Abstract
Parkinson's disease (PD) is a progressive neurodegenerative movement disorder characterized by the selective loss of dopaminergic neurons and the presence of Lewy bodies. The pathogenesis of PD is not fully understood, but it appears to involve both genetic susceptibility and environmental factors. Treatment for PD that prevents neuronal death progression in the dopaminergic system and abnormal protein deposition in the brain is not yet available. Recently, mutations in the leucine-rich repeat kinase 2 (LRRK2) gene have been identified to cause autosomal-dominant late-onset PD and contribute to sporadic PD. Here, we review the recent models for LRRK2-linked Parkinsonism and their utility in studying LRRK2 neurobiology, pathogenesis, and potential therapeutics.
1. Introduction
Parkinson's disease (PD) is the second most common neurodegenerative disorder with movement, cognitive, and emotional dysfunction, affecting 2% of the population over the age of 60 years [1]. PD is characterized by tremors, rigidity, bradykinesia/akinesia, and postural instability resulting from the loss of dopamine neurons in the substantia nigra and other regions of the brain [2–5]. The pathological hallmark of PD is the presence of proteinaceous cytoplasmic inclusions termed Lewy bodies [5, 6]. PD is similar to other neurodegenerative diseases in that it presents with neuronal cell death and protein aggregation, though the relation between them is uncertain [6, 7]. The pathogenesis of PD remains incompletely understood, but it appears to involve both genetic susceptibility and environmental factors. Treatment for PD that prevents neuronal death progression in the dopaminergic system and abnormal protein deposition in the brain is not yet available.
Recently, mutations in the LRRK2 gene have been identified to cause autosomal dominant PD and contribute to sporadic PD [8–10]. To date, more than 50 variants including at least 16 disease-causing mutations have been reported [11–22]. This paper highlights the recent models for LRRK2-linked Parkinsonism and their utility in studying LRRK2 neurobiology, pathogenesis, and potential therapeutics. For other aspects of LRRK2 please refer to several recent excellent review papers [23–26]. Due to the length of this review, we apologize that we did not include all LRRK2 publications.
2. LRRK2 Gene and Protein
The LRRK2 gene spans a genomic region of 144 Kb, with 51 exons encoding 2527 amino acids. The LRRK2 mRNA is expressed throughout the brain and other organs [9]; in situ hybridization in mice reveals that expression predominates within regions of the basal ganglia, which are associated with motor dysfunction in PD, and within nonmotor areas such as the hippocampus [27–31]. The LRRK2 gene is conserved across species from invertebrates to human. Caenorhabditis elegans and Drosophila melanogaster each have only one LRRK2 ortholog [9].
The LRRK2 protein contains several predicted domains (Figure 1) including Roc (Ras in complex proteins, belonging to the Ras/GTPase family), COR (C terminal of Roc), LRR, a leucine-rich repeat, consisting of twelve repetitions of a 22–28 amino acid motif, MAPKKK, a protein kinase catalytic domain which may be involved in serine/threonine phosphorylation, a WD40 domain and ankyrin repeats. The LRR and WD40 domains may be involved in protein-protein interactions [32]. The LRRK2 protein is expressed in all tissues examined, although at low levels. In the brain, LRRK2 is expressed in neurons, astrocytes, and microglia. Recent studies have detected LRRK2 in specific brain regions including the cortex, striatum, hippocampus, cerebellum and in dopaminergic neurons of the substantia nigra [30, 31, 33, 34]. However, the expression levels of LRRK2 in the dopaminergic neurons of the SNpc are very low. LRRK2 protein can be detected in Lewy neurites [35] and in Lewy bodies of sporadic PD [36]. In the subcellular level, it was found mainly in the cytoplasm and associated with lipid rafts, lysosomes, endosomes, mitochondria, and Golgi transport vesicles [9, 30, 33, 34, 37–39]. Several studies show that LRRK2 is enriched at the membrane of cells [30, 31, 40, 41] and that the membrane-associated fraction of LRRK2 may display greater kinase and GTP-binding activities than cytosolic LRRK2 [41]. Another study shows the recruitment of LRRK2 to the endosomal-autophagic pathway suggesting the functional involvement of LRRK2 in this pathway [42, 43].
Figure 1.
LRRK2 domain structure and PD-linked point mutations. The predicted domain boundaries are indicated by the residue numbers beneath. The position of the putatively pathogenic amino acid substitutions are shown in purple. Substitutions segregating with PD are shown in green. The kinase null and no-GTP binding alterations are shown in red.
Patients with LRRK2 mutations typically have a relatively late onset of PD with asymmetric rest tremor, bradykinesia, rigidity, and a good response to L-DOPA treatment [9, 44]. The pathological heterogeneity of affected individuals examined ranges from pure nigral degeneration without Lewy bodies to nigral degeneration associated with Lewy bodies, widespread Lewy bodies consistent with diffuse Lewy body disease, or neurofibrillary tau-positive tangles [8, 9, 45, 46]. Point mutations have been identified in almost all of the predicted domains of LRRK2 (Figure 1) [1, 21, 22, 47–49]. The most common mutation, G2019S, contributes to 5-6% of autosomal-dominant PD [50, 51] and 1-2% of sporadic PD [52]. The distribution of mutations across several different LRRK2 domains, the lack of deletions or truncations, and the dominant pattern of inheritance, are consistent with a gain-of-function mechanism for LRRK2-associated PD.
The normal function of LRRK2 is still unclear. Loss-of-function studies indicate that the Drosophila LRRK2 homologous protein (CG5483) is critical for the integrity of dopaminergic neurons in the fly [53] and Zebrafish LRRK2 homology is important for neuronal development [54]. Suppression of LRRK2 with siRNAs or a dominant inhibitory allele leads to increased neurite process length and complexity [55]. Based on the multidomain structure and various identified LRRK2 mutations, LRRK2 is predicted to serve as an upstream central integrator of multiple signaling pathways that are crucial for proper neuronal functioning. The presence of LRR and WD40 (protein interaction domains) and Roc and MAPKKK (enzymatic domains) within LRRK2 suggests that this protein may serve as a scaffold for the assembly of a multiprotein signaling complex. LRRK2 associates with various protein partners that are involved in several cellular pathways including chaperone machinery, cytoskeleton arrangement, protein translational machinery, synaptic vesicle endocytosis, the MAPK signaling cascades, ubiquitin/autophage protein degradation pathways, and other unidentified processes [23].
3. In Vitro Models and LRRK2 Biology
Studies using in vitro models (Table 1) reveal that LRRK2 is a kinase and a GTPase and identify various interaction partners, suggesting that LRRK2 may play important roles in protein aggregation and neuronal degeneration.
Table 1.
LRRK2 in vitro cell models.
| Genes | Cell type | Toxicity | Protein aggregation | Kinase activity | GTPase activity | References |
|---|---|---|---|---|---|---|
| WT, R1441C, Y1699C, G2019S |
HEK293T SH-SY5Y Primary neurons |
+ | ND | ND | ND | [79] |
| WT, I2020T | HEK293 | ND | ND | + | ND | [38] |
| WT, G2019S, R1441C | HEK293 SH-SY5Y |
ND | ND | + | ND | [39] |
| WT, G2019S, I2020T | Primary neuron | + | + | + | ND | [55] |
| WT, R1441C, Y1699C, G2019S |
SH-SY5Y | + | + | + | ND | [56] |
| WT, G2019S, G2019S- K1906A, G2019S- D1994N, G2019S- DY2017-2018AL, WT- K1347A, G2019S- K1347A |
HEK-293 SH-SY5Y Primary neuron |
+ | ND | + | + | [57] |
| WT, K1906M, G2019S, R1441C, R1441G, I1371V, I1122V, R1514Q, Y1699C, G2385R, I2012T, I2020T |
HEK293FT SH-SY5Y Primary neuron |
+ | ND | + | + | [60] |
| WT, G2019S, A2016T, WT/A2016T, G2019S/A2016T, R1441C, Y1699C |
HEK-293 Swiss-3T3 Human lymphoblastoid cells |
ND | ND | + | ND | [80] |
| WT, T1343G, K1906M, T2035A, R1398Q |
HEK-293 Neuro-2a |
ND | ND | − | ND | [76] |
| WT, R1441C/G, T1398N | HEK-293T | ND | ND | ND | + | [67] |
| WT, G2019S | HEK-293 Primary neuron |
ND | ND | + | ND | [81] |
WT: wild type; ND: not determined.
3.1. LRRK2 Kinase Activity
In vitro studies demonstrate that LRRK2 is predominately a serine/threonine protein kinase, which can phosphorylate itself and a generic substrate, myelin basic protein (MBP) [39, 55–60]. A LRRK2 variant with three potential sites of autophosphorylation altered to alanines (T2031A, S2032A and T2035A), does not display autophosphorylation activity and cannot phosphorylate the generic substrate, MBP [61, 62]. Further in vitro studies demonstrate that S2031 and T2032 are the critical residues required for LRRK2 autophosphorylation, and T2035 is important for catalytic activity, but does not serve as a phosphate acceptor [58]. Additional studies show that dimeric LRRK2 undergoes intramolecular autophosphorylation and that an intact C-terminus is required for kinase activity [61]. One recent report shows that T1343 also is an autophosphorylation site [63]. Moreover, S910 and S935 are also potential phosphorylation sites that may be involved in 14-3-3 proteins binding with LRRK2 [64–66]
Several pathogenic mutations of LRRK2 in PD have been found within the protein kinase domain active segment (e.g., G2019S), suggesting that these mutations may cause pathology through altering the kinase activity of LRRK2 [23]. The results from the most common mutation G2019S support this notion to increase LRRK2 kinase activity in assays to measure autophosphorylation or phosphorylation of generic substrates [39, 55–59]. However, controversy remains regarding whether other PD mutations alter LRRK2 kinase activity. For example, several studies demonstrated that the I1122V, R1441C, R1441G, R1514Q, Y1699C, and I2020T familial PD linked mutations of LRRK2 increased kinase activity [35, 37, 57, 67, 68]. Additionally, other mutations either did not influence or inhibit kinase activity [59, 60, 69]. Currently, LRRK2 kinase assays use in vitro autophosphorylation or phosphorylation of generic substrate or a phosphopeptide. Accordingly, the kinase activity results of some mutants vary among various laboratories, in part due to lack of sensitivity in the kinase assay of choice and various expression constructs. Identifying a physiologic substrate of LRRK2 and resolving the question of whether pathogenic mutations affect phosphorylation of this substrate is critically important to determine the mechanism by which LRRK2 induces PD see Figure 2.
Figure 2.
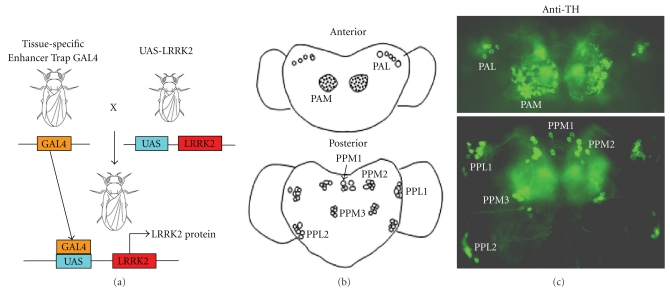
UAS/GAL4 system and fly brain dopaminergic neurons. A. Diagram of GAL4/UAS system to illustrate that tissue-specific expression of GAL4 leads to transcriptional activation of LRRK2. B. Diagram of DA neuron clusters in the medial and lateral areas of the adult fly brain as in previous publications [34, 67]. Five clusters: PPM1 (unpaired), PPM2 (paired), PPM3 (paired; protocerebral posterior medial), and PPL1 and PPL2 (paired; protocerebral posterolateral) on the posterior side. (Center) Two DA clusters: PAL (protocerebral anterolateral) and PAM (paired anterolateral medial) on the anterior side. C. Images of whole-mount-immunostaining of dopaminergic neurons in a adult fly brain using anti-TH antibodies followed by green fluorescent-conjugated second antibody detection.
To date, the physiological substrate(s) of LRRK2 remains unclear [23]. A search for proteins that are phosphorylated by the PD-linked mutant LRRK2-G2019S using rat brain extracts reveals that moesin is a substrate [59]. Moesin is a protein that anchors the actin cytoskeleton to the plasma membrane. Denatured moesin is efficiently phosphorylated by LRRK2 at Thr558, the residue previously identified as an in vivo phosphorylation site that regulates the ability of moesin to bind actin. LRRK2 also phosphorylates ezrin and radixin, which are involved in moesin binding actin [59]. Collapsing response mediator protein-2 (CRMP-2) has also been identified as a weak LRRK2 substrate, which is involved in the regulation of growth cones, microtubule dynamics and neurogenesis [70]. Recent reports also show that 4E-BP [71] and mitogen-activated protein kinase can also be phosphorylated by LRRK2 [72–74]. Additional studies are required to establish the physiological significance of these proteins as LRRK2 substrates.
3.2. LRRK2 GTP Binding and GTPase Activity
LRRK2 is a member of the recently defined ROCO family [75] and harbors a GTP-binding regulatory domain (ROC-COR) [76, 77]. LRRK2 is a GTP/GDP-binding protein, as measured by specific binding to GTP-agarose and radio-labeled GTP [57, 60, 76]. Both wild-type and PD-linked mutant LRRK2 bind to GTP and GDP. LRRK2-K1347A, which bears a mutation that alters the predicted GTP-binding site, does not appreciably bind to GTP and reduces kinase activity. This finding is further confirmed by the recent report showing that the crystal structure of the LRRK2 ROC domain in complex with GDP-Mg (2+) at 2.0-A resolution [78]. The crystal structure displays a dimer of the ROC domain. Two PD-associated pathogenic residues, R1441 and I1371, are located at the interface of two monomers that may alter the ROC dimerization and regulate LRRK2 GTPase and/or kinase activity. LaVoie's recent study further suggests that LRRK2 dimerization is associated with membrane binding and increased GTPase activity [41]. Familial-linked mutations in LRRK2 within the ROC and COR domains (I1371V, R1441C, R1441G, and Y1699C) appear to increase GTP-binding as measured by binding to GTP-agarose, whereas mutations outside these domains did not affect GTP binding compared with wild-type LRRK2 [60]. However, other studies have shown that R1441C mutation do not increase GTP binding [67, 69].
The ROC domain of LRRK2 shares sequence homology with all five subfamilies of the Ras-related small GTPase super family (Ras, Rho, Rab, Sar/Arf and Ran) and contains conserved motifs for GTPase activity. Three independent groups have demonstrated that LRRK2 has intrinsic GTPase activity and undergoes intrinsic GTP hydrolysis [67–69, 82, 83]. The purified full-length LRRK2 has only weak GTPase activity, suggesting that if it is active in the cell it may require accessory proteins. Notably, the ROC domain of LRRK2 is sufficient for its intrinsic GTPase activity. LRRK2 binds and hydrolyzes GTP similarly to other Ras-related small GTPases. Based on in vitro assays, R1441C/G and Y1699C PD-linked mutations appear to decrease in the rate of GTP hydrolysis compared to the wild-type LRRK2, suggesting that these mutants spend more time in the activated GTP-bound state [69, 78, 84].
Several studies have demonstrated that GTPase domain activity may regulate LRRK2 kinase activity [57, 58, 67, 82] since GTP binding stimulating LRRK2 kinase activity [58, 67] although there is still some evidence against the GTP binding activation model [85]. It is hypothesized that LRRK2, like other Ras-related GTPases, may serve as a molecular switch to regulate diverse cellular functions by cycling between GTP-bound (active) and GDP-bound (inactive) conformations. Based on the putative dimeric structure of LRRK2, it is predicted that the dimeric ROC or ROC-COR domains act as binary switches to regulate kinase activation [78, 84]. In this model, at the GTP-bound conformation, the dimerization of ROC or ROC-COR domains further induces self-association of the kinase domains, thus allowing for autophosphorylation and subsequent activation of downstream kinase activity [61, 78, 84]. LRRK2 may regulate its own activity, as well as perhaps fulfilling a signaling role by regulating other proteins in the cell [41, 61, 86–88]. Multiple reports have shown that the kinase domain of wild-type LRRK2 phosphorylates several sequences within the GTP-binding ROC domain [61, 63, 89, 90], suggesting that the kinase domain may also regulate overall LRRK2 function. The PD-linked mutations do not identically display the same kinase or GTP domain activities, suggesting that there may be some interesting mechanistic differences between different mutations in the same domain, however the caveat that these observations could also be due to methodological differences between assays [91]. Nevertheless, mutations may prompt the protein to enter a GTP-bound state or slow the protein's return to the GDP-bound state.
3.3. Mutant LRRK2 Induces Toxicity
Patients with LRRK2 mutations exhibit neuronal degeneration in the brain [8, 9, 45–47]. PD-associated mutations of LRRK2 induce cell toxicity in multiple cell lines and rodent primary neurons with reduction of cell viability ranging from 10–40% (Table 1). Expression of mutant LRRK2 variants (I1122V, R1441C, Y1699C, G2019S, and I2020T) strikingly decreases neuronal cell viability by 2–5-folds. However, overexpression of wild type LRRK2 does not significantly decrease cell viability [55–58, 60, 79, 92].
Mutant LRRK2-mediated cell toxicity appears to involve apoptotic mechanisms as measured by TUNEL staining and caspase activation [57, 58, 79]. The mitochondria-dependent apoptotic pathway, in which cytochrome c is released and caspase-3 is activated, is thought to mediate mutant LRRK2 toxicity in neuronal cells. This seems to be dependent on Apaf1, a scaffold protein participating in apoptosome formation [83]. Another study has also shown that LRRK2 interacts with the death adaptor Fas-associated protein at the death domain (FADD), which may play a role in apoptotic neuronal death [93]. Since kinase activity is a critical component of LRRK2, significant efforts have been made to determine whether kinase activity is responsible for LRRK2 toxicity. Abolishing LRRK2 kinase activity diminishes the toxicity of all PD mutants tested in cell culture [56, 57]. Genetic alterations of LRRK2 with D1994N (a predicted proton acceptor) abolishes the predicted active site, K1906A (a ATP binding site) abolishes a putative ATP-binding site, and/or DY2017-2018AL altering the predicted DYG kinase active conserve motif significantly reduces LRRK2 kinase activity. Importantly, these constructs reduce mutant LRRK2-induced neuronal degeneration [56, 57]. Additionally, one study has shown that overexpression of the kinase domain, the ROC-COR-kinase fragment or the ROC-COR-kinase-WD40 fragments containing G2019S and R1441C mutations can reduce cell viability [94]. A recent report further supports this notion that inhibitors of Raf kinase GW5074, sorafenib and Raf inhibitor IV inhibit LRRK2 autophosphorylation and MBP phosphorylation result in reducing mutant LRRK2 toxicity [95].
R1441C and Y1699C mutants are associated with reduced LRRK2 GTPase activity [68, 69, 82, 83] suggesting that GTPase activity may contribute to LRRK2 toxicity. In addition, the K1347A alteration abolishes GTP binding and reduces kinase activity thereby reducing the mutant LRRK2-induced neuronal toxicity in cell culture [57, 60]. A recent study shows that the cytotoxic effect of ROC-ROC-kinase fragment in yeast was increased in a GTPase dead background or after the induction of R1441C mutation, which reduced GTPase activity [83]. This toxicity can be reduced by introduction of GTPase stimulating alterations (T1343G/R1398Q or R1398L). However, in cell culture, only the augmentation of the toxicity effect caused by ROC-ROC-kinase fragment can be replicated [83]. Thus, the contribution of GTPase domain activity in LRRK2 toxicity still warrants further investigation.
Recent studies have also shown that deletion of the LRR and WD40 domains can rescue G2019S and/or R1441C-LRRK2-induced toxicity [92, 96], likely via kinase activity as the deletion of the WD40 domain or even shorter C-terminal sequences renders LRRK2 kinase inactive [59, 96]. Given that the LRR and WD40 are putative protein-protein interaction domains, it is suggested that LRRK2 protein interactions may also contribute to its toxicity. However, this needs further study. Several pathogenic mutations (I1122V, R1441C, Y1699C, G2019S, and I2020T) increase the tendency of LRRK2 to form inclusion bodies [33, 56] suggesting that LRRK2 kinase activity may also contribute to protein aggregation [56, 79]. Together, these findings suggest that LRRK2 protein kinase activity plays an important role in both neuronal degeneration and protein aggregation, but the cellular pathways underlying these functions need further study.
3.4. LRRK2 Interaction Partners and Potential Cellular Pathways
There is a growing number of LRRK2 interaction partners that are identified and involved in several cellular pathways including chaperone machinery, cytoskeleton arrangement, protein translational machinery, synaptic vesicle endocytosis, the MAPK signaling cascades, ubiquitin/autophage protein degradation pathways, and other unidentified processes (Table 2).
Table 2.
LRRK2 potential interaction proteins.
| Pathway | LRRK2 fragment | Link with LRRK2 | Method | References |
|---|---|---|---|---|
| Apoptosis | Full length | FADD | Co-IP (HEK293T cells, mouse brain) |
[93] |
| Full length | TRADD, RIP1 | Co-IP (HEK293T cells) |
[93] | |
| Synaptic vesicle endocytosis | Full length LRR |
Rab5b | YTH, pulldown, Co-IP |
[111] |
| MAPK signaling | Full length, COR, Kinase domain |
MKK3 | Co-IP (HEK293T cells) |
[72, 73] |
| Full length | MKK4 | Co-IP (HEK293T cells) |
[72] | |
| Full length, COR, Kinase domain |
MKK6 | Co-IP (HEK293T cells), C. elegans |
[72, 73] | |
| Full length, COR, Kinase domain |
MKK7 | Co-IP (HEK293T cells) |
[72, 73] | |
| Full length | JIP1–3 | Co-IP (HEK293T cells) |
[74] | |
| Full length | JIP4 | Co-IP (HEK293T cells) |
[74] | |
| Chaperone machinery | Full length, Kinase domain, N-term |
Hsp90 | Co-IP (HEK293T cells, mouse brain), YTH |
[81, 88, 97–99] |
| Full length, Kinase domain, |
p50CDC37 | Co-IP (HEK293T cells, mouse brain) |
[38, 81] | |
| Full length, ROC, N-term |
CHIP | Co-IP (HEK293T cells, mouse brain), YTH |
[98, 99] | |
| cytoskeleton | Full length, ROC |
a-tubulin | pulldown | [102] |
| Full length, ROC |
b-tubulin | Co-IP (HEK293T cells, mouse brain), pulldown |
[102, 105] | |
| Full length | EF1A | Co-purification (insect cells), Co-IP (HEK293T cells) |
[113] | |
| — | moesin | in vitro, in vivo | [59, 101] | |
| Full length, ROC-COR |
DVL1/2/3 | YTH, Co-IP (HEK293T cells) |
[109] | |
| Full length | Sgg/GSK3b | Drosophila | [114] | |
| Full length | Actin cytoskeleton proteins | QUICK, Co-IP (NIH3T3 cells) |
[115] | |
| Protein translation | — | 4E-BP | in vitro, Drosophila | [71, 116] |
| PD related proteins and others | Full length, COR | Parkin | Co-IP (HEK293T cells, SH-SY5Y cells, primary neurons), |
[79] |
| Full length | 14-3-3 isoforms | Co-IP (HEK293T cells, Swiss 3T3 cells, mouse brain, kidney, spleen) |
[64–66, 80] | |
FADD: Fas-associated protein with death domain; TRADD: tumor necrosis factor receptor type 1-associated death domain protein; LRR: leucine-rich repeat; YTH: yeast two-hybrid; ROC: Ras of complex protein; COR: C-terminal of ROC; MKK: mitogen activated protein kinase kinase; JIP: JNK interacting protein; Hsp: Heat shock protein; CHIP: C-terminus of Hsp70 interacting protein; EF1A: elongation factor 1α; DVL: dishevelled family of proteins; Sgg: glycogen synthase kinase 3β homolog Shaggy; QUICK: quantitative immunoprecipitation combined with knockdown.
LRRK2 interacts with proteins involved in chaperon pathways including Hsp90, Hsp90//p50cdc37, HSp60, Hsp 70, and the c-terminal Hsp70 interacting protein (CHIP) [38, 81, 88, 97–99]. The Hsp60 interacts with recombinant human LRRK2 kinase domain in E. coli, and Hsp90/p50cdc37 interacts with full-length LRRK2 in mammalian cells [38, 58]. These chaperone proteins may help to maintain the proper folding of LRRK2. The HSP90/p50cdc37 chaperone complex binds to LRRK2 and may assist with the activation of other protein kinases [38]. In these studies, the Hsp90/p50cdc37 proteins do not serve as substrates but rather associate as chaperones assisting in proper folding and activation of the kinase. It has been shown that inhibition of Hsp90 disrupts the association of this chaperone with LRRK2 leading to proteasomal degradation of LRRK2, suggesting that Hsp90 inhibitors may be useful therapeutically to limit mutant LRRK2-mediated toxicity in neurons [81, 98]. CHIP binds ubiquitinates and promotes the ubiquitin proteasomal degradation of LRRK2 [98]. Overexpression of CHIP protects against mutant LRRK2-induced toxicity whereas knockdown of CHIP exacerbates toxicity mediated by mutant LRRK2 via reducing degradation of LRRK2 proteins.
LRRK2 associates with various cytoskeleton proteins including alpha/beta-tublin, F-actin, moesin-related ezrin-radixin-moesin (ERM) family members, and the dishevelled family proteins [100], suggesting that LRRK2 may play a critical role in the regulation of microtubule and actin dynamics. LRRK2 associated with actin dynamics is evidenced by the following studies. MacLeod et al. first associated LRRK2 with the maintenance of neuronal process [55] and demonstrated that the neurons expressing the G2019S mutation but not wtLRRK2 had shorter neurites. Suppression of LRRK2 expression by shRNAs led to an increase in neurite length. Moreover expression of G2019S mutation led to tau-positive inclusions, which also colocalized with tau in these inclusions [55]. Biochemical studies show that LRRK2 phosphorylates denatured moesin and associates with other actin-binding ERM proteins: ezrin and radixin [59]. Further studies indicate that LRRK2 may connect with actin dynamics through phosphorylation of ERM proteins [101]. In developing LRRK2 G2019S neurons, the numbers of pERM and F-actin enriched filopodia were significantly increased, which correlates with the retardation of neurite outgrowth in these neurons. Conversely, the levels of pERM and F-actin within the filopodia of LRRK2 knockout neurons were significantly decreased and neurite outgrowth was promoted. These observations suggest a physiological link between LRRK2 and pERM in neuron development and neurite outgrowth [100].
Increasing evidence links LRRK2 with microtubule dynamics. For instance, LRRK2 colocalizes [38, 102] and interacts with tubulin through the LRRK2 ROC domain [102, 103]. LRRK2 phosphorylates β-tubulin at Thr107 in mouse brain, and this phosphorylation is significantly enhanced by G2019S mutation [104]. In vitro studies shows that tubulin phosphorylation by LRRK2 enhances microtubule stability in the presence of microtubule-associated proteins [105]. Moreover, levels of soluble β-tubulin are dramatically decreased in brains of LRRK2 expression mice [103, 106] and are significantly increased in the brains of LRRK2 KO mice [105]. The maintenance of microtubule dynamics is critical for neuronal development, axonal trafficking as well as synaptic formation and maintenance. The G2019S-enhanced tubulin phosphorylation may thus result in deregulation of microtubule dynamics that may in turn interfere with proper neuronal function [105]. Microtubules and microtubule-axonal transport has been reported to play a critical role in maintaining Golgi structure and integrity [107, 108]. Increased fragmentation of the Golgi apparatus was reported in transgenic mice overexpressing LRRK2, and this strongly suggests that the enhancement of tubulin polymerization affects the organization of microtubule in neurons leading to Golgi disruption [103]. Other studies also show that LRRK2 interacts with the dishevelled family of phosphoproteins (DVL1-3) and Rab5b suggesting that the interactions may play an important role in axon guidance and maintaining synaptic function [109, 110] by modulating the endocytosis of synaptic vesicles, further supporting a role for LRRK2 in trafficking [111]. Further investigation still remains to determine whether LRRK2 kinase and GTPase activities are involved in regulation of microtubule and actin dynamics in neuron development, neurite outgrowth and trafficking.
LRRK2 associates with proteins in other kinase cascades. LRRK2 kinase domain shares homology with MLKs and RIPKs, which are involved in signaling events in response to cellular stress insults. Similar to MLKs, LRRK2 has been shown to bind MKK3, 6 and 7 and to phosphorylate MKK3, 4, 6 and 7 [72, 73]. LRRK2 also interacts with the JNK-interacting proteins (JIPs) 1–4 which are scaffolding proteins that bring together MKKs and MAPKs activating the downstream kinases, JNK and p38 [74]. However, it is still unclear whether all the PD-linked mutations alter the interactions with MKKs, JIPs and their linked kinase cascades in PD pathology. Our unpublished data show that genetic or pharmacological suppression of JNK pathway suppressed PD-like Parkinsonism in LRRK2 transgenic flies. In addition, LRRK2 may also interact with ERK1/2 MAPK pathway since the ERK inhibitor U0126 can rescue LRRK2 G2019S-induced neurite shortening and cytotoxicity in culture cells [112]. A report also shows that LRRK2 may interact with oxidative stress via ERK phosphorylation [94]. Like RIPK1, LRRK2 interacts with FADD to induce death signaling resulting in caspase activation and apoptosis [93]. Taken together, these studies suggest that LRRK2 may act as an upstream kinase and interact with multiple cellular stress and cell death signaling pathways.
LRRK2 also associated with other PD-linked proteins. Co-immunoprecipitation studies have shown that LRRK2 associates with the PD-associated protein parkin [79] although there is a conflicting report using a different tagged LRRK2 construct that can not co-IP with parkin [110]. But further Drosophila studies including our own observations show that parkin suppressed LRRK2-induced PD-like phenotypes, suggesting that parkin is associated with LRRK2 in vivo [117, 118]. Although LRRK2 cannot directly bind α-synuclein, DJ-1, or pink-1, there are genetic interactions between LRRK2 and these genes in Drosophila [79, 118], C. elegans [119, 120], cell cultures [121] and mouse models [122]. This is illustrated by studies showing that expression of mutant LRRK2 promotes α-synuclein pathology in mice [103]. Since LRRK2 interacts with 14-3-3 proteins [64–66], which also interact with α-synuclein and negatively regulate cell death pathways, it is suggested that LRRK2 may indirectly interact with α-synculein via other proteins such as 14-3-3 to converge in PD. Given LRRK2 is a large and complex protein, further identification and characterization of LRRK2 interaction partners and their linked pathways is necessary to decipher the main functional roles of LRRK2 in PD pathogenesis.
4. Animal Models for LRRK2-Linked Parkinsonism (Table 3)
Table 3.
LRRK2 animal models.
| Drosophila model | |||||
|---|---|---|---|---|---|
| Transgene | Loss of TH positive neurons | Lewy body | Motor deficits | Suitability for testing disease modifying therapy | Reference |
| LRRKWT, LRRKR1069C (R1441C) LRRKP1, LRRKex1 (loss-of-function line) |
ND | ND | + | ND | [53] |
| dLRRK(−/−), dLRRK(+/−), dLRRK RNAi, dLRRK Tg, R1069G(R1441G), Y1383C(Y1699C), I1915T(I2020T) |
+ | ND | + | ND | [71] |
| dLRRKe03680, dLRRK-WT, dLRRK- I1915T, dLRRKdf |
ND | ND | + | ND | [140] |
| WT, G2019S, Y1699C, G2385R |
+ | ND | + | + | [117] |
| hLRRK2(WT), hLRRK2(I1122V), hLRRK2(Y1699C), hLRRK2(I2020T) |
+ | ND | + | + | [118] |
| WT, G2019S | + | ND | + | ND | [141] |
| dLRRK-WT, dLRRK-mutant(e03680) | + | ND | ND | Maybe | [142] |
| Caenorhabditis elegans model | |||||
| LRRK2+: lrk-1(km17), LRRK2+: lrk-1(km41), G2019S+: lrk-1(km41) LRRK2+: lin-15(765ts) R1441C+: lin-15(765ts) G2019S+: lin-15(765ts) K1347A+: lin-15(765ts) |
+ | ND | + | ND | [119] |
| wlzIs2(WT), km4, N2(WT) | ND | ND | ND | ND | [73] (Interact with MKK6) |
| wlzIs1(WT), wlzIs2(WT), wlzIs3(G2019S), wlzIs4(G2019S), wlzIs5(R1441C), wlzIs6(KD), wlzIs7(R1441C/KD), wlzIs2 : wlzIs4(LRRK2/DAT::GFP) |
+ | ND | ND | + | [143] |
| LGI, lrk-1(tm1898, km41); LGII, pink-1(tm1779); LGX, lqIs4 (ceh-10::gfp, lin-15(n765)), N2 |
ND | ND | ND | ND | [120] (Interact with PINK1) |
| N2(WT), N2(G2019S) | ND | ND | ND | + | [144] |
| LRK-1-K1726A(hLRRK2-I2020T) LRK-1-I1877T)(hLRRK2-I2020T) |
ND | ND | ND | ND | [145] (trans-Golgi network) |
| Rodent model | |||||
| BAC(WT) | + | − | − | ND | [27] |
| BAC(G2019S) | ND | ND | + | ND | [146] |
| LRRK2R1441G BAC | + | − | + | Maybe | [147] |
| R1441C KI | ND | ND | +(AMPH-induced) | − | [148] |
| BAC(WT) BAC(G2019S) |
+ | ND | ND | Maybe | [149] |
| BAC(WT) BAC(G2019S) |
+ | ND | + | ND | [150] |
| WT, A53T, G2019S, KD, A53T/LRRK2WT, A53T/LRRK2G2019S, LRRK2−/− | ND | + | ND | ND | [103] |
| LRRK2 null | − | ND | ND | ND | [151] |
| LRRK2−/− | − | +(Kidney) | ND | ND | [122] |
| HSV-WTHSV-G2019SHSV-G2019S/D1994A | + | ND | ND | + | [152] |
| LRRK2 conditional G2019S | ND | ND | ND | Maybe | [81] (Hsp90 and LRRK2 stability) |
WT: wild type; ND: not determined; GFP: Green fluorescent protein; KI, knocking in.
4.1. LRRK2 Drosophila Model
Drosophila melanogaster is an excellent model organism for studying pathogenesis and therapeutics of neuronal degenerative diseases [123, 124]. Use of the fly system has led to the unveiling of molecular and cellular pathophysiology of neurodegeneration, and has potential in discovering novel drug targets for long-sought therapeutics. Fly models have been successfully used to study the roles of α-synuclein, parkin, pink-1, DJ-1 and stress factors as well as provide important insights into disease pathogenesis [125–138]. Approximately 75% of the disease-related loci in humans have at least one Drosophila homologue, indicating a high degree of conservation from flies to human [139]. Adult fly brains have 13 dopaminergic neuron clusters with more than 1000 neurons that can be labeled with antityrosine hydroxylase (TH) antibodies, as illustrated in Figure 1. The fly has one homologue (CG5483) of human LRRK2. Several groups have generated transgenic or loss-of-function mutants LRRK2 fly models using UAS-GAL4 system (Table 3). This system takes advantage of the findings that the yeast GAL4 transcription factor binds very specifically to an upstream activation sequence (UAS). LRRK2 transgenes can be expressed either in various tissues or in a small group of specific cells under the control of the given promoter (promoter-GAL4).
Loss-of-function mutant studies indicate that CG5483 protein is critical for the integrity of fly DA neurons [53] and control of synaptic overgrowth [140]. Drosophila lines expressing either fly LRRK (dLRRK) [53, 71] or human LRRK2 [117, 118, 141] resemble some features of LRRK2-linked Parkinsonism. Inactivation of dLRRK kinase activity is not essential for fly development [142]. Although the neurochemical and behavioral phenotypes of these LRRK2 flies differ considerably from various groups. Transgenic expression of Drosophila wild-type LRRK2 homology protein (CG5483) and a mutation (R1069C) corresponding to the human “R1441C” mutation does not show any significant defects [53]. However, this mutation in the context of Drosophila CG5483 may not be as pathogenic as the same R1441C change in the context of the human LRRK2 patients. Alternatively, the expression level of this mutant allele may not reach the pathology threshold in the fly. Overexpressing the human wild-type LRRK2 and the most common mutant form LRRK2-G2019S led to a selective loss of dopaminergic neurons in the brain, early mortality and locomotor impairment as reported by our group [141]. Moreover, LRRK2-G2019S increased autophosphorylation activity and caused a more severe parkinsonism-like phenotype than did wild-type LRRK2. Treatment with L-DOPA improved the mutant LRRK2-induced locomotor impairment, but did not prevent the loss of dopaminergic neurons, similar to what is seen in LRRK2-linked human PD. In support of this line of findings, several groups [117, 118, 141] have shown loss of dopamine and of dopaminergic neurons accompanied by behavioral deficits in their LRRK2 fly models. Coexpression of human parkin in LRRK2 G2019S-expressing flies provides significant protection against DA neurodegeneration that occurs with age or in response to rotenone [117]. Imai et al. reported that both human LRRK2 and the Drosophila orthologue of LRRK2 phosphorylate eukaryotic initiation factor 4E-(eIF4E-) binding protein (4E-BP), a negative regulator of eIF4E-mediated protein translation is a key mediator of various stress responses and suggest that 4E-BP may be a potential LRRK2 substrate [71]. Tain et al. have shown that loss of the Drosophila LRRK2 homolog activated 4E-BP and is able to suppress Pink-1 and parkin pathology [153]. Additionally, a recent study reports that LRRK2 interacts with 4E-BP at the postsynapse, whereas LRRK2 phosphorylates and negatively regulates the microtubule (MT-) binding protein Futsch at the presynapse [140].
LRRK2 also interacts with the microRNA (miRNA) pathway to regulate protein synthesis. Drosophila e2f1 and dp messenger RNAs are translationally repressed by let-7 and miR-184, respectively. Pathogenic LRRK2 antagonizes these miRNAs, leading to the overproduction of E2F1/DP, previously implicated in cell cycle and survival control and shown here to be critical for LRRK2 pathogenesis. LRRK2 associates with Drosophila Argonaute-1 (dAgo1) or human Argonaute-2 (hAgo2) of the RNA-induced silencing complex (RISC). In aged fly brain, dAgo1 protein level is negatively regulated by LRRK2. Furthermore, pathogenic LRRK2 promotes the association of phospho-4E-BP1 with hAgo2. These studies suggest that deregulated synthesis of E2F1/DP caused by the miRNA pathway impairment is a key event in LRRK2 pathogenesis [154]. With an outstanding battery of genetic tools for gene manipulation as well as the ability to carry out large-scale genetic screens inexpensively and rapidly for mutations affecting the disease process, the LRRK2 fly model provides a powerful tool to screen for LRRK2 interaction partners and LRRK2 substrates. Furthermore, the LRRK2 fly model can be used to conduct preclinical therapeutic screens to prevent neuronal loss and to rescue locomotor dysfunction in PD.
4.2. LRRK2 Caenorhabditis Elegan Models
LRK-1 is the Caenorhabditis elegans ortholog of human LRRK2, and transgenic as well as deletion mutants have been created in the worm [73, 119, 120, 143, 144, 155]. In LRK-1 deletion mutants, synaptic vesicle proteins mislocalize to both presynaptic and dendritic endings in neurons, suggesting that LRK-1 is involved in determining polarized sorting of synaptic vesicle proteins to axons by excluding these proteins from the dendrite-specific transport machinery in the Golgi [145]. In Wolozin et al.'s earlier studies, overexpression of wild-type and LRRK2 (G2019S) in C. elegans was protective against rotenone toxicity, whereas knockdown of endogenous LRK-1 by RNAi promoted toxicity, suggesting a role for LRRK2 in mitochondrial regulation [144]. In contrast, a recent study shows that the transgenic C. elegans overexpressing human LRRK2 wild type, R1441C and G2019S in dopaminergic (DA) neurons causes age-dependent DA neurodegeneration, behavioral deficits, and locomotor dysfunction that are accompanied by a reduction of dopamine levels in vivo. In comparison, R1441C and G2019S mutants cause more severe phenotypes than the wild-type protein. Interestingly, treatment with exogenous dopamine rescues the LRRK2-induced behavioral and locomotor phenotypes. In contrast, expression of the GTP-binding defective mutant, K1347A, or knockout of the C. elegans LRRK2 homolog, LRK-1, prevents the LRRK2-induced neurodegeneration and behavioral abnormalities. These results provide strong support for the critical role of GTPase/kinase activity in LRRK2-linked pathologies [119].
4.3. Zebrafish Model
Zebrafish have a homolog of human LRRK2 (XM_682700). The blockage of zebrafish LRRK2 (zLRRK2) protein by morpholinos caused embryonic lethality and severe developmental defects such as growth retardation and loss of neurons. In contrast, the deletion of the WD40 domain of zLRRK2 by morpholinos targeting splicing did not induce severe embryonic developmental defects; rather it caused Parkinsonism-like phenotypes, including loss of dopaminergic neurons in the diencephalon and locomotion defects. These neurodegenerative and locomotion defects could be rescued by overexpressing zLRRK2 or hLRRK2 mRNA [54]. The zLRRK2-ΔWD40 deletion also caused a significant reduction and disorganization of axon tracts, more prominently in the midbrain. These studies suggest that zLRRK2 may play an important role in neuronal development and provide a useful small vertebrate model for PD research.
4.4. LRRK2 Rodent Models
The LRRK2 protein expressed in mice shares 86% homology with the human protein (Genbank: NM-25730). Several groups generated LRRK transgenic and knockout models but they are not very robust PD models (Table 3). LRRK2 transgenic mice show some neurochemical and behavioral abnormalities but lack selective loss of dopaminergic neurons in substantia nigra [95, 103, 146–150]. Knockout of LRRK2 in mice also lack the obvious abnormality in DA neurons in brains [122, 151].
Conditional expression of LRRK2 WT and LRRK2 G2019S failed to exhibit neurodegeneration of DA neurons, but LRRK2 was expressed at low levels in DA neurons due to the use of the calcium/calmodulin dependent protein kinase II (CamKII) promoter [81, 103]. When the R1441C mutation is expressed under the control of the endogenous regulatory elements, by knock in of the R1441C mutation, there is no degeneration of DA neurons, but they show reductions in amphetamine-(AMPH-) induced locomotor activity [148]. Bacterial artificial chromosome (BAC) transgenic mice expressing LRRK2 WT, LRRK2 R1441G, LRRK2 G2019S have some evidence of neurodegeneration [147, 150], which is demonstrated by measuring the dopamine content after pharmacologically blocking the dopamine uptake. Li et al. report LRRK2R1441G BAC transgenic mice display hyperphosphorylation of tau and motor deficits. Two groups recently report that G2019S Lrrk2 BAC mice display abnormal dopamine neurotransmission as evident by a decrease in extracellular dopamine levels [149, 150]. However, Li et al. shows that the wild-type LRRK2 BAC mice revealed increases in dopamine release thereby contributing to hyperactivity phenotypes, while Melrose et al. shows wild-type LRRK2 mice also decrease dopamine levels but a bit less than G2019S-LRRK2 BAC mice. Moreover, they later also show that G2019S-LRRK2 BAC mice display changes in localization and increased phosphorylation of microtubule binding protein tau, suggesting that LRRK2 may impact tau processing [150].
Mutations in α-synuclein and Leucine-rich repeat kinase 2 (LRRK2) are linked to autosomal dominant forms of Parkinson's disease (PD). Recently, Lin et al. shows that there is a potential pathophysiological interplay between these two PD-related genes by generating a double transgenic mouse model coexpressing both human α-synuclein and LRRK2 genes [103]. Overexpression of LRRK2 alone did not cause neurodegeneration but the presence of excess LRRK2 greatly accelerated the progression of neuropathological abnormalities developed in PD-related A53T α-synuclein transgenic mice. Moreover, LRRK2 promoted the abnormal aggregation and somatic accumulation of α-synuclein in A53T mice, which likely results from the impairment of microtubule dynamics, Golgi organization, and the ubiquitin-proteasome pathway. Conversely, genetic ablation of LRRK2 preserved the Golgi structure and suppressed the aggregation and somatic accumulation of α-synuclein, thereby delaying the progression of neuropathology in A53T mice. These findings suggest that overexpression of LRRK2 enhances α-synuclein-mediated cytotoxicity [103]. Currently, there are no mouse models that overexpression mutant LRRK2 in parkin, pink-1 or DJ-1 knockout backgrounds.
LRRK2 knockout (KO) mice [151] are viable, have no major abnormalities and live to adulthood. Moreover, there is no significant difference in the susceptibility of LRRK2 KO and wild type mice to MPTP suggesting that LRRK2 may play a minor role in the development and the survival of DA neurons. Alternatively, the roles of LRRK2 may be compensated by LRRK1 since LRRK1 shares high homology with LRRK2 and is expressed in the brains. However, a recent study shows that there is an age-dependent kidney abnormality in LRRK2 KO mice. The kidneys of these mice, develop striking accumulation and aggregation of α-synuclein and ubiquitinated proteins, and may be involved in the autophagy-lysosomal defects [122]. The kidneys also display apoptotic cell death, oxidative damage and inflammatory response, suggesting that LRRK2 may play an important peripheral role during aging at least in kidney.
Most of the current LRRK2 transgenic mice have abnormalities in the nigrostriatal system, such as stimulated DA neurotransmission, decrease dopamine levels, or behavioral deficits, which probably represent some of the earliest neuronal dysfunction that is set in motion by pathogenic LRRK2 mutations. Reasons are not clear why mouse LRRK2 transgenic models do not exhibit more substantial pathology and why LRRK2 KO mice do not display abnormality in nigrostriatal system. It may relate to the fact that LRRK2 mutations in humans are only partially penetrant and that there may need to be other genetic and/or environmental hits that are required for degeneration of DA neurons. The BAC and knock in models express mutant LRRK2 during development and thus there may be compensatory mechanisms in the mouse that prevent loss of DA neurons by LRRK1 or other genes with the similar functions. Current mouse LRRK2 models can be used for early mechanism studies of LRRK2 but are less than ideal to test the neuronprotection therapies. The rodent models need to be improved by combining other PD risk factors, or by other approaches to express LRRK2 in the nigrostriatal system. A recent promising mouse model is using AAV-mediated expression of mutant LRRK2 in middle brain causes remarkable dopaminergic neuron degeneration [152], which can be potentially used to test protective therapeutics of LRRK2-linked diseases.
5. Conclusion Remarks
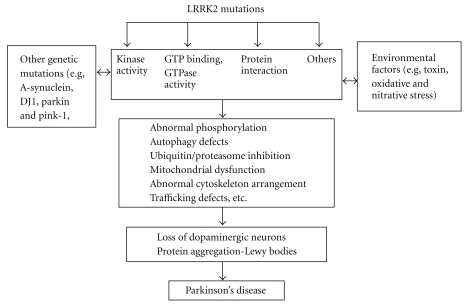
In summary, the current findings in LRRK2 indicate that kinase activity and GTPase domain activity are the key components of LRRK2 functions and are associated with LRRK2-induced neuronal degeneration. Mutations within LRRK2 may potentially perturb protein conformation or protein-protein interactions with accessory proteins necessary for kinase and GTPase domain activity. It is important to note, however, that the increase in kinase activity seen with PD-associated mutations of LRRK2 must be interpreted with caution until these observations are confirmed with physiologically relevant substrates. Nevertheless, these current findings in LRRK2 kinase and GTPase are consistent with a model in which LRRK2 cycles between an active and an inactive conformation potentially integrating multiple signaling pathways and subsequently lead to protein aggregation and neurodegeneration. LRRK2 may also serve as a scaffold protein to recruit other signaling molecules through its protein-protein interaction domains. Thus, LRRK2 kinase, GTPase domain and scaffold activities may function together with other PD-related players to elicit disease pathology as depicted in Figure 3.
Figure 3.
Potential pathways associated with LRRK2 in PD.
In addition, mutant LRRK2 may directly or indirectly interact with environmental factors and other genetic PD causes to converge on the pathways that induce protein aggregation and neuronal death. These interactions may occur at various levels, such as altering LRRK2 GTP-binding, GTPase and/or kinase activity, modulating LRRK2 kinase substrates, or influencing the function of LRRK2 interaction partners among others yet to be identified. Thus, identifying the putative LRRK2-interacting proteins, physiological substrates of LRRK2 kinase, regulators and downstream effectors of LRRK2 GTPase, as well as establishing how mutations lead to the familial and sporadic forms of PD through interactions between genetic factors and environmental toxins will likely provide crucial insights into the pathways involved in PD pathogenesis. Such investigations will facilitate the development of LRRK2 cell and animal models as well as enable the formulation of novel pharmacological interventions for the treatment of PD. The current findings in LRRK2 are beginning to pave the way for better-designed therapeutic options. The discovery of chemical inhibitors of LRRK2 kinase and GTPase domain activities may likely involve optimizing strategies that prevent dopaminergic neuron degeneration and to treat LRRK2-linked PD. Recently, several groups already report some potential LRRK2 kinase inhibitors in preventing neuronal death [85, 95, 121]. With more research into the genetics and biochemistry of LRRK2 and more LRRK2 animal models available, identifying LRRK2 kinase and GTPase domain inhibitors might lead us to effective new therapeutic approaches for the treatment of PD.
Acknowledgments
This research was funded by NINDS (NS55684), National Parkinson Foundation, and American Parkinson Disease Association Inc. and the Michael J. Fox Foundation to W. W. Smith.
Abbreviations
- PD:
Parkisnon's disease
- LB:
Lewy body
- LRRK2:
Leucine-rich repeat kinase 2
- PAL:
Protocerebral anterior lateral
- PAM:
Paired anterolateral medial
- PPL:
Protocerebral posterior lateral
- PPM:
Protocerebral posterior medial
- TH:
Tyrosine hydroxylase.
References
- 1.Gasser T. Genetics of Parkinson’s disease. Current Opinion in Neurology. 2005;18(4):363–369. doi: 10.1097/01.wco.0000170951.08924.3d. [DOI] [PubMed] [Google Scholar]
- 2.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39(6):889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 3.Forno LS. Neuropathology of Parkinson’s disease. Journal of Neuropathology and Experimental Neurology. 1996;55(3):259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Mouradian MM. Recent advances in the genetics and pathogenesis of Parkinson disease. Neurology. 2002;58(2):179–185. doi: 10.1212/wnl.58.2.179. [DOI] [PubMed] [Google Scholar]
- 5.Dawson TM, Dawson VL. Molecular Pathways of Neurodegeneration in Parkinson’s Disease. Science. 2003;302(5646):819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 6.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296(5575):1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 7.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature Medicine. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 8.Paisán-Ruíz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Cookson MR, Dauer W, Dawson T, Fon EA, Guo M, Shen J. The roles of kinases in familial Parkinson’s disease. Journal of Neuroscience. 2007;27(44):11865–11868. doi: 10.1523/JNEUROSCI.3695-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein C, Lohmann-Hedrich K. Impact of recent genetic findings in Parkinson’s disease. Current Opinion in Neurology. 2007;20(4):453–464. doi: 10.1097/WCO.0b013e3281e6692b. [DOI] [PubMed] [Google Scholar]
- 12.Hedrich K, Winkler S, Hagenah J, et al. Recurrent LRRK2 (park8) mutations in early-onset Parkinson’s disease. Movement Disorders. 2006;21(9):1506–1510. doi: 10.1002/mds.20990. [DOI] [PubMed] [Google Scholar]
- 13.Funayama M, Li Y, Tomiyama H, et al. Leucine-rich repeat kinase 2 G2385R variant is a risk factor for Parkinson disease in Asian populatio. NeuroReport. 2007;18(3):273–275. doi: 10.1097/WNR.0b013e32801254b6. [DOI] [PubMed] [Google Scholar]
- 14.Zabetian CP, Samii A, Mosley AD, et al. A clinic-based study of the LRRK2 gene in Parkinson disease yields new mutations. Neurology. 2005;65(5):741–744. doi: 10.1212/01.wnl.0000172630.22804.73. [DOI] [PubMed] [Google Scholar]
- 15.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends in Neurosciences. 2006;29(5):286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Taylor JP, Hulihan MM, Kachergus JM, et al. Leucine-rich repeat kinase 1: a paralog of LRRK2 and a candidate gene for Parkinson’s disease. Neurogenetics. 2007;8(2):95–102. doi: 10.1007/s10048-006-0075-8. [DOI] [PubMed] [Google Scholar]
- 17.Kachergus J, Mata IF, Hulihan M, et al. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. American Journal of Human Genetics. 2005;76(4):672–680. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lesage S, Dürr A, Brice A. LRRK2: a link between familial and sporadic Parkinson's disease? Pathologie Biologie. 2007;55(2):107–110. doi: 10.1016/j.patbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Berg D, Schweitzer K, Leitner P, et al. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson’s disease. Brain. 2005;128(12):3000–3011. doi: 10.1093/brain/awh666. [DOI] [PubMed] [Google Scholar]
- 20.Di Fonzo A, Tassorelli C, De Mari M, et al. Comprehensive analysis of the LRRK2 gene in sixty families with Parkinson’s disease. European Journal of Human Genetics. 2006;14(3):322–331. doi: 10.1038/sj.ejhg.5201539. [DOI] [PubMed] [Google Scholar]
- 21.Farrer M, Stone J, Mata IF, et al. LRRK2 mutations in Parkinson disease. Neurology. 2005;65(5):738–740. doi: 10.1212/01.wnl.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- 22.Paisán-Ruíz C, Lang AE, Kawarai T, et al. LRRK2 gene in Parkinson disease: mutation analysis and case control association study. Neurology. 2005;65(5):696–700. doi: 10.1212/01.wnl.0000167552.79769.b3. [DOI] [PubMed] [Google Scholar]
- 23.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nature Reviews Neuroscience. 2010;11(12):791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan EK, Schapira AH. LRRK2 as a therapeutic target in Parkinson’s disease. European Journal of Neurology. 2011;18(4):545–546. doi: 10.1111/j.1468-1331.2010.03305.x. [DOI] [PubMed] [Google Scholar]
- 25.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66(5):646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webber PJ, West AB. LRRK2 in Parkinson’s disease: function in cells and neurodegeneration. FEBS Journal. 2009;276(22):6436–6444. doi: 10.1111/j.1742-4658.2009.07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melrose HL, Kent CB, Taylor JP, et al. A comparative analysis of leucine-rich repeat kinase 2 (Lrrk2) expression in mouse brain and Lewy body disease. Neuroscience. 2007;147(4):1047–1058. doi: 10.1016/j.neuroscience.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 28.Melrose H, Lincoln S, Tyndall G, Dickson D, Farrer M. Anatomical localization of leucine-rich repeat kinase 2 in mouse brain. Neuroscience. 2006;139(3):791–794. doi: 10.1016/j.neuroscience.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Simón-Sánchez J, Herranz-Pérez V, Olucha-Bordonau F, Pérez-Tur J. LRRK2 is expressed in areas affected by Parkinson’s disease in the adult mouse brain. European Journal of Neuroscience. 2006;23(3):659–666. doi: 10.1111/j.1460-9568.2006.04616.x. [DOI] [PubMed] [Google Scholar]
- 30.Biskup S, Moore DJ, Celsi F, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Annals of Neurology. 2006;60(5):557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 31.Higashi S, Biskup S, West AB, et al. Localization of Parkinson’s disease-associated LRRK2 in normal and pathological human brain. Brain Research. 2007;1155(1):208–219. doi: 10.1016/j.brainres.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 32.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Current Opinion in Structural Biology. 2001;11(6):725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 33.Gaiter D, Westerlund M, Carmine A, Lindqvist E, Sydow O, Olson L. LRRK2 expression linked to dopamine-innervated areas. Annals of Neurology. 2006;59(4):714–719. doi: 10.1002/ana.20808. [DOI] [PubMed] [Google Scholar]
- 34.Taymans JM, Van Den Haute C, Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. Journal of Neurochemistry. 2006;98(3):951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- 35.Giasson BI, Covy JP, Bonini NM, et al. Biochemical and pathological characterization of Lrrk2. Annals of Neurology. 2006;59(2):315–322. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Siedlak SL, Smith MA, Perry G, Chen SG. LRRK2 protein is a component of Lewy bodies. Annals of Neurology. 2006;60(5):617–618. doi: 10.1002/ana.20928. [DOI] [PubMed] [Google Scholar]
- 37.Darios F, Corti O, Lücking CB, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Human Molecular Genetics. 2003;12(5):517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 38.Gloeckner CJ, Kinkl N, Schumacher A, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Human Molecular Genetics. 2006;15(2):223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 39.West AB, Moore DJ, Biskup S, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(46):16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehmensiek V, Tan EM, Schwarz J, Storch A. Expression of mutant α-synucleins enhances dopamine transporter-mediated MPP+ toxicity in vitro. NeuroReport. 2002;13(10):1279–1283. doi: 10.1097/00001756-200207190-00013. [DOI] [PubMed] [Google Scholar]
- 41.Berger Z, Smith KA, Lavoie MJ. Membrane localization of LRRK2 is associated with increased formation of the highly active lrrk2 dimer and changes in its phosphorylation. Biochemistry. 2010;49(26):5511–5523. doi: 10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alegre-Abarrategui J, Wade-Martins R. Parkinson disease, LRRK2 and the endocytic-autophagic pathway. Autophagy. 2009;5(8):1208–1210. doi: 10.4161/auto.5.8.9894. [DOI] [PubMed] [Google Scholar]
- 43.Alegre-Abarrategui J, Christian H, Lufino MMP, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Human Molecular Genetics. 2009;18(21):4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng H, Le W, Guo YI, Hunter CB, Xie W, Jankovic J. Genetic and clinical identification of Parkinson’s disease patients with LRRK2 G2019S mutation. Annals of Neurology. 2005;57(6):933–934. doi: 10.1002/ana.20510. [DOI] [PubMed] [Google Scholar]
- 45.Ross OA, Toft M, Whittle AJ, et al. Lrrk2 and Lewy body disease. Annals of Neurology. 2006;59(2):388–393. doi: 10.1002/ana.20731. [DOI] [PubMed] [Google Scholar]
- 46.Rajput A, Dickson DW, Robinson CA, et al. Parkinsonism, Lrrk2 G2019S, and tau neuropathology. Neurology. 2006;67(8):1506–1508. doi: 10.1212/01.wnl.0000240220.33950.0c. [DOI] [PubMed] [Google Scholar]
- 47.Funayama M, Hasegawa K, Ohta E, et al. An LRRK2 mutation as a cause for the Parkinsonism in the original PARK8 family. Annals of Neurology. 2005;57(6):918–921. doi: 10.1002/ana.20484. [DOI] [PubMed] [Google Scholar]
- 48.Lewthwaite AJ, Nicholl DJ. Genetics of Parkinsonism. Current Neurology and Neuroscience Reports. 2005;5(5):397–404. doi: 10.1007/s11910-005-0064-6. [DOI] [PubMed] [Google Scholar]
- 49.Morris HR. Genetics of Parkinson’s disease. Annals of Medicine. 2005;37(2):86–96. doi: 10.1080/07853890510007269. [DOI] [PubMed] [Google Scholar]
- 50.Nichols WC, Pankratz N, Hernandez D, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson’s disease. Lancet. 2005;365(9457):410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 51.Di Fonzo A, Rohé CF, Ferreira J, et al. A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson’s disease. Lancet. 2005;365(9457):412–415. doi: 10.1016/S0140-6736(05)17829-5. [DOI] [PubMed] [Google Scholar]
- 52.Gilks WP, Abou-Sleiman PM, Gandhi S, et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365(9457):415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 53.Lee SB, Kim W, Lee S, Chung J. Loss of LRRK2/PARK8 induces degeneration of dopaminergic neurons in Drosophila. Biochemical and Biophysical Research Communications. 2007;358(2):534–539. doi: 10.1016/j.bbrc.2007.04.156. [DOI] [PubMed] [Google Scholar]
- 54.Sheng D, Qu D, Kwok KHH, et al. Deletion of the WD40 domain of LRRK2 in zebrafish causes parkinsonism-like loss of neurons and locomotive defect. PLoS Genetics. 2010;6(4) doi: 10.1371/journal.pgen.1000914. Article ID e1000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52(4):587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 56.Greggio E, Jain S, Kingsbury A, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiology of Disease. 2006;23(2):329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nature Neuroscience. 2006;9(10):1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 58.Luzón-Toro B, de la Torre ER, Delgado A, Pérez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson’s disease-associated G2019S LRRK2 mutation. Human Molecular Genetics. 2007;16(17):2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- 59.Jaleel M, Nichols RJ, Deak M, et al. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. Biochemical Journal. 2007;405(2):307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.West AB, Moore DJ, Choi C, et al. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Human Molecular Genetics. 2007;16(2):223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 61.Greggio E, Zambrano I, Kaganovich A, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. Journal of Biological Chemistry. 2008;283(24):16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Moore DJ, Xiong Y, Dawson TM, Dawson VL. Reevaluation of phosphorylation sites in the parkinson disease-associated leucine-rich repeat kinase 2. Journal of Biological Chemistry. 2010;285(38):29569–29576. doi: 10.1074/jbc.M110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greggio E, Taymans JM, Zhen EY, et al. The Parkinson’s disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochemical and Biophysical Research Communications. 2009;389(3):449–454. doi: 10.1016/j.bbrc.2009.08.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichols RJ, Dzamko N, Morrice NA, et al. 14-3-3 Binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochemical Journal. 2010;430(3):393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudenko IN, Cookson MR. 14-3-3 proteins are promising LRRK2 interactors. Biochemical Journal. 2010;430(3):e5–e6. doi: 10.1042/BJ20101200. [DOI] [PubMed] [Google Scholar]
- 66.Yacoubian TA, Slone SR, Harrington AJ, et al. Differential neuroprotective effects of 14-3-3 proteins in models of Parkinson’s disease. Cell Death and Disease. 2010;1(1, article e2) doi: 10.1038/cddis.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. Journal of Neurochemistry. 2007;103(1):238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daniëls V, Vancraenenbroeck R, Law BMH, et al. Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. Journal of Neurochemistry. 2011;116(2):304–315. doi: 10.1111/j.1471-4159.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase thatstimulates kinase activity. Experimental Cell Research. 2007;313(16):3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stenmark P, Ogg D, Flodin S, et al. The structure of human collapsin response mediator protein 2, a regulator of axonal growth. Journal of Neurochemistry. 2007;101(4):906–917. doi: 10.1111/j.1471-4159.2006.04401.x. [DOI] [PubMed] [Google Scholar]
- 71.Imai Y, Gehrke S, Wang HQ, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO Journal. 2008;27(18):2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. Journal of Neurochemistry. 2009;109(4):959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- 73.Hsu CH, Chan D, Greggio E, et al. MKK6 binds and regulates expression of Parkinson’s disease-related protein LRRK2. Journal of Neurochemistry. 2010;112(6):1593–1604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu CH, Chan D, Wolozin B. LRRK2 and the stress response: interaction with MKKs and JNK-interacting proteins. Neurodegenerative Diseases. 2010;7(1–3):68–75. doi: 10.1159/000285509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bosgraaf L, Van Haastert PJM. Roc, a Ras/GTPase domain in complex proteins. Biochimica et Biophysica Acta. 2003;1643(1–3):5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 76.Ito G, Okai T, Fujino GO, et al. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry. 2007;46(5):1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 77.Lewis PA. The function of ROCO proteins in health and disease. Biology of the Cell. 2009;101(3):183–191. doi: 10.1042/BC20080053. [DOI] [PubMed] [Google Scholar]
- 78.Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR. Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith WW, Pei Z, Jiang H, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(51):18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dzamko N, Deak M, Hentati F, et al. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser910/Ser935, disruption of 14-3-3 binding and altered cytoplasmic localization. Biochemical Journal. 2010;430(3):405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang L, Xie C, Greggio E, et al. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. Journal of Neuroscience. 2008;28(13):3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochemical and Biophysical Research Communications. 2007;357(3):668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiong Y, Coombes CE, Kilaru A, et al. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genetics. 2010;6(4) doi: 10.1371/journal.pgen.1000902. Article ID e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gotthardt K, Weyand M, Kortholt A, Van Haastert PJM, Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO Journal. 2008;27(16):2239–2249. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu M, Dobson B, Glicksman MA, Yue Z, Stein RL. Kinetic mechanistic studies of wild-type leucine-rich repeat kinase2: characterization of the kinase and GTPase activities. Biochemistry. 2010;49(9):2008–2017. doi: 10.1021/bi901851y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klein CL, Rovelli G, Springer W, Schall C, Gasser T, Kahle PJ. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. Journal of Neurochemistry. 2009;111(3):703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- 87.Lu B, Zhai Y, Wu C, Pang X, Xu Z, Sun F. Expression, purification and preliminary biochemical studies of the N-terminal domain of leucine-rich repeat kinase 2. Biochimica et Biophysica Acta. 2010;1804((9):1780–1784. doi: 10.1016/j.bbapap.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 88.Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. Journal of Biological Chemistry. 2009;284(52):36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamikawaji S, Ito G, Iwatsubo T. Identification of the autophosphorylation sites of LRRK2. Biochemistry. 2009;48(46):10963–10975. doi: 10.1021/bi9011379. [DOI] [PubMed] [Google Scholar]
- 90.Gloeckner CJ, Boldt K, Von Zweydorf F, et al. Phosphopeptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. Journal of Proteome Research. 2010;9(4):1738–1745. doi: 10.1021/pr9008578. [DOI] [PubMed] [Google Scholar]
- 91.Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro. 2009;1(1) doi: 10.1042/AN20090007. Article ID e00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laccarino C, Crosio C, Vitale C, Sanna G, Carrì MT, Barone P. Apoptotic mechanisms in mutant LRRK2-mediated cell death. Human Molecular Genetics. 2007;16(11):1319–1326. doi: 10.1093/hmg/ddm080. [DOI] [PubMed] [Google Scholar]
- 93.Ho CCY, Rideout HJ, Ribe E, Troy CM, Dauer WT. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. Journal of Neuroscience. 2009;29(4):1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heo HY, Park JM, Kim CH, Han BS, Kim KS, Seol W. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Experimental cell research. 2010;316(4):649–656. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 95.Lee BD, Shin J-H, Vankampen J, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nature Medicine. 2010;16(9):998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jorgensen ND, Peng Y, Ho CC, et al. The WD40 domain is required for LRRK2 neurotoxicity. PloS One. 2009;4(12, article e8463) doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dächsel JC, Taylor JP, Mok SUS, et al. Identification of potential protein interactors of Lrrk2. Parkinsonism and Related Disorders. 2007;13(7):382–385. doi: 10.1016/j.parkreldis.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ko HS, Bailey R, Smith WW, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ding X, Goldberg MS. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0005949. Article ID e5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parisiadou L, Cai H. LRRK2 function on actin and microtubule dynamics in Parkinson disease. Communitative and Integrative Biology. 2010;3(5):396–400. doi: 10.4161/cib.3.5.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parisiadou L, Xie C, Hyun JC, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. Journal of Neuroscience. 2009;29(44):13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. Journal of Neuroscience Research. 2008;86(8):1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin X, Parisiadou L, Gu XL, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant α-synuclein. Neuron. 2009;64(6):807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cairns NJ, Lee VM-Y, Trojanowski JQ. The cytoskeleton in neurodegenerative diseases. Journal of Pathology. 2004;204(4):438–449. doi: 10.1002/path.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability—a point of convergence in Parkinsonian neurodegeneration? Journal of Neurochemistry. 2009;110(5):1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- 106.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proceedings of the National Academy of Sciences of the United States of America. 1975;72(5):1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cole NB, Lippincott-Schwartz L. Organization of organelles and membrane traffic by microtubules. Current Opinion in Cell Biology. 1995;7(1):55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- 108.Lane J, Allan V. Microtubule-based membrane movement. Biochimica et Biophysica Acta. 1998;1376(1):27–55. doi: 10.1016/s0304-4157(97)00010-5. [DOI] [PubMed] [Google Scholar]
- 109.Sancho RM, Law BMH, Harvey K. Mutations in the LRRK2 Roc-COR tandem domain link Parkinson’s disease to Wnt signalling pathways. Human Molecular Genetics. 2009;18(20):3955–3968. doi: 10.1093/hmg/ddp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dächsel JC, Mata IF, Ross OA, et al. Digenic parkinsonism: investigation of the synergistic effects of PRKN and LRRK2. Neuroscience Letters. 2006;410(2):80–84. doi: 10.1016/j.neulet.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 111.Shin N, Jeong H, Kwon J, et al. LRRK2 regulates synaptic vesicle endocytosis. Experimental Cell Research. 2008;314(10):2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 112.Plowey ED, Cherra SJ, III, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. Journal of Neurochemistry. 2008;105(3):1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gillardon F. Interaction of elongation factor 1-alpha with leucine-rich repeat kinase 2 impairs kinase activity and microtubule bundling in vitro. Neuroscience. 2009;163(2):533–539. doi: 10.1016/j.neuroscience.2009.06.051. [DOI] [PubMed] [Google Scholar]
- 114.Lin C-H, Tsai P-I, Wu R-M, Chien C-T. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3β. Journal of Neuroscience. 2010;30(39):13138–13149. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meixner A, Boldt K, Van Troys M, et al. A QUICK screen for Lrrk2 interaction partners—leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Molecular and Cellular Proteomics. 2011;10(1) doi: 10.1074/mcp.M110.001172. Article ID M110.001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kumar A, Greggio E, Beilina A, et al. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2010;5(1) doi: 10.1371/journal.pone.0008730. Article ID e8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ng CH, Mok SZS, Koh C, et al. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. Journal of Neuroscience. 2009;29(36):11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Venderova K, Kabbach G, Abdel-Messih E, et al. Leucine-rich repeat kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson's disease. Human Molecular Genetics. 2009;18(22):4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- 119.Yao C, El Khoury R, Wang W, et al. LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson’s disease. Neurobiology of Disease. 2010;40(1):73–81. doi: 10.1016/j.nbd.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sämann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. Journal of Biological Chemistry. 2009;284(24):16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu M, Poulose S, Schuman E, et al. Development of a mechanism-based high-throughput screen assay for leucine-rich repeat kinase 2-Discovery of LRRK2 inhibitors. Analytical Biochemistry. 2010;404(2):186–192. doi: 10.1016/j.ab.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tong Y, Yamaguchi H, Giaime E, et al. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of α-synuclein, and apoptotic cell death in aged mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(21):9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cauchi RJ, Van Den Heuvel M. The fly as a model for neurodegenerative diseases: is it worth the jump? Neurodegenerative Diseases. 2007;3(6):338–356. doi: 10.1159/000097303. [DOI] [PubMed] [Google Scholar]
- 124.Marsh JL, Thompson LM. Drosophila in the study of neurodegenerative disease. Neuron. 2006;52(1):169–178. doi: 10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 125.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 126.Haass C, Kahle PJ. Parkinson’s pathology in a fly. Nature. 2000;404(6776):341–343. doi: 10.1038/35006188. [DOI] [PubMed] [Google Scholar]
- 127.Auluck PK, Meulener MC, Bonini NM. Mechanisms of suppression of α-synuclein neurotoxicity by geldanamycin in Drosophila. Journal of Biological Chemistry. 2005;280(4):2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 128.Auluck PK, Chan HYE, Trojanowski JQ, Lee VM-Y, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295(5556):865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 129.Chen L, Feany MB. α-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nature Neuroscience. 2005;8(5):657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 130.Meulener MC, Xu K, Thompson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang Y, Gehrke S, Imai Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(28):10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clark IE, Dodson MW, Jiang C, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 133.Park J, Lee SB, Lee S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441(7097):1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 134.Park J, Sung YK, Cha GH, Lee SB, Kim S, Chung J. Drosophila DJ-1 mutants show oxidative stress-sensitive locomotive dysfunction. Gene. 2005;361(1-2):133–139. doi: 10.1016/j.gene.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 135.Yang Y, Gehrke S, Haque ME, et al. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(38):13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pesah Y, Pham T, Burgess H, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131(9):2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 137.Yang Y, Nishimura I, Imai Y, Takahashi R, Lu B. Parkin suppresses dopaminergic neuron-selective neurotoxicity induced by Pael-R in Drosophila. Neuron. 2003;37(6):911–924. doi: 10.1016/s0896-6273(03)00143-0. [DOI] [PubMed] [Google Scholar]
- 138.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Research. 2001;11(6):1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee S, Liu H-P, Lin W-Y, Guo H, Lu B. LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. Journal of Neuroscience. 2010;30(50):16959–16969. doi: 10.1523/JNEUROSCI.1807-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu Z, Wang X, Yu YI, et al. A Drosophila model for LRRK2-linked parkinsonism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang D, Tang B, Zhao G, et al. Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Molecular Neurodegeneration. 2008;3(1, article no. 3) doi: 10.1186/1750-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saha S, Guillily MD, Ferree A, et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. Journal of Neuroscience. 2009;29(29):9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wolozin B, Saha S, Guillily M, Ferree A, Riley M. Investigating convergent actions of genes linked to familial Parkinson’s disease. Neurodegenerative Diseases. 2008;5(3-4):182–185. doi: 10.1159/000113697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Current Biology. 2007;17(7):592–598. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- 146.Winner B, Melrose HL, Zhao C, et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiology of Disease. 2011;41(3):706–716. doi: 10.1016/j.nbd.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li Y, Liu W, Oo TF, et al. Mutant LRRK2 BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nature Neuroscience. 2009;12(7):826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tong Y, Pisani A, Martella G, et al. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(34):14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Melrose HL, Dächsel JC, Behrouz B, et al. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiology of Disease. 2010;40(3):503–517. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li X, Patel JC, Wang J, et al. Enhanced striatal dopamine transmission and motor performance with LRRK2 overexpression in mice is eliminated by familial Parkinson’s disease mutation G2019S. Journal of Neuroscience. 2010;30(5):1788–1797. doi: 10.1523/JNEUROSCI.5604-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Andres-Mateos E, Mejias R, Sasaki M, et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) Journal of Neuroscience. 2009;29(50):15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee BD, Shin J-H, Vankampen J, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nature Medicine. 2010;16(9):998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nature Neuroscience. 2009;12(9):1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466(7306):637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Feigin A, Zgaljardic D. Recent advances in Huntington’s disease: Implications for experimental therapeutics. Current Opinion in Neurology. 2002;15(4):483–489. doi: 10.1097/00019052-200208000-00013. [DOI] [PubMed] [Google Scholar]