfq. In addition, in vitro studies do not support a requirement for PNPase for Hfq to bind sRNAs (Mikulecky et al. 2004; Fender et al. 2010). If PNPase was required to displace already bound sRNAs, then we would have expected that inducing new synthesis of Hfq just prior to inducing the expression of an sRNA would have suppressed the observed defect in sRNA regulation in an pnp mutant. This was not the case (Fig. 5B,C), and in vitro studies have shown that sRNAs can rapidly displace one another from purified Hfq in the absence of additional proteins by a concentration-mediated effect (Fender et al. 2010). Therefore, we conclude that the defect in sRNA regulation in a pnp deletion strain was not via lowering the expression level of Hfq or by reducing the ability of Hfq to bind sRNAs.
A model that is consistent with our data is that PNPase protects sRNAs prior to pairing by limiting access of RNase E to Hfq-bound sRNAs. The C terminus of RNase E, a scaffold for binding of RhlB, enolase, and PNPase, also interacts with Hfq. The site of interaction of Hfq appears to overlap with the RhlB/enolase binding site, present in rneΔ10 but deleted in rne-131 and rneΔ14 (Morita et al. 2005). In this model, the loss-of- function mutations in pnp allow Hfq-bound sRNAs to be accessed by the RNase E prematurely, leading to the inappropriate rapid degradation of these sRNAs. However, in the rne-131 or the rneΔ14 mutant, the interaction of RNase E with the Hfq-bound sRNA is reduced, whether or not PNPase is present, due to the loss of the RNase E domain involved in the normal interaction. As a result, there is reduced degradation of the sRNA in the presence or absence of PNPase. In addition, the C-terminal deletion in RNase E may also contribute to the suppression by reducing the general degradation activity of RNase E. PNPase may also be decreasing the rate of degradation of sRNAs by negatively regulating RNase E activity through allosteric effects achieved from binding to the C-terminal scaffold domain, or by blocking the binding of other proteins to the C-terminal scaffold domain that may enhance its activity. This may explain the additive effects of the hfq and pnp deletion in increasing the degradation of sRNAs (Fig. 3A,B). Consistent with a general regulation of the catalytic activity of RNase E by the C-terminal scaffold domain, previous studies by Leroy et al. (2002) demonstrated that deletion of the whole C terminus (rne-131) significantly reduced the ability of RNase E to degrade a model untranslated substrate in vivo, while deletion of just the PNPase binding site (rneΔ10) increased activity. In another study, a deletion similar to rneΔ10, rne-225, increased the turnover of lpp, while longer deletions reversed this effect (Ow et al. 2000).
Why does truncation of the C-terminal scaffold domain that includes the RhlB and enolase binding site of RNase E suppress the cold sensitivity of the PNPase mutant? PNPase is up-regulated at low temperatures (Mathy et al. 2001). The assumption has been that PNPase directly participates in the degradation of structured mRNAs at low temperature. In the absence of PNPase, the turnover of mRNAs encoding several cold-shock proteins is decreased, and the stabilization of these mRNAs, which allows continued expression of cold-shock proteins, presumably causes the growth arrest of pnp mutants at low temperatures (Yamanaka and Inouye 2001). However, our finding that rne truncations suppressed cold sensitivity suggests that the loss of PNPase is harmful at low temperature due instead to the resulting overdegradation or unregulated degradation of some sRNAs and mRNAs.
There have been previous reports on the effect of a pnp deletion on the turnover of sRNAs, with different results from the result here. In our previous work (Massé et al. 2003), we found that a pnp mutant did not affect RyhB-dependent regulation of sodB. We retested the pnp::Tn5 strain used, EM1368, and found that it did not show cold sensitivity and that the kanamycin resistance marker was not linked to the pnp region (data not shown). Therefore, it seems likely that the Tn5 transposon moved elsewhere during construction of this strain. In the current work, we observed significant growth defects for pnp mutants under all conditions, highlighting some of the difficulties of working with these strains and the possibility of selecting suppressors. Whether the other differences with published results reflect different roles for PNPase for different sRNAs or are due to different strains and different growth conditions is not yet known. Viegas et al. found that introduction of a pnp deletion into Salmonella enterica serovar Typhimurium increased the half-life of the MicA and SraL sRNAs threefold during stationary phase (Viegas et al. 2007). In related work, Andrade and Arraiano showed that the half-life of the MicA sRNA is more than three times longer in a pnp deletion mutant than in a wild-type strain of E. coli (Andrade and Arraiano 2008). These authors also found that introduction of a pnp deletion into a strain of E. coli substantially decreased the ompA mRNA level, and this decrease was at least partially dependent upon MicA. These results suggest that the increase in the stability of the MicA sRNA in the pnp mutant was responsible for the lower level of the ompA mRNA that was observed. These papers used stationary phase cultures. Andrade and Arraiano found no significant difference in the half-life of the MicA sRNA in a pnp mutant during exponential phase, the condition under which we examined the half-lives of multiple sRNAs.
We note that we did not isolate any mutations in rne itself. The failure to find such mutations probably reflects the conditions of our selection/screening, in which repression of translation by the sRNAs was sufficient to give the wild-type phenotype. As confirmation of this, we reconstructed the selection strain with an rne-131 mutation. The cirA-lacZ fusion is still repressed by OmrB in that host (data not shown). Thus, in spite of the clear role of the RNase E degradosome in degradation of both the mRNA targets and the sRNA, such degradation is not always essential for Hfq-dependent gene regulation (Maki et al. 2008).
Our model leaves a number of questions unanswered. What changes upon pairing, in terms of RNase E access, allowing degradation under these conditions? Possibly pairing to an mRNA directly activates RNase E. Alternatively, pairing leads to Hfq displacement or more efficient competition by other sRNAs for the bound Hfq. Thus, our results suggest that the trafficking of Hfq on and off RNAs and the interaction with RNase E are more complex than expected. This will be a useful direction for further work.
MATERIALS AND METHODS
Bacterial strains and plasmids
All strains used in this study, other than the DY330 strain used for recombineering, are derivatives of E. coli K-12 strain MG1655. Strains and plasmids used in this study are listed in Table 2. The primers and 5′-biotinylated probes used in this study are listed in Table 3 and were supplied by Integrated DNA Technologies, Inc. All transductions were performed using phage P1vir as described by Miller (1992). The yjfP deletion strain NRD336 was generated by λ Red recombinase-mediated gene replacement using the PCR product generated from the template plasmid pKD3, using yjfPKO For and yjfPKO Rev primers. Replacement of the entire pnp coding region by a chloramphenicol or kanamycin cassette in strain NRD463 or NRD465, respectively, was accomplished via λ Red recombinase-mediated gene replacements using the PCR product generated from the template plasmid pKD3 or pKD4, respectively, using the primers pnpKO For and pnpKO Rev.
TABLE 2.
Strains and plasmids used in this study
TABLE 3.
Primers and probes used in this study
Plasmid pNRD412 was constructed by amplification of pnp from strain MG1655 using pnp-SacI For and pnp-HindIII Rev primers and subsequently cloning the PCR product into the vector pBAD30 digested with SacI and HindIII. Plasmid pNRD414 was generated by amplification of hfq from strain MG1655 using hfq-EcoRI For and hfq-HindIII Rev primers and then cloning the PCR product into pBAD30 digested with EcoRI and HindIII. Finally, plasmid pNRD416 was made by amplification of rne from strain MG1655 using primers rne-EcoRI For and rne-XbaI Rev and subsequently cloning the PCR product into pBAD30 digested with EcoRI and XbaI. All of the plasmids were sequenced to verify the fidelity of the cloned gene.
Culture media and growth conditions
Strains were grown in liquid medium or agar plates containing Lennox broth or M63 supplemented with vitamin B1 at a final concentration of 0.001% and glucose or succinate at a final concentration of 0.2%. When medium was supplemented with arabinose, arabinose was used at a final concentration of 0.1%, unless otherwise specified. Antibiotics were used at the following concentrations: ampicillin, 100 mg L−1; chloramphenicol, 10 mg L−1; kanamycin, 25 mg L−1; rifampicin, 250 mg L−1; and tetracycline, 25 mg L−1. 2,2′-Dipyridyl (dipyridyl) or α-methyl-D-glucoside (α-MG) was added to Lennox broth liquid medium or agar plates at a final concentration of 250 μM or 0.5%, respectively. XG and IPTG were used at a final concentration of 20 mg L−1 and 100 μM, respectively.
Mutagenesis
Chemical mutagenesis of strain MG1193 harboring plasmid pBR-plac-OmrB was performed using 2-aminopurine as described by Miller (1992). Briefly, 50 μL of a 10−4 dilution of an overnight culture of strain MG1193 harboring pBr-plac-OmrB was added to three tubes containing 3.0 mL of LB liquid medium with ampicillin and 2-aminopurine (700 mg L−1). After 24 h incubation at 37°C, the cultures were serially diluted 10-fold, and the dilutions were plated on minimal M63 agar plates containing ampicillin, vitamin B1, and succinate and incubated at 37°C. Colonies that formed on the succinate minimal plates were restreaked onto glucose minimal plates containing ampicillin, IPTG, and X-Gal. In the absence of mutagenesis, mutations with the desired phenotype (growth on succinate, blue on X-Gal) arose at around 1/109. After mutagenesis, the rate was increased more than 100-fold, to better than 1/107. A total of 26 mutants from three independent pools of mutagenesis and one spontaneous mutant that survived the selection and passed the screen were further characterized.
Generation of a pool of random transposants
Strain MG1193 was transformed with plasmid pNK2884. An overnight culture of strain MG1193 harboring pNK2884 was diluted 200-fold into fresh LB medium containing ampicillin and grown at 37°C to an OD600 of 0.3–0.4. Transposition of the mini-Tn10 cam was induced for 1 h with IPTG, and the cells were then pelleted at 4500 rpm for 10 min. The supernatant was removed, and the cells were suspended in fresh LB to a final OD600 between 0.1 and 0.2. Phage P1vir was then added to the culture, and a P1 lysate was generated as described by Miller (1992).
Mapping of mutations
Preliminary experiments demonstrated that loss of hfq activity would allow cells to pass the selection/screening procedure. Thus, the initial step in mapping was to identify any hfq mutants that were obtained. All mutants, originally selected for growth on succinate, were transduced with a P1 lysate grown on strain NRD336 containing yjfP::cat, a transposon linked to hfq+, and transductants were selected on M63 agar plates containing vitamin B1, glucose, and chloramphenicol at 37°C. The chloramphenicol-resistant transductants were then tested for growth on succinate by restreaking onto M63 agar plates with vitamin B1 and either glucose or succinate and incubated at 37°C. For those mutants that produced a significant number of chloramphenicol-resistant transductants that no longer grew on succinate minimal plates, suggesting the presence of an hfq mutation enabling succinate growth in the original mutant strain, the hfq allele from the original mutant was amplified and sequenced using the primers hfq For and hfq Rev.
Those mutants that yielded only chloramphenicol-resistant transductants that had retained the ability to grow on minimal succinate media were concluded to be somewhere other than hfq and were treated as follows to identify a transposon linked to the mutation (Kleckner et al. 1991). The mutants were transduced with a P1vir lysate grown on a pool of mini-Tn10 cam transposants generated in the wild-type parental strain MG1193. Transductants were plated on MacConkey-lactose agar plates with ampicillin and chloramphenicol and incubated at 37°C and the rare (1/200) Lac− transductants identified; in these strains, the negative regulation of the cirA-lacZ fusion by OmrB is restored. Thus, the transposon insertion in these transductants should be cotransducible with the wild-type allele of the original unmapped mutation. The cotransduction frequency of the transposon and the Lac− (wild type) phenotype was determined by transducing the original mutant again, this time with a P1vir lysate grown on a Lac− transductant, and selecting for the acquisition of the transposon and screening for the loss of the Lac+ (mutant) phenotype on MacConkey-lactose agar plates with ampicillin and chloramphenicol. Those transductants that acquired the Lac− phenotype were also tested for the ability to grow on minimal plates containing succinate as the sole carbon source to assess whether the ability of RyhB to negatively regulate the sdhCDAB mRNA was also restored.
The location of the transposon linked to the Lac− phenotype was identified by thermal asymmetric interlaced (TAIL) PCR from 200 ng of purified genomic DNA, according to Liu et al., using primer Tn10cam487 along with primer AD1, AD2, or AD3 (Liu et al. 1995). The resulting PCR product was diluted 40-fold in ultrapure water and was subsequently used as a template for a second PCR reaction using the primers Tn10cam580 and AD1, AD2, or AD3. The product of this subsequent PCR was then purified using a Qiagen MinElute PCR purification kit and then sequenced using the Tn10cam580 primer. After the insertion site of the transposon was identified by sequencing, sequencing was then performed on the most promising candidate genes in the vicinity of the transposon to identify the mutation responsible for the Lac+ phenotype in the mutant. Three of these transposon insertions, present in NRD429, NRD430, and NRD437, were used in further analysis.
Northern blotting
The effects of hfq, pnp, and rne mutations on levels and turnover of sRNAs and mRNAs were tested by Northern blotting. For the determination of RyhB and sodB turnover, overnight cultures were diluted 200-fold into fresh LB medium and incubated at 37°C. A 700-μl sample was removed from each culture when an OD600 between 0.3 and 0.4 was reached, and dipyridyl was added to each culture. Additional samples were removed 2, 5, 10, and 15 min after dipyridyl addition. Sixteen minutes after dipyridyl addition, each culture was treated with rifampicin, and additional samples were removed at appropriate intervals after treatment. RNA was extracted from each sample by the hot phenol method previously described by Massé et al. (2005). To examine the effect of overexpression of Hfq on the turnover of RyhB and the sodB mRNA, overnight cultures of strains harboring pBAD30 or pNRD414 (pBAD30-hfq+) were diluted 200-fold into fresh LB medium containing ampicillin. When cultures reached an OD600 between 0.3 and 0.4, arabinose was added to each culture to a final concentration of 0.2%. A 700-μL sample was removed from each culture 5 min after arabinose addition, and dipyridyl was then added to each culture. Additional samples were removed 2, 5, 10, and 15 min after dipyridyl addition. Rifampicin was added to each culture 16 min after dipyridyl addition, and samples were removed and RNA extracted as described above.
To examine the turnover of SgrS and ptsG mRNA, overnight cultures were diluted 200-fold into fresh LB medium and incubated at 37°C. A 700-μL sample was removed from each culture when an OD600 between 0.3 and 0.4 was reached, and α-MG was added to each culture. Additional samples were removed 2, 5, 10, and 15 min after α-MG addition. Sixteen minutes after α-MG was added, rifampicin was added to each culture, and samples were removed 2, 5, 10, and 20 min after rifampicin treatment. RNA was extracted from each sample as described above. To determine the turnover of CyaR and the ompX and ompX′-′lacZ mRNA, overnight cultures were diluted 200-fold into fresh LB medium and incubated at 37°C. When the OD600 of each culture was between 0.3 and 0.4, a 700-μL sample was taken, and then IPTG was added to each culture. Additional samples were taken from each culture 2, 5, 10, and 15 min after IPTG addition, and 16 min after IPTG addition, rifampicin was added to each culture. Samples were removed after rifampicin treatment, and RNA was extracted as described above for RyhB and SgrS.
Northern blot analysis of RyhB, SgrS, and CyaR expression and turnover was performed by fractionating 3 μg of RNA from each sample on a Bio-Rad Criterion 10% Tris-borate-EDTA (TBE)-urea polyacrylamide gel in 1× TBE at 55 V for 3 h after prerunning the gel at 55V for 30 min. The fractionated RNA was then transferred by electroblotting to a Zeta-Probe GT membrane (Bio-Rad) at 200 mA for 2 h in 0.5× TBE. The RNA was cross-linked to the membrane by UV irradiation, and the membrane was then probed with the appropriate sRNA probe in ULTRAhyb solution (Ambion) at 42°C overnight. The blot was then developed using the Brightstar Biodetect kit (Ambion) as per the manufacture's instructions.
Northern blot analysis of mRNA turnover or rne expression level was performed by fractionating 10 μg of RNA from each sample on a 1.2% agarose gel that was prerun for 5 min at 12 V/cm in a 1× morpholinepropanesulfonic acid (MOPS) buffer and subsequently run at 5 V/cm for 2 h. The RNA was transferred to a Zeta-Probe GT membrane (Bio-Rad) by capillary action overnight, and cross-linked to the membrane by UV irradiation. The membrane was probed with the appropriate mRNA probe in ULTRAhyb solution (Ambion) at 42°C overnight and then developed as described above. For a given experiment, all of the RNA samples fractionated were loaded with an equivalent amount of RNA, run on the same agarose gel, transferred to the same membrane, and developed for the same amount of time to allow direct comparisons between different genetic backgrounds.
β-galactosidase assays
To determine the expression of the ompX′-′lacZ translational fusion, overnight cultures of strains harboring the vector pBr-plac or pNRD405 were diluted 200-fold into fresh LB medium containing ampicillin and IPTG and grown to an OD600 between 0.3 and 0.4. Samples were then removed from each culture, and a β-galactosidase assay was performed as described by Miller (1992).
Western blotting
To determine the expression level of Hfq or RNase E, overnight cultures of the relevant strains were diluted 200-fold into fresh LB medium and incubated at 37°C. A 1.0-mL sample was removed from each culture when the OD600 reached 1 (for Hfq) or between 0.3 and 0.4 (for RNase E). For the RpoS Western blots, overnight cultures of each strain were diluted 200-fold into fresh LB medium with ampicillin and incubated at 37°C. When cells were between 0.3 and 0.4, IPTG was added for 20 min to induce the sRNAs and a 1.0-mL sample was removed. All samples were immediately added to a tube containing 110 μL of an ice-cold 50% solution of trichloroacetic acid (TCA). The sample was incubated on ice for 10 min, and subsequently the precipitated protein was pelleted by centrifugation at 13,000 rpm for 10 min. The supernatant was removed, and the pellet was suspended in an 80% acetone solution. The washed protein was pelleted by centrifugation, the supernatant was removed, and the sample was allowed to air dry for 10 min. The protein was then suspended in 1× SDS sample buffer (New England Biolabs).
Western blot analysis of Hfq was performed by fractionating the collected protein on a NuPAGE Novex 12% Bis-Tris polyacrylamide gel (Invitrogen) in NuPAGE MES SDS running buffer (Invitrogen) at 180 V. The fractionated protein was then transferred to a 0.2 μm-nitrocellulose membrane by electroblotting at 40 V for 75 min in NuPAGE transfer buffer (Invitrogen). The membrane was rinsed with 1× phosphate-buffered saline (pH 7.4) with 0.1% TWEEN 20 (PBST), blocked overnight in PBST with 3% nonfat dry milk and washed 3 times in PBST for 10 min. The membrane was probed with rabbit anti-Hfq antisera diluted 5000-fold in PBST with 0.3% nonfat dry milk for 1 h and subsequently, after three additional PBST washes, was probed with alkaline phosphatase-linked anti-rabbit IgG antibody (Cell Signaling Technology) diluted 1000-fold in PBST with 0.3% nonfat dry milk for 1 h. After 3 washes with PBST, the membrane was then developed using the Lumi-Phos WB chemiluminescent alkaline phosphatase substrate (Thermo Scientific).
Western blot analysis of RNase E levels was performed by fractionating harvested protein on a NuPAGE 4%–12% Bis-Tris polyacrylamide gel (Invitrogen) in NuPAGE MOPS running buffer at 120 V for 2 h. The fractionated protein was subsequently transferred to a 0.2-μm nitrocellulose membrane by electroblotting and treated as described above, except rabbit anti-FLAG monoclonal antibody (Sigma-Aldrich) was used as the primary antibody at a dilution of 1:1000 in PBST with 0.3% nonfat dry milk.
Western blot analysis of RpoS was performed by fractionating the collected protein on a NuPAGE Novex 10% Bis-Tris polyacrylamide gel (Invitrogen) at 180 V for 1 h in NuPAGE MOPS SDS running buffer (Invitrogen). The fractionated protein was transferred to a 0.2-μm nitrocellulose membrane by electroblotting and treated as described above, except rabbit anti-RpoS antisera was used as the primary antibody at a dilution of 1:5000 in PBST with 0.3% nonfat dry milk.
ACKNOWLEDGMENTS
We thank Gisela Storz and members of our laboratory including N. Majdalani, P. Milanesio, D. Schu, K. Moon, A. Battesti, and T. Song for their comments on this manuscript and their advice. We thank H. Aiba and M. Guillier for providing us with strains. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2531211.
REFERENCES
- Andrade JM, Arraiano CM 2008. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM 2007. Hfq structure, function, and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Brown L, Elliott T 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J Bacteriol 178: 3763–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM 1994. Copurification of E. coli RNAase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76: 889–900 [DOI] [PubMed] [Google Scholar]
- Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65: 157–177 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S 2009. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol 191: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Figueroa NV, Pina-Escobedo A, Schroeder I, Simons RW, Garcia-Mena J 2006. Polynucleotide phosphorylase interacts with ribonuclease E through a ββαββα domain. Biochimie 88: 725–735 [DOI] [PubMed] [Google Scholar]
- Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG 2010. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev 24: 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Hayward WS, August JT 1972. Bacterial proteins required for replication of phage Qbeta ribonucleic acid: Purification and properties of host factor I, a ribonucleic acid-binding protein. J Biol Chem 247: 824–831 [PubMed] [Google Scholar]
- Guillier M, Gottesman S 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59: 231–247 [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36: 6781–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrige A, Brechemier-Baey D, Mathy N, Duche O, Portier C 2002. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J Mol Biol 321: 397–409 [DOI] [PubMed] [Google Scholar]
- Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P 2008. Down-regulation of outer membrane proteins by noncoding RNAs: Unraveling the cAMP-CRP- and σE-dependent CyaR-ompX regulatory case. J Mol Biol 383: 1–9 [DOI] [PubMed] [Google Scholar]
- Kajitani M, Ishihama A 1991. Identification and sequence determination of the host factor gene for bacteriophage Q beta. Nucleic Acids Res 19: 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H 2005. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev 19: 328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Bender J, Gottesman S 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol 204: 139–180 [DOI] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci 95: 12456–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy A, Vanzo NF, Sousa S, Dreyfus M, Carpousis AJ 2002. Function in Escherichia coli of the non-catalytic part of RNase E: Role in the degradation of ribosome-free mRNA. Mol Microbiol 45: 1231–1243 [DOI] [PubMed] [Google Scholar]
- Liou GG, Chang HY, Lin CS, Lin-Chao S 2002. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J Biol Chem 277: 41157–41162 [DOI] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS 1989. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol 171: 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lopilato J, Bortner S, Beckwith J 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet 205: 285–290 [DOI] [PubMed] [Google Scholar]
- Luttinger A, Hahn J, Dubnau D 1996. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol Microbiol 19: 343–356 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S 2001. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol Microbiol 39: 1382–1394 [DOI] [PubMed] [Google Scholar]
- Maki K, Uno K, Morita T, Aiba H 2008. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc Natl Acad Sci 105: 10332–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S 2009. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol 72: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29: 3094–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March JB, Colloms MD, Hart-Davis D, Oliver IR, Masters M 1989. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol Microbiol 3: 903–910 [DOI] [PubMed] [Google Scholar]
- Massé E, Gottesman S 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N, Jarrige AC, Robert-Le Meur M, Portier C 2001. Increased expression of Escherichia coli polynucleotide phosphorylase at low temperatures is linked to a decrease in the efficiency of autocontrol. J Bacteriol 183: 3848–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen CA, Benhammou JN, Majdalani N, Gottesman S 2010. Mechanism of positive regulation by DsrA and RprA sRNAs: Pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol 192: 5559–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miczak A, Kaberdin VR, Wei CL, Lin-Chao S 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci 93: 3865–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11: 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol 54: 905–920 [DOI] [PubMed] [Google Scholar]
- Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem 278: 15608–15614 [DOI] [PubMed] [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, Aiba H 2004. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol 54: 1063–1075 [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H 2005. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R 1996. The RNA-binding protein HF-1, known as a host factor for phage QBeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev 10: 1143–1151 [DOI] [PubMed] [Google Scholar]
- Ow MC, Liu Q, Kushner SR 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol Microbiol 38: 854–866 [DOI] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol 68: 890–906 [DOI] [PubMed] [Google Scholar]
- Py B, Causton H, Mudd EA, Higgins CF 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol 14: 717–729 [DOI] [PubMed] [Google Scholar]
- Py B, Higgins CF, Krisch HM, Carpousis AJ 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381: 169–172 [DOI] [PubMed] [Google Scholar]
- Sun X, Wartell RM 2006. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry 45: 4875–4887 [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C 2004. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol Microbiol 51: 1525–1533 [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54: 1076–1089 [DOI] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev 12: 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM 2007. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res 35: 7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31: 6435–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15: 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G 2008. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Inouye M 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J Bacteriol 183: 2808–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci 97: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Deutscher MP 1997. An essential function for the phosphate-dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J Bacteriol 179: 4391–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]