Abstract
Xpo1p (Crm1p) is the nuclear export receptor for proteins containing a leucine-rich nuclear export signal (NES). Xpo1p, the NES-containing protein, and GTP-bound Ran form a complex in the nucleus that translocates across the nuclear pore. We have identified Yrb1p as the major Xpo1p-binding protein in Saccharomyces cerevisiae extracts in the presence of GTP-bound Gsp1p (yeast Ran). Yrb1p is cytoplasmic at steady-state but shuttles continuously between the cytoplasm and the nucleus. Nuclear import of Yrb1p is mediated by two separate nuclear targeting signals. Export from the nucleus requires Xpo1p, but Yrb1p does not contain a leucine-rich NES. Instead, the interaction of Yrb1p with Xpo1p is mediated by Gsp1p-GTP. This novel type of export complex requires the acidic C-terminus of Gsp1p, which is dispensable for the binding to importin β-like transport receptors. A similar complex with Xpo1p and Gsp1p-GTP can be formed by Yrb2p, a relative of Yrb1p predominantly located in the nucleus. Yrb1p also functions as a disassembly factor for NES/Xpo1p/Gsp1p-GTP complexes by displacing the NES protein from Xpo1p/Gsp1p. This Yrb1p/Xpo1p/Gsp1p complex is then completely dissociated after GTP hydrolysis catalyzed by the cytoplasmic GTPase activating protein Rna1p.
INTRODUCTION
The nuclear envelope separates the nucleus and the cytoplasm but contains transport gates termed nuclear pore complexes (NPCs) that allow for selective exchange of proteins and RNAs. Although macromolecules of up to 40–60 kDa in size can diffuse through the aqueous channel of the NPC, most macromolecules are transported across the NPC in a receptor-mediated and energy-dependent manner (for reviews see Ohno et al., 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Cole, 2000). The mediators of the translocation are soluble receptors of the importin β family, which can be divided into importins and exportins. They recognize specific transport signals within their cargo, migrate through the NPC, and return in their cargo-free form after release of the transport substrate. Importin β-like proteins vary in size between 95 and 135 kDa and are characterized by an aminoterminal Ran-binding domain and a large carboxyterminal domain containing tandem repeats (Chook and Blobel, 1999; Cingolani et al., 1999; Vetter et al., 1999a).
The yeast Saccharomyces cerevisiae contains 14 importin β-like proteins (reviewed by Weis, 1998; Schlenstedt and Solsbacher, 1999), which transport a variety of cargoes. Importin β (Kap95p) mediates the import of importin α (Srp1p), which in turn binds to proteins containing classical nuclear localization signals (NLSs). Kap104p, the homologue of mammalian transportin, mediates import of several RNA-binding proteins (Aitchison et al., 1996). Pse1p and Yrb4p (Kap123p) are the importins for some ribosomal proteins (Rout et al., 1997; Schlenstedt et al., 1997). The mRNA-binding protein Npl3p is imported by Mtr10p (Pemberton et al., 1997; Senger et al., 1998). In addition to these importins, four exportins have been identified: Los1p, the homologue of exportin t, is involved in tRNA export (Hellmuth et al., 1998); Msn5p exports a subset of phosphorylated proteins (Kaffman et al., 1998; DeVit and Johnston, 1999); Cse1p, the homologue of human CAS, is the specific exportin for Srp1p (Solsbacher et al., 1998; Künzler and Hurt, 1998; Hood and Silver, 1998); and Xpo1p (Crm1p) mediates export of proteins containing a leucine-rich nuclear export signal (NES) (Stade et al., 1997; Neville et al., 1997).
A number of transport cargoes have been identified for the mammalian CRM1 protein (reviewed by Görlich and Kutay, 1999; Kaffman and O'Shea, 1999) including the inhibitor of cAMP-dependent protein kinases (PKI) and the human immunodeficiency virus Rev protein. A typical NES contains four characteristically spaced leucines or other hydrophobic amino acids. This motif is often degenerate or even absent as in snurportin, the import adapter for U snRNPs (Paraskeva et al., 1999). The export of the S. cerevisiae proteins Hog1p, Yap1p, Dbp5p, and Ssb1p requires Xpo1p, because these proteins all accumulate in the nucleus of temperature-sensitive xpo1 mutants (Ferrigno et al., 1998; Yan et al., 1998; Kuge et al., 1998; Hodge et al., 1999; Shulga et al., 1999). However, direct binding of these proteins to Xpo1p has not been demonstrated.
The Ran GTPase (Gsp1p in S. cerevisiae) is an abundant, mostly nuclear protein that switches between two conformations, the GTP-bound and the GDP-bound state. Ran is the key coordinator of receptor-mediated transport across the NPC (reviewed by Ohno et al., 1998; Moore, 1998; Nakielny and Dreyfuss, 1999; Görlich and Kutay, 1999). Ran binds to all importin β-like proteins and is exported from the nucleus in a complex with these receptors. The only known regulators of the Ran GTPase cycle are the cytoplasmic GTPase activating protein RanGAP1/Rna1p, which catalyzes GTP hydrolysis by Ran, and the chromatin-bound GDP-GTP exchange factor RCC1/Prp20p, which converts Ran-GDP to Ran-GTP inside the nucleus. The asymmetric distribution of the Ran regulators is thought to ensure that Ran is mainly bound to GTP in the nucleoplasm but is rapidly converted to the GDP-bound state in the cytoplasm. Binding of Ran-GTP to import receptors in the nucleus triggers substrate release, whereas Ran-GTP is required for substrate binding to exportins. The low affinity for Ran-GTP in the absence of the transport cargo and the highly cooperative association with Ran-GTP and the export substrate are characteristic features of all exportins. While bound to an importin β-like protein, Ran is unable to hydrolyze GTP even in the presence of RanGAP/Rna1p. This GAP resistance is thought to prevent premature disassembly of export complexes.
The dissociation of export complexes in the cytoplasm is facilitated by Ran binding protein 1 (RanBP1), a 23-kDa cytoplasmic protein that functions as a coactivator of the Ran GTPase (Bischoff et al., 1995a). RanBP1 binds with high affinity specifically to Ran-GTP but contains a Ran-binding domain different from that of importin β. Yrb1p, the S. cerevisiae homologue of RanBP1, is an essential cytoplasmic protein required for bidirectional transport across the NPC (Schlenstedt et al., 1995). Together, RanGAP1/Rna1p and RanBP1/Yrb1p mediate the disassembly of all Ran-GTP-containing export complexes analyzed thus far (Deane et al., 1997; Floer et al., 1997; Görlich et al., 1997; Kutay et al., 1997; Lounsbury and Macara, 1997; Schlenstedt et al., 1997; Kehlenbach et al., 1998; Solsbacher et al., 1998). RanBP1 binds to Ran-GTP complexed to a receptor to form a disassembly intermediate. This intermediate is then dissociated by Ran-GTP hydrolysis which is induced either by RanBP1-facilitated access of RanGAP to Ran-GTP or by transient release of RanBP1/Ran-GTP (Bischoff and Görlich, 1997).
Yrb1p accumulates in the nucleus after expression of dominant-negative YRB4 or LOS1 mutants (Schlenstedt et al., 1997; Hellmuth et al., 1998) and in xpo1–1 mutants (Künzler et al., 2000) indicating that Yrb1p can enter the nucleus. Xpo1p is required for the nuclear export of Yrb1p (Künzler et al., 2000), but it is unclear whether export of Yrb1p is mediated by an NES.
Besides Yrb1p, S. cerevisiae contains two other proteins with a Ran-binding domain (RBD) of the RanBP1-type, the nucleoporin Nup2p and the soluble 36-kDa protein Yrb2p. Nup2p is involved in NLS protein import (Solsbacher et al., 2000) and Srp1p export (Booth et al., 1999). Yrb2p interacts with Gsp1p as well as with Prp20p and Rna1p. In contrast to Yrb1p, Yrb2p is not essential for vegetative growth and is located predominantly in the nucleus (Taura et al., 1997; Noguchi et al., 1997). Cells deleted for YRB2 show a specific defect in Xpo1p-mediated NES protein export (Taura et al., 1998; Noguchi et al., 1999), but the exact role of Yrb2p in NES export is unclear.
In this study, we identify Yrb1p as a major binding partner of Xpo1p in the presence of Gsp1p-GTP in yeast extracts. We show that the binding of Yrb1p to Xpo1p does not involve a leucine-rich NES but is mediated by Gsp1p. Thus, the Yrb1p/Gsp1p-GTP/Xpo1p complex can be regarded as a kinetically stable disassembly intermediate. Consequently, this complex is dissociated by Rna1p. In addition, we show that Yrb2p forms a very similar complex with Xpo1p and Gsp1p-GTP.
MATERIALS AND METHODS
Strains and Plasmids
The strain containing the integrated fusion of GFP to YRB1 (MATa ura3 leu2 his3 trp1 GFP-YRB1, GSY641) was constructed by homologous recombination using the pop-in/pop-out strategy (Scherer and Davis, 1979). Plasmid pGS589, encoding a fusion of GFP to the N-terminus of Yrb1p, contains the PCR-derived coding sequence of GFP (a S65T, V163A mutant) inserted between the YRB1-promoter and the coding region of YRB1 in pRS306 (Sikorski and Hieter, 1989). The wild-type strain GSY562 was transformed with HindIII-linearized pGS589, Ura+ transformants were plated on medium containing 5-fluoro-orotic acid (5-FOA), and the pop-out strains expressing GFP-YRB1 were identified microscopically. The correct integration of GFP-YRB1 was confirmed by PCR and immunoblotting with Yrb1p-specific antibodies. The growth rate of strain GSY641 in YPD medium was slightly reduced compared with the isogenic wild-type strain. Strain GSY641 was crossed to an xpo1–1 mutant (KWY121; Stade et al., 1997), and strain GSY658 (MATa ura3 leu2 his3 trp1 GFP-YRB1 XPO1::LEU2 + CEN HIS3 xpo1–1) was isolated after tetrad dissection.
Plasmid pRS313-ZZ contains two copies of the IgG-binding domain of Staphylococcus aureus protein A (Z domains) as an EcoRI/BamHI PCR fragment, which derived from plasmid pTL27 (Lafontaine and Tollervey, 1996), inserted into pRS313 (Sikorski and Hieter, 1989). Plasmid pKW577, encoding an in-frame fusion of two Z domains to the C-terminus of Xpo1p, was created by cloning a SalI/EcoRI fragment derived from pKW466 (Stade et al., 1997), which contains the XPO1-promoter and the complete coding sequence of XPO1, into pRS313-ZZ. Plasmid pGEX-2TEV-YRB1 (pGS552) encoding GST-Yrb1p contains the complete YRB1 coding sequence (Schlenstedt et al., 1995) as a 715-bp BamHI fragment inserted into pGEX-2TEV (pGS560), which derives from pGEX-2T (Pharmacia) and contains a TEV (tobacco etch virus protease) cleavage site in place of the thrombin cleavage site. GAL1-promoter driven YRB1 truncations fused to GFP were cloned by PCR with Pwo polymerase (Roche Molecular Biochemicals) as BamHI fragments and inserted into the BamHI site of YCpGAL1-GFP (pGS372). A 675-bp SphI/HindIII fragment containing the complete coding sequence of GSP1 was amplified by PCR and inserted into pQE9 (Qiagen), creating an N-terminal fusion to a 6-histidine tag (pKW581), or into pGEX-4T-1, creating an N-terminal fusion to GST (pGS785). The GSP1Q71L mutation was introduced by site-directed mutagenesis with PCR. The PCR product was cloned via NgoMIV and BstEII into pKW581 to generate pKW586. The mutation in pKW586 was verified by DNA sequencing. Similarly, the GSP1ΔC truncation was constructed by PCR, replacing the 7 C-terminal codons by a Leu codon. The complete YRB2 coding sequence (Taura et al., 1997) was inserted into pQE70 (Qiagen) as a 991-bp BamHI fragment, creating a C-terminal fusion to a 6-histidine tag (pGS558), or inserted into pGEX-4T-1 as a 1096-bp BamHI/SalI fragment, creating an N-terminal fusion to GST (pGS270). SSB1-C, containing the C-terminal 90 codons of SSB1, was amplified by PCR from genomic DNA as a 452-bp BamHI/XhoI fragment and inserted into pGEX-4TEV (pGS804), yielding pGS864, which encodes an N-terminal fusion to GST. Plasmid pGS488 contains the 1500-bp EcoRI/XhoI insert of pKW430 (Stade et al., 1997) in pGEX-4T-1 and encodes GST-PKI-NES-2GFP. Plasmid pGS489 was constructed similarly but contains the P12-NES mutant derived from pKW431 (Stade et al., 1997). The coding sequences of XPO1, KAP95, PSE1, KAP104, and YRB4 were amplified by PCR from genomic DNA. Plasmid pQE70-XPO1 (pGS512) contains a 3265-bp BamHI fragment. Plasmid pGEX-4TEV-KAP95 (pGS962) carries a 2593-bp BamHI insert. Plasmid pQE9-PSE1 (pGS1006) contains a 3280-bp BamHI fragment. Plasmid pGEX-4TEV-KAP104 (pGS1010) has a 2874-bp BamHI insert. Plasmid pQE30-YRB4 (pGS1011) contains a 3550-bp BamHI/SalI fragment in pQE30 (Qiagen).
Protein Analysis
Yeast extracts were prepared by grinding frozen yeast cells with a pestle in the presence of liquid nitrogen. The extract was treated with protease inhibitors, dialyzed against binding buffer (20 mM Tris-HCl pH 7.4, 100 mM potassium acetate, 5 mM magnesium acetate, 10% glycerol, 1 mM DTT), and centrifuged for 30 min at 20,000g. The total protein concentration of the extract was adjusted to 5 mg/ml. To purify Xpo1p-ZZ and any bound proteins, 10 ml of extract were applied to 50 μl IgG Sepharose beads (Pharmacia) either in the absence or presence of 200 μg Gsp1pQ71L and incubated over night at 4°C. After extensive washes with ∼200 column volumes of binding buffer, bound proteins were eluted with binding buffer containing 500 mM KCl. Fractions were precipitated with methanol/chloroform and analyzed by SDS PAGE.
The purifications of Yrb1p (Schlenstedt et al., 1995), Srp1p, GST-Srp1p and Cse1p (Solsbacher et al., 1998), and S. pombe Rna1p (Bischoff et al., 1995b) were all described before. Xpo1p-6His, 6His-Pse1p, 6His-Yrb4p, Yrb2p-6His, and 6His-Gsp1p were purified with nickel-nitrolotriacetic agarose (Qiagen). Xpo1p, Pse1p, and Yrb4p were further purified by Mono Q (Pharmacia) chromatography. The GDP-bound and GTP-bound forms of 6His-Gsp1p and 6His-Gsp1pQ71L were separated on a Mono S column (Pharmacia) and the GTP/GDP contents were determined by HPLC analysis (Görlich et al., 1996). The Gsp1pQ71L-GTP preparation contained 97% GTP-bound and 3% GDP-bound protein, the Gsp1p-GTP preparation contained 50% GTP-bound protein, and the Gsp1pQ71L-GDP and Gsp1p-GDP fractions contained 100% GDP-bound protein (not shown). The various GST fusion proteins were purified using glutathione Sepharose (Pharmacia). PKI-NESp and mutant PKI-NESp were separated from GST after thrombin cleavage (Schlenstedt et al., 1995). Yrb1p, Kap95p, and Kap104p were cleaved from the respective GST fusion proteins by TEV protease and further purified on Mono Q.
Gel filtration chromatography and solution binding assays were performed in PBSKMT buffer (25 mM Na-phosphate, 150 mM NaCl, 3 mM KCl, 1 mM MgCl2, 0.1% Tween 20, pH 7.3). Solution binding assays were as described before (Solsbacher et al., 1998). Purified GST fusion proteins were incubated with glutathione Sepharose (Pharmacia) for 30 min at 4°C. After three washes with 1 ml PBSKMT, proteins or peptides were added to a volume of 300 μl as indicated in the figure legends. After washing, bound proteins were eluted with SDS sample buffer. PKI-NES peptides (GGGNELALKLAGLDINKT) and mutant PKI-NES peptides (GGGNELALKLAGADANKT) were a gift of Elena Conti (EMBL, Heidelberg). NS2 peptides (CVDEMTKKFGTLTIHDTEK) and NS2 mutant peptides (CVDEMTKKFGTATAHDTEK) were described previously (Askjaer et al., 1999). GTPase assays were performed as described before (Bischoff et al., 1995b; Solsbacher et al., 1998) with the following modifications. Enzymatic reactions were performed at 15°C in 20 mM HEPES-NaOH pH 7.4, 120 mM K-acetate, 1 mM Mg-acetate, 0.5% hydrolyzed gelatin, 0.02% NaN3. If not indicated otherwise, 30-s GTPase reactions were started by addition of 40 nM Rna1p.
RESULTS
Yrb1p Interacts with Xpo1p and Gsp1p-GTP in Yeast Extracts
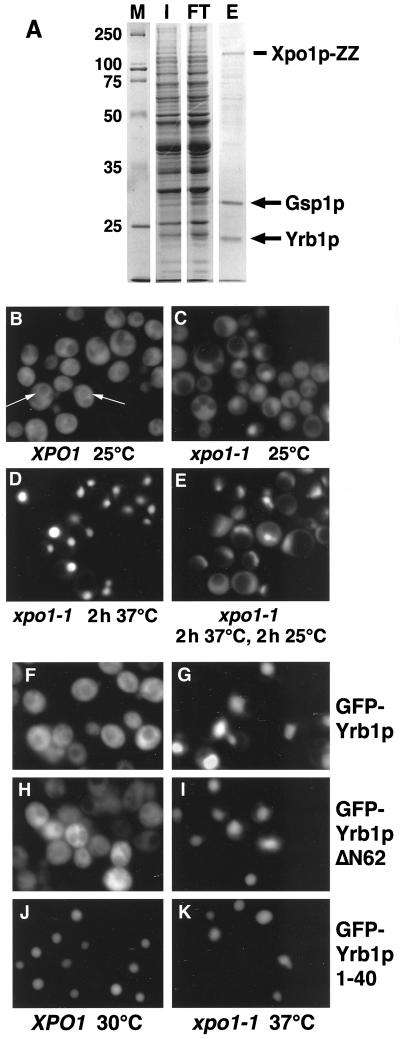
In order to identify export cargoes of Xpo1p, a Xpo1p-ZZ fusion protein was generated (see Materials and Methods). The fusion protein was functional since it was able to rescue a strain deleted for XPO1 at all temperatures tested (data not shown). Xpo1p-ZZ was purified from yeast extracts either in the absence or presence of recombinant Gsp1p-GTP, and proteins bound to immobilized Xpo1p-ZZ were eluted with potassium chloride and analyzed (see Material and Methods). Gsp1p-GTP was added to promote formation of trimeric cargo/Xpo1p/Gsp1p-GTP complexes. Because wild-type Gsp1p rapidly hydrolyzes GTP in the presence of cellular extracts, GTPase-deficient Gsp1pQ71L loaded with GTP was used. The two major proteins detected (Figure 1A) were analyzed using matrix-assisted laser desorption/ionization (MALDI) after in-gel digestion with trypsin of the excised bands (Shevchenko et al., 1996a, 1996b). This unambiguously identified the proteins migrating with apparent molecular weights of ∼30 and ∼23 kDa as Gsp1p and Yrb1p, respectively. The identity of Gsp1p and Yrb1p in the eluate was confirmed by immunoblotting (not shown). The interaction between Yrb1p and Xpo1p was specific and dependent on the presence of Gsp1pQ71L, because Yrb1p was not detected when Gsp1pQ71L was omitted or when control extracts were mock-purified in the absence of Xpo1p-ZZ (not shown).
Figure 1.
(A) Yrb1p is a major Xpo1p-binding protein. Xpo1p-ZZ was purified from yeast extracts in the presence of recombinant Gsp1pQ71L-GTP as described in Materials and Methods, and proteins bound to immobilized Xpo1p were eluted with 500 mM KCl. The input (I), flow-through (FT), and eluate (E) were analyzed by SDS PAGE and Coomassie blue staining. The two major proteins in the eluate were identified by MALDI and Western blotting and correspond to Gsp1p and Yrb1p. Molecular weight markers (M) are in kDa. (B-E) Yrb1p shuttles between the cytoplasm and the nucleus. Wild-type cells (B) or xpo1–1 cells (C-E) expressing GFP-YRB1 were grown in liquid medium at 25°C. The cultures were kept at 25°C (C), shifted to 37°C for 2 h (D), or shifted to 37°C for 2 h and then shifted back to 25°C for 2 h (E). To inhibit protein synthesis, cycloheximide (final concentration 0.1 mg/ml) was added to the cultures before the temperature shift. The cells were viewed by fluorescence microscopy to visualize GFP-Yrb1p. The perinuclear staining in wild-type cells is indicated by arrows. (F-K) Yrb1p has two nuclear targeting sequences. Wild-type cells (F, H, and J) or xpo1–1 mutants (G, I, and K) were transformed with plasmids encoding GFP-Yrb1p, GFP-Yrb1ΔN62p, or GFP-Yrb1p 1–40, as indicated. The cultures were incubated at 30°C (XPO1 cells) or 25°C (xpo1–1 cells) in raffinose-containing medium. Expression of the GFP fusions was induced by 2% galactose and repressed by addition of 2% glucose after 1 h. Cells were kept at 30°C (XPO1) or shifted to 37°C for 2 h (xpo1–1) and viewed by fluorescence microscopy.
Shuttling of Yrb1p between the Cytoplasm and the Nucleus
Yrb1p is located predominantly in the cytoplasm of wild-type cells (Schlenstedt et al., 1995). However, it was recently shown that Yrb1p accumulates in the nucleus of temperature-sensitive xpo1–1 mutants (Künzler et al., 2000) that are defective in NES-mediated protein export (Stade et al., 1997). In order to test directly whether Yrb1p shuttles between the cytoplasm and the nucleus in living cells, strains were used in which YRB1 was genetically replaced by a functional GFP-YRB1 fusion (see Material and Methods). In wild-type cells analyzed by fluorescence microscopy, GFP-Yrb1p localized predominantly to the cytoplasm (Figure 1B). In agreement with previous results obtained by immunofluorescence microscopy (Schlenstedt et al., 1995), GFP-Yrb1p was also concentrated around nuclei (Figure 1B, arrows). In xpo1–1 mutant cells shifted to the restrictive temperature of 37°C, GFP-Yrb1p was exclusively nuclear (Figure 1C and D). When the cells were returned to the permissive temperature of 25°C to restore Xpo1p-dependent export, most GFP-Yrb1p was exported and found in the cytoplasm within 2 h (Figure 1E). These results show that Yrb1p shuttles between the nucleus and the cytoplasm.
The Ran-binding domain (RBD) of Yrb1p comprises approximately residues 65–193 (Schlenstedt et al., 1995; Vetter et al., 1999b). In comparison to human RanBP1, Yrb1p contains a 40-residue N-terminal extension and lacks a C-terminal domain harboring a leucine-rich NES (Richards et al., 1996; Pasquinelli et al., 1997; Plafker and Macara, 2000). In an attempt to define the regions within Yrb1p responsible for nuclear import and export, fragments of Yrb1p were fused to GFP and expressed in wild-type and xpo1–1 cells. Both a GFP fusion to full length Yrb1p and GFP-Yrb1ΔN62p, a mutant lacking the 62 N-terminal residues, were cytoplasmic in wild-type cells but completely nuclear in xpo1–1 mutants (Figure 1F-I). In contrast, GFP fused to the 40 N-terminal residues of Yrb1p was found in the nucleus of both wild-type and xpo1–1 cells (Figure 1J and K). This confirms the observation that the RBD alone can mediate both import and export of Yrb1p (Künzler et al., 2000). Moreover, it suggests that Yrb1p contains a second nuclear import signal that resides within the 40 N-terminal residues.
Yrb1p Associates with Xpo1p through Gsp1p-GTP
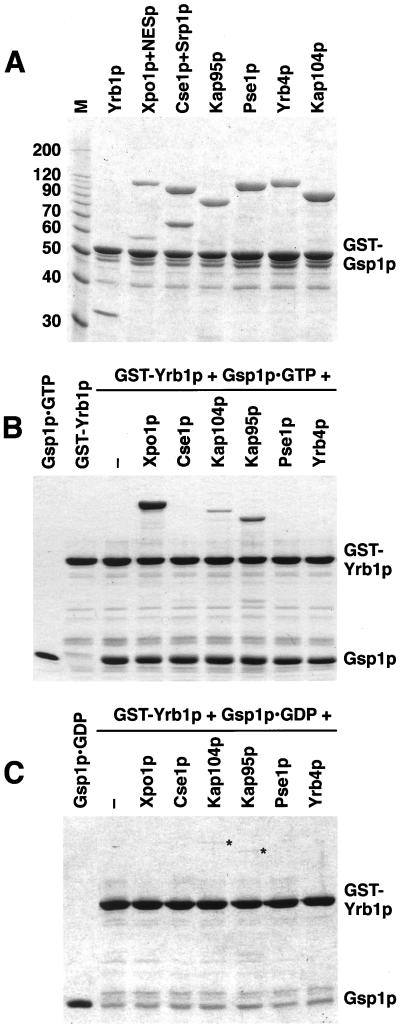
It was recently shown with purified recombinant proteins that Xpo1p (Künzler et al., 2000) but not Cse1p (Solsbacher et al., 1998) associates with Yrb1p and Gsp1p-GTP. Using solution binding assays, we tested whether other importin β-like proteins form complexes with Yrb1p and Gsp1p. First, we incubated Xpo1p and Cse1p as well as the importins Kap95p, Kap104p, Pse1p, and Yrb4p with immobilized GST-Gsp1p-GTP. Like Yrb1p, all importins bound efficiently to Gsp1p-GTP (Figure 2A), whereas the exportins Xpo1p and Cse1p did not (see Figure 3A and Solsbacher et al., 1998). However, Xpo1p and Cse1p associated with Gsp1p in the presence of their respective export substrates, a PKI-NES-GFP fusion protein or Srp1p (Figure 2A). We detected no binding of the receptors to immobilized GST-Yrb1p in the absence of Gsp1p (not shown). However, in the presence of Gsp1p-GTP, a strong interaction of Yrb1p with Xpo1p and a somewhat weaker binding to Kap95p and Kap104p was observed (Figure 2B). On the other hand, Cse1p, Pse1p, and Yrb4p (Figure 2B) and Los1p (not shown) formed no complex with Yrb1p and Gsp1p-GTP. We then incubated the importin β-like proteins with Gsp1p-GDP and immobilized Yrb1p, which resulted in only a weak binding of Kap95p and Kap104p (Figure 2C). These interactions did not result from a contamination of Gsp1p-GDP with Gsp1p-GTP, because the Gsp1p-GDP preparation contained no GTP (see Material and Methods) and because Xpo1p did not bind under these conditions (Figure 2C). The complexes in the presence of Gsp1p-GDP are similar to previously described complexes of mammalian importin β, Ran-GDP, and RanBP1 (Chi et al., 1996; Deane et al., 1997). Taken together, the data indicate that Xpo1p is able to form a stable interaction with Yrb1p only in the presence of Gsp1p-GTP. Similar stoichiometric complexes could not be formed by the other importin β-like proteins tested.
Figure 2.
Association of importin β-like proteins with Gsp1p-GTP and Yrb1p. (A) GST-Gsp1p-GTP (5 μg per reaction) was immobilized on glutathione Sepharose and incubated for 30 min at 4°C with 3 μg of Yrb1p or 12 μg of Xpo1p, Cse1p, Kap95p, Pse1p, Yrb4p, or Kap104p, as indicated. The reactions containing the export receptors Xpo1p and Cse1p received additionally 4 μg of PKI-NES-2GFP (NESp) or 5 μg of importin α (Srp1p). Molecular weight markers (M) are in kDa. (B and C) GST-Yrb1p (4 μg per reaction) was immobilized on glutathione Sepharose and incubated for 30 min at 4°C with 12 μg of each importin β-like protein and with 10 μg of Gsp1pQ71L loaded with either GTP (B) or GDP (C). The load of Gsp1p (50% of the input) is shown in the left lane. Asterisks indicate weak binding of Kap104p and Kap95p to Yrb1p in the presence of Gsp1p-GDP. The bound material of all reactions was washed three times and analyzed by SDS PAGE and Coomassie blue staining.
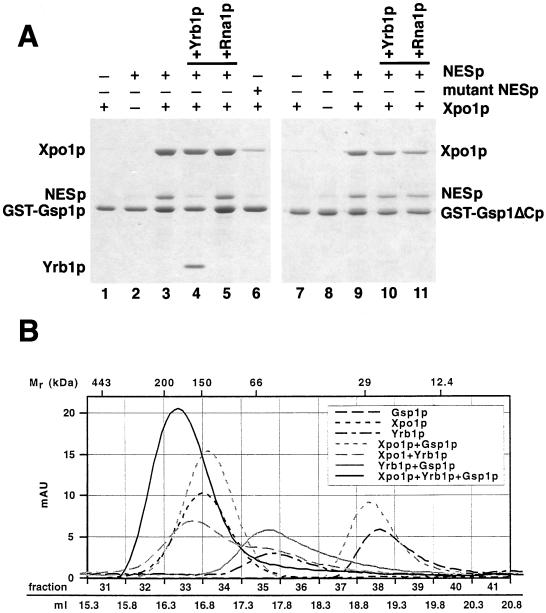
Figure 3.
(A) Yrb1p but not an NES protein requires the acidic C-terminus of Gsp1p for complex formation. GST-Gsp1p-GTP (lanes 1–6) and GST-Gsp1ΔCp-GTP (lanes 7–11)(4 μg per reaction) were immobilized on glutathione Sepharose and incubated for 30 min at 4°C with 8 μg of PKI-NES-2GFP (NESp), 8 μg of P12-NES-2GFP (mutant NESp), and/or 12 μg of Xpo1p, as indicated. Bound material was washed three times and either eluted with SDS sample buffer (lanes 1–3 and 6–9) or incubated further for 15 min at 4°C with 4 μg of Yrb1p (lanes 4 and 10) or 1 μg of Rna1p (lanes 5 and 11) and then washed again three times. All samples were analyzed by SDS PAGE and Coomassie blue staining. (B) Xpo1p, Yrb1p, and Gsp1p-GTP form a stable 1:1:1 complex. Xpo1p (125 kDa), Yrb1p (23 kDa), and 6His-Gsp1pQ71L-GTP (26 kDa) were mixed as indicated and incubated for 30 min at 4°C. The reactions were gelfiltrated with a Superose 6 column (Pharmacia) in PBSKMT buffer (see Materials and Methods) at a flow rate of 0.4 ml/min. Absorption units at 280 nm were recorded using the Äkta Explorer 100 software (Pharmacia) and plotted against the fraction numbers and the volume after sample injection. Molecular weight markers (Sigma): apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), serum albumin (66 kDa), carbonic anhydrase (29 kDa), cytochrome c (12.4 kDa).
To investigate the formation and the disassembly of the NES/Xpo1p/Gsp1p complex, we immobilized GST-Gsp1p-GTP and incubated it with Xpo1p and the PKI-NES protein. These proteins individually showed no significant binding, but together they bound efficiently to Gsp1p-GTP (Figure 3A, lanes 1–3), demonstrating the strong cooperativity in binding of transport substrates and Gsp1p to Xpo1p. We performed a number of specificity controls. A mutant NES protein containing the P12 point mutation (Wen et al., 1995; Stade et al., 1997) bound only inefficiently to Xpo1p (Figure 3A, lane 6). In addition, essentially no binding to Gsp1p-GDP was detected, and Xpo1p deletion mutants lacking either the N-terminal 122 residues or the C-terminal 198 residues formed no complexes with Gsp1p-GTP and the NES protein or Yrb1p (not shown). In the presence of Rna1p, Yrb1p disassembles export complexes containing Gsp1p and transport receptors (Schlenstedt et al., 1997; Hellmuth et al., 1998; Solsbacher et al., 1998). The PKI-NESp/Xpo1p/Gsp1p-GTP complex was resistant to Rna1p (Figure 3A, lane 5), indicating that Xpo1p inhibits the Rna1p-mediated activation of the Gsp1p GTPase. Remarkably, in the absence of Rna1p, Yrb1p released the NES protein and bound itself to Xpo1p/Gsp1p-GTP (Figure 3A, lane 4). Thus, in contrast to the Srp1p/Cse1p/Gsp1p-GTP export complex, which is entirely dissociated by Yrb1p in the absence of Rna1p (Solsbacher et al., 1998), Yrb1p displaces the NES protein from the Xpo1p export complex.
To test whether Yrb1p binds directly to Xpo1p or whether the interaction is mediated by Gsp1p, a Gsp1p mutant was used that lacks the seven C-terminal amino acid residues (Asp-Glu-Asp-Asp-Ala-Asp-Leu), which are essential for the interaction of Ran-GTP with RanBP1 (Richards et al., 1995; Hieda et al., 1999; Vetter et al., 1999b) but not required for binding to importin β-like proteins (Lounsbury et al., 1996; Kutay et al., 1997; Hieda et al., 1999). Gsp1ΔCp-GTP did not bind to Yrb1p but bound to Kap95p, Kap104p, Pse1p, Yrb4p, and Cse1p/Srp1p in control reactions (not shown) and also associated efficiently with Xpo1p and the NES protein (Figure 3A, lane 9). However, although the NESp/Xpo1p/Gsp1ΔCp complex was still resistant to Rna1p, it was no longer dissociated by Yrb1p (Figure 3A, lanes 10 and 11). The requirement for the C-terminus of Gsp1p demonstrates that the interaction between Yrb1p and Xpo1p is different from the binding of Xpo1p to NES-containing proteins and indicates that Yrb1p associates with Xpo1p via Gsp1p.
In order to investigate the stoichiometry of the different complexes between Xpo1p, Yrb1p, and Gsp1p-GTP, the proteins were mixed and examined by gel filtration chromatography. The peak fractions eluting from a Superose 6 column were analyzed by measuring the absorption at 280 nm (Figure 3B) and by Coomassie blue staining of SDS gels (not shown). Both Xpo1p and Gsp1p migrated as monomers when loaded alone or when combined and preincubated before gel filtration. Yrb1p formed homodimers and thus behaves as human RanBP1, which also dimerizes (Bischoff et al., 1995a). Xpo1p and Yrb1p did not form a complex, but Gsp1p and Yrb1p together migrated as a heterodimer, suggesting that binding of Gsp1p to Yrb1p induces the dissociation of Yrb1p homodimers. Xpo1p, Yrb1p, and Gsp1p combined eluted at a position corresponding to ∼175 kDa (Figure 3B), indicating that the three proteins form a stable 1:1:1 complex.
Characterization of the Stability of Xpo1p Complexes
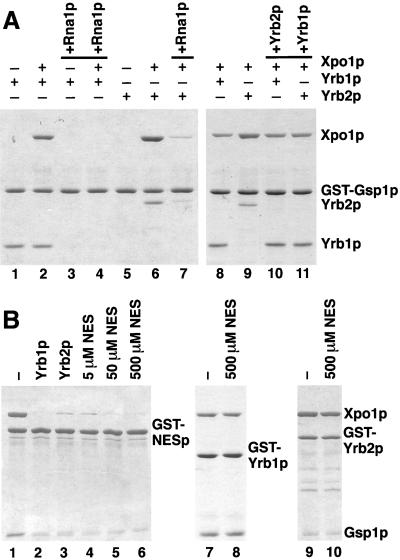
We next investigated the effect of Rna1p on the Yrb1p/Gsp1p-GTP/Xpo1p complex. As expected, Rna1p-mediated GTP hydrolysis by Gsp1p resulted in the release of Yrb1p from Gsp1p alone (Figure 4A, lanes 1 and 3). Rna1p also dissociated the Yrb1p/Gsp1p-GTP/Xpo1p complex (Figure 4A, lanes 2 and 4). We also tested Yrb2p in this assay, since Yrb2p is the only other soluble yeast protein besides Yrb1p that contains a RBD of the RanBP1-type. Yrb2p alone bound only very weakly to Gsp1p-GTP (Figure 4A, lane 5), which indicates that Yrb2p has a much lower affinity for Gsp1p than does Yrb1p. However, Yrb2p and Xpo1p together bound efficiently to Gsp1p-GTP (Figure 4A, lane 6). Like Yrb1p, Yrb2p could not bind to Gsp1ΔCp in the presence or absence of Xpo1p (not shown). Thus, Xpo1p cooperatively binds to Gsp1p in the presence of either Yrb1p or Yrb2p. Furthermore, Xpo1p greatly increases the affinity of Yrb2p for Gsp1p. As for Yrb1p, Rna1p also dissociated the Yrb2p/Gsp1p-GTP/Xpo1p complex (Figure 4, lane 7). We conclude that, unlike an NES protein, Yrb1p and Yrb2p do not inhibit GTPase activation of Xpo1p-bound Gsp1p under these conditions. This observation and the requirement for the acidic C-terminus of Gsp1p characterize the Yrb1p/Gsp1p-GTP/Xpo1p and Yrb2p/Gsp1p-GTP/Xpo1p complexes as stable disassembly intermediates.
Figure 4.
The association of Yrb1p and Yrb2p with Xpo1p/Gsp1p-GTP is sensitive to Rna1p but resistant to excess amounts of NES peptides. (A) GST-Gsp1p-GTP (4 μg per reaction) was immobilized on glutathione Sepharose and incubated for 30 min at 4°C with 5 μg of Yrb1p, 5 μg of Yrb2p, and/or 12 μg of Xpo1p, as indicated. Bound material was washed three times and either eluted with SDS sample buffer (lanes 1, 2, 5, 6, 8, and 9) or incubated further for 15 min at 4°C with 1 μg of Rna1p (lanes 3, 4, and 7), 10 μg of Yrb2p (lane 10), or 10 μg of Yrb1p (lane 11), and then washed again three times. All samples were analyzed by SDS PAGE and Coomassie blue staining. (B) GST fusions (6 μg per reaction) to PKI-NES-2GFP (GST-NESp)(lanes 1–6), Yrb1p (lanes 7 and 8), or Yrb2p (lanes 9 and 10) were immobilized on glutathione Sepharose and incubated for 30 min at 4°C with 10 μg of Gsp1p-GTP and 12 μg of Xpo1p. After three washes, the reactions were further incubated for 15 min at 4°C with buffer alone (lanes 1, 7, and 9), with 6 μg of Yrb1p (lane 2), with 6 μg of Yrb2p (lane 3), or with the indicated amounts of peptides corresponding to the PKI-NES (lanes 4–6, 8, and 10). After three washes, bound material was analyzed by SDS PAGE and Coomassie blue staining.
We next incubated a preformed Yrb2p/Gsp1p-GTP/Xpo1p complex (Figure 4A, lane 9) with Yrb1p. Remarkably, addition of Yrb1p resulted in the complete displacement of Yrb2p and replacement by Yrb1p (Figure 4A, lane 11). Titration experiments showed that roughly equimolar amounts of Yrb1p over bound Yrb2p were sufficient to remove Yrb2p from Gsp1p/Xpo1p (not shown). In contrast, Yrb2p did not affect the stability of the preformed Yrb1p/Gsp1p-GTP/Xpo1p complex (Figure 4A, lanes 8 and 10). Even a tenfold molar excess of Yrb2p did not displace Yrb1p from Gsp1p/Xpo1p (not shown). This indicates that Yrb1p and Yrb2p compete for Gsp1p-GTP-binding but Yrb1p has a much higher affinity.
The PKI-NES/Xpo1p/Gsp1p-GTP complex also formed when the NES protein was immobilized on Sepharose beads (Figure 4B, lane 1). When this complex was incubated with Yrb1p or with Yrb2p, either protein released Xpo1p and Gsp1p from the NES protein (Figure 4B, lanes 2 and 3). Thus, both Yrb1p and Yrb2p can act as dissociation factors for NES export complexes in vitro. Peptides corresponding to the PKI-NES also displaced Xpo1p/Gsp1p from the NES protein (Figure 4B, lanes 4–6). The majority of bound Xpo1p and Gsp1p was released by a 10-fold molar excess of the peptides over Xpo1p (Figure 4B, lane 4), whereas mutant peptides did not affect the stability of the complex (not shown). The PKI-NES peptide had no effect on the Yrb1p/Gsp1p-GTP/Xpo1p and Yrb2p/Gsp1p-GTP/Xpo1p complexes, even when a 1000-fold excess of peptide was employed (Figure 4B, lanes 7–10). This further supports the conclusion that Yrb1p and Yrb2p do not bind to the NES binding site of Xpo1p.
We attempted to characterize genuine yeast proteins containing export signals for their interaction with Xpo1p and Gsp1p. The proteins Hog1p and Yap1p, which accumulate in the nuclei of xpo1 mutants (Ferrigno et al., 1998; Yan et al., 1998), and the mRNA export factor Gle1p, which contains a leucine-rich NES-like sequence (Murphy and Wente, 1996), were purified and analyzed in solution binding assays. Surprisingly, none of these proteins interacted with Xpo1p in our assays (not shown). The direct interaction of these proteins with Xpo1p and Gsp1p may require additional factors or modifications not present in bacterially expressed proteins. However, we did detect binding of Xpo1p to Ssb1p, a member of the 70-kDa heat shock protein family, which contains an NES at the C-terminus (Shulga et al., 1999). Xpo1p and Gsp1p-GTP cooperatively bound to Ssb1p, and Yrb1p displaced Ssb1p from Xpo1p/Gsp1p (not shown, see Figure 5B).
Figure 5.
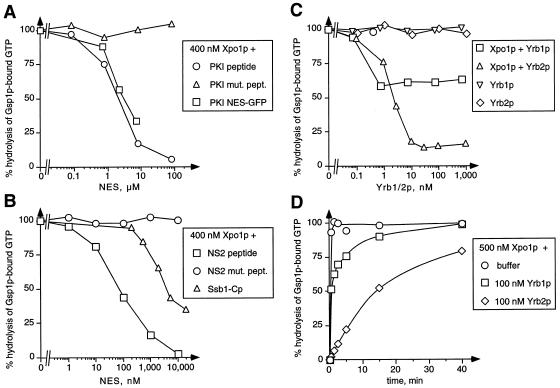
(A-C) Cooperative binding of NES, Yrb1p, and Yrb2p to Xpo1p/Gsp1p-GTP. Gsp1p[γ-32P]GTP (50 pM) was incubated with 400 nM Xpo1p and the indicated concentrations of NES peptides or proteins (A and B) or Yrb1p or Yrb2p (C). Reactions containing Gsp1p[γ-32P]GTP and Yrb1p or Yrb2p alone served as controls. After 30 min, the GTPase reactions were started by addition of 40 nM Rna1p. Hydrolysis of GTP by Gsp1p was determined as released [32P]phosphate after 30 s by the charcoal method. (D) Kinetics of Rna1p-induced complex disassembly. Gsp1p[γ-32P]GTP (50 pM) was incubated for 30 min with 500 nM Xpo1p alone, with Xpo1p and 100 nM Yrb1p, or with Xpo1p and 100 nM Yrb2p. After Rna1p addition, the reactions were incubated further for the indicated time periods. GTP hydrolysis was then determined by the charcoal method.
To analyze the formation of Xpo1p/NES substrate complexes in more detail, we performed GTPase inhibition assays, which allow quantification of the stability of the complexes by measuring the inhibitory effect of importin β-like proteins on Rna1p-mediated GTPase activation of Gsp1p. Typically, the complexes are allowed to form during an incubation for 30 min. Then Rna1p is added and the rate of GTP hydrolysis by Gsp1p is measured after further incubation for 30 s. Both the PKI-NES protein and PKI-NES peptides, but not mutant PKI-NES peptides, formed complexes with Xpo1p and Gsp1p-GTP and thus inhibited GTP hydrolysis by Gsp1p in a concentration-dependent manner (Figure 5A). Surprisingly, the calculated dissociation constants of roughly 2 μM were ∼ 1,000-fold higher than those of complexes containing Gsp1p-GTP and Kap95p, Pse1p, Yrb4p, Cse1p, or Los1p (Schlenstedt et al., 1997; Solsbacher et al., 1998; Hellmuth et al., 1998). This indicates that the PKI-NES has a relative low affinity for Xpo1p in the presence of Gsp1p-GTP. Hog1p, Yap1p, and Gle1p did not affect the Rna1p-mediated GTPase activation (not shown). However, the C-terminal domain of Ssb1p containing the NES as well as peptides containing the NES of the minute virus of mice NS2 protein (Askjaer et al., 1999; Bodendorf et al., 1999), but not NS2 mutant peptides, also inhibited GTP hydrolysis in the presence of Xpo1p (Figure 5B). The calculated kD of the NS2 complex was ∼ 70 nM. Therefore, the NS2-NES has a roughly 30-fold higher affinity for Xpo1p than the NESs of PKI or Ssb1p. Xpo1p alone did not inhibit Rna1p-mediated GTPase activation up to a concentration of 2 μM (not shown), which confirms that the binding of the NES substrate and Gsp1p to Xpo1p is highly cooperative.
We then used the same assay to analyze the effect of Yrb1p and Yrb2p on GTPase activation. Binding of Yrb2p to Xpo1p and Gsp1p-GTP inhibited Rna1p-induced hydrolysis of Gsp1p-bound GTP (Figure 5C). This complex formation was highly cooperative with a kD of 2 nM, which is roughly 30 times lower than the kD of the NS2 complex. The maximum GTPase inhibition (10% GTP hydrolysis compared with samples containing no Xpo1p) was observed at 10 nM Yrb2p. Interestingly, the GTPase inhibition was not further increased at higher concentrations of Yrb2p. Surprisingly, a similar effect was observed with Yrb1p. Even at micromolar concentrations of Yrb1p, this GTPase block could not be increased over the 40% inhibition already obtained at 1 nM Yrb1p. Therefore, we sought to analyze the kinetic stability of the Yrb1p/Gsp1p-GTP/Xpo1p and Yrb2p/Gsp1p-GTP/Xpo1p complexes. The half-life of the Yrb1p/Gsp1p-GTP/Xpo1p complex was ∼20 s in the presence of Rna1p, whereas the Yrb2p/Gsp1p-GTP/Xpo1p complex was much more stable, with a half-life of 14 min (Figure 5D). In contrast, Rna1p-induced disassembly of the Yrb1p/Gsp1p-GTP complex was complete at the first measurable time point (10 s). As a result, 60% of the Yrb1p/Gsp1p-GTP/Xpo1p complexes and 10% of the Yrb2p/Gsp1p-GTP/Xpo1p complexes were disassembled after the incubation with Rna1p for 30 s (Figure 5C). These results show that Rna1p disassembles Yrb1p/Gsp1p-GTP/Xpo1p complexes, and particularly Yrb2p/Gsp1p-GTP/Xpo1p complexes, less efficiently than complexes of Gsp1p-GTP and Yrb1p alone or complexes of Gsp1p-GTP and some importins like Pse1p or Yrb4p in the presence of Yrb1p (Schlenstedt et al., 1997).
Yrb1p and Rna1p are thought to function as disassembly factors for export complexes in the cytoplasm. We therefore asked whether Yrb1p could dissociate export complexes in a nuclear environment, i.e. in the absence of Rna1p. Due to its high affinity for Gsp1p-GTP, nuclear Yrb1p is expected to be quantitatively bound to Gsp1p. Therefore, we incubated the Srp1p/Cse1p/Gsp1p-GTP complex with a preformed Yrb1p/Gsp1p-GTP dimer (Figure 6). We chose the Cse1p export complex because its dissociation is mediated by Yrb1p alone and does not require Rna1p in vitro (Solsbacher et al., 1998). Figure 6 shows that the Cse1p export complex is sensitive to Yrb1p but resistant to the Yrb1p/Gsp1p-GTP dimer. We conclude that Gsp1p neutralizes the potential inhibitory effect of Yrb1p in the nucleus. The formation and stability of nuclear export complexes is therefore unlikely to be affected by Yrb1p so long as its concentration does not exceed that of Gsp1p. This is consistent with the previous observations that high amounts of RanBP1 injected into Xenopus oocyte nuclei inhibited nuclear transport (Izaurralde et al., 1997) but increased levels of nuclear Yrb1p could be tolerated in yeast (Künzler et al., 2000).
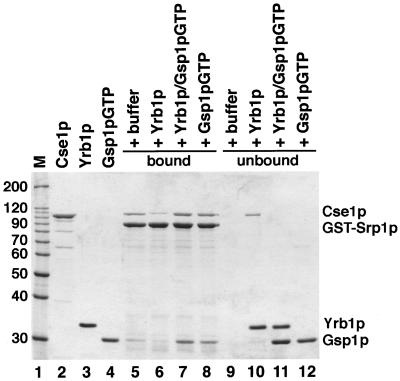
Figure 6.
The Srp1p/Cse1p/Gsp1p-GTP complex is sensitive to Yrb1p but resistant to Yrb1p complexed to Gsp1p-GTP. GST-Srp1p (8 μg per reaction) was immobilized on glutathione Sepharose and incubated with 12 μg of Cse1p and 4 μg of Gsp1pQ71L-GTP for 30 min at 4°C. After three washes, bound material was incubated with buffer alone (lane 5), with 5 μg of Yrb1p (lane 6), with a preincubated mixture of Yrb1p and Gsp1pQ71L-GTP (lane 7), or with 10 μg of Gsp1pQ71L-GTP (lane 8). After further incubation for 30 min at 4°C, bound (lanes 5–8) and unbound (lanes 9–12) material was separated by SDS PAGE and analyzed by Coomassie blue staining. The molecular weight markers (M) and protein loads are shown in lanes 1–4.
DISCUSSION
We show in this study that Yrb1p, a protein that is predominantly cytoplasmic at steady-state, shuttles between the cytoplasm and the nucleoplasm. Nuclear export is mediated by the export receptor Xpo1p. Yrb1p not only accumulates in xpo1–1 mutant cells but also forms a specific and stable complex with Xpo1p and Gsp1p-GTP. Previous evidence for nucleocytoplasmic shuttling of Yrb1p came from its nuclear accumulation upon overexpression of Yrb4p or Los1p mutants that are deficient in Gsp1p-binding and thus act as dominant-negative inhibitors of bidirectional transport (Schlenstedt et al., 1997; Hellmuth et al., 1998). Yrb1p is actively imported into the nucleus, because Yrb1p fusion proteins that are larger than 100 kDa, which are not expected to pass the NPC by diffusion, also accumulate in the nucleus in xpo1–1 cells (data not shown and Künzler et al., 2000). Import requires an intact RBD and depends on the region between residues 121 and 131 of Yrb1p. This region by itself, however, is not sufficient to mediate import (Künzler et al., 2000). We show here that Yrb1p has two distinct import signals. In addition to the RBD, the unique N-terminal extension can also mediate nuclear import, and this region may be required for efficient transport in vivo.
Several findings indicate that the interaction of Yrb1p with Xpo1p is mediated by Gsp1p. First, binding of Xpo1p to Yrb1p, but not to the NES, requires the acidic C-terminus of Gsp1p. Second, the Yrb1p/Gsp1p-GTP/Xpo1p complex but not NES/Xpo1p/Gsp1p-GTP is disassembled by Rna1p. Third, the association of Xpo1p with Yrb1p correlates strictly with Gsp1p-binding to Yrb1p. This was shown by examining various Yrb1p deletions and point mutants (data not shown and Künzler et al., 2000). We show by GTPase inhibition assays that the affinity of Yrb1p for Xpo1p/Gsp1p-GTP is several orders of magnitudes higher than that of the PKI-NES. This explains the failure of PKI-NES peptides to affect the Yrb1p/Gsp1p/Xpo1p complex even at high peptide concentrations. The difference in the affinity of various NESs for Xpo1p/CRM1, which also has been observed by others (Askjaer et al., 1999; Paraskeva et al., 1999), suggests that the strength of a particular NES may determine the export rate.
Yrb1p/RanBP1 functions as the dissociation factor for export complexes containing importin β-like receptors and Ran-GTP. As expected, Yrb1p also disassembles the NES/Xpo1p/Gsp1p-GTP complex. Remarkably, Yrb1p displaces the NES protein and binds to Gsp1p-GTP/Xpo1p. This is expected to occur in the cytoplasm where the Yrb1p/Gsp1p-GTP/Xpo1p disassembly intermediate will be efficiently dissociated by Rna1p. However, the Yrb1p/Gsp1p-GTP/Xpo1p complex is also generated in the nucleus. Because Gsp1p is in the GTP-bound state in the nucleus, it binds to nuclear Yrb1p. Subsequently, Yrb1p/Gsp1p-GTP is recognized by Xpo1p and exported rapidly. We characterized the Yrb1p/Gsp1p-GTP/Xpo1p complex as a kinetically stable disassembly intermediate, which represents a novel type of export complex. In contrast to other export complexes, the transport cargo is a Ran effector and not a receptor substrate. The high efficiency of Xpo1p-mediated Yrb1p export can be explained by the unusual stability of the Yrb1p/Gsp1p-GTP/Xpo1p complex. It remains to be determined which properties of Yrb1p, Gsp1p, or Xpo1p support this stability at the molecular level. In contrast to other transport receptor/Ran complexes, the association of Xpo1p with Yrb1p-bound Gsp1p may involve additional interactions and Xpo1p may directly contact Yrb1p.
It was observed previously that the stability of disassembly intermediates is different for various receptors. In the case of CAS/Cse1p, RanBP1/Yrb1p alone dissociates the export complex in vitro (Kutay et al., 1997; Solsbacher et al., 1998). Efficient disassembly of the transportin/Ran-GTP/RanBP1 complex occurs only in the presence of RanGAP1 (Bischoff and Görlich, 1997). Importin β dissociation additionally requires importin α, which forms a complex with Ran-free importin β (Bischoff and Görlich, 1997; Floer et al., 1997). Similar to Xpo1p, we observed that Kap104p (yeast transportin) and Kap95p (yeast importin β) also form complexes with Gsp1p/Yrb1p. As shown for mammalian importin β, the Yrb1p/Gsp1p-GTP/Kap95p complex was also Rna1p-resistant. Rna1p-mediated disassembly of the Yrb1p/Gsp1p-GTP/Kap104p complex was delayed, suggesting the requirement for another cytoplasmic component (Bischoff, Maurer, and Schlenstedt, unpublished results). However, we can exclude the possibility that Kap95p and Kap104p function as exportins for Yrb1p, because Yrb1p does not accumulate in the nucleus of kap95 and kap104 mutants (our unpublished results).
Our data show that Yrb2p also forms a remarkably stable complex with Xpo1p and Gsp1p-GTP, which is similar to the Yrb1p/Xpo1p/Gsp1p complex. Yrb2p also does not contain a leucine-rich NES and interacts with Xpo1p via Gsp1p. In contrast to Yrb1p, the affinity of Yrb2p for Gsp1p is greatly enhanced in the presence of Xpo1p, but GTPase assays show that Yrb1p binds with a higher affinity to Xpo1p/Gsp1p than does Yrb2p. Consequently, Yrb2p/Xpo1p/Gsp1p complexes can be disassembled by Yrb1p, suggesting that these complexes could also be a target of Yrb1p in the cytoplasm. It was suggested that Yrb2p could productively increase the rate of Xpo1p-mediated NES export (Taura et al., 1998). Our data make this model unlikely, because PKI-NES-binding and Yrb2p-binding to Xpo1p are mutually exclusive. Our biochemical results would suggest that Yrb2p is a shuttling protein. Free Yrb2p can form a stable complex with Xpo1p and Gsp1p in the nucleus that, like the Yrb1p/Xpo1p/Gsp1p complex, could be exported to the cytoplasm. However, Yrb2p was reported not to shuttle between the nucleus and the cytoplasm (Taura et al., 1998). We therefore would have to postulate an unknown factor that either prevents efficient formation of the Yrb2p/Xpo1p/Gsp1p-GTP complex in the nucleus or impedes its export to the cytoplasm.
It was recently shown that RanBP1 is also imported into the nuclei of interphase cells by an active transport mechanism (Plafker and Macara, 2000). Thus, the shuttling of RanBP1/Yrb1p has been conserved from yeast to mammals. A requirement to export RanBP1 after completion of mitosis cannot explain the rapid shuttling of Yrb1p, because S. cerevisiae does not exhibit nuclear envelope breakdown during mitosis. Our data suggest that Yrb1p does not disassemble export complexes within the nucleus. At present, we can only speculate about the nuclear function of Yrb1p. We detected the low affinity-formation of complexes containing Yrb1p, Gsp1p-GDP, and Kap95p or Kap104p that could be imported into the nucleus. However, it is unclear whether these complexes are physiologically relevant. Yrb1p could also be involved in nuclear processes different from nucleocytoplasmic transport, such as spindle assembly during mitosis (Ouspenski, 1998; Kalab et al., 1999). It is also possible that RanBP1/Yrb1p has to be present within the NPC to neutralize Gsp1p-GTP, which should inhibit nuclear import in its monomeric form.
ACKNOWLEDGMENTS
We are grateful to Rebecca Heald for her comments on the manuscript. We thank Richard Zimmermann for helpful discussions and Sandra Ruprecht, Ellen Roth and Silke Guthörl for expert technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft to G.S. and the National Institute of Health to K.W. (GM 58065).
Footnotes
Corresponding authors. E-mail addresses: bcgsch@med-rz.uni-sb.de and kweis@uclink4.berkeley.edu.
REFERENCES
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995a;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase activating protein RanGap1. is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995b;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bodendorf U, Cziepluch C, Jauniaux JC, Rommelaere J, Salome N. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J Virol. 1999;73:7769–7779. doi: 10.1128/jvi.73.9.7769-7779.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth JW, Belanger KD, Sannella MI, Davis LI. The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J Biol Chem. 1999;274:32360–32367. doi: 10.1074/jbc.274.45.32360. [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-β2-Ran-GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Müller CW. Structure of importin-β bound to the IBB domain of importin-α. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Cole CN. mRNA export: the long and winding road. Nature Cell Biol. 2000;2:E55–E58. doi: 10.1038/35008681. [DOI] [PubMed] [Google Scholar]
- Deane R, Schäfer W, Zimmermann HP, Müller L, Görlich D, Prehn S, Ponstingl H, Bischoff FR. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-β but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- Ferrigno P, Posas F, Koepp D, Saito H, Silver PA. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–670. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieda M, Tachibana T, Yokoya F, Kose S, Imamoto N, Yoneda Y. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J Cell Biol. 1999;144:645–655. doi: 10.1083/jcb.144.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JK, Silver PA. Cse1p is required for export of Srp1p/importin-α from the nucleus in Saccharomyces cerevisiae. J Biol Chem. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, Kobbe Cv, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kaffman A, O'Shea EK. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Toda T, Iizuka N, Nomoto A. Crm1 (Xpo1) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells. 1998;3:521–532. doi: 10.1046/j.1365-2443.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- Künzler M, Hurt EC. Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- Künzler M, Gerstberger T, Stutz F, Bischoff FR, Hurt E. Yeast Ran-binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (XPO1)-dependent pathway. Mol Cell Biol. 2000;20:4295–4308. doi: 10.1128/mcb.20.12.4295-4308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3471. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury KM, Richards SA, Perlhungher RR, Macara IG. Ran binding domains promote the interaction of Ran with p97/β-karyopherin, linking the docking and translocation steps of nuclear import. J Biol Chem. 1996;271:2357–2360. doi: 10.1074/jbc.271.5.2357. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Saitoh Y, Sazer S, Nishimoto T. Disruption of the YRB2 gene retards nuclear protein export, causing a profound mitotic delay, and can be rescued by overexpression of XPO1/CRM1. J Biochem. 1999;125:574–585. doi: 10.1093/oxfordjournals.jbchem.a022323. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ouspenski II. A RanBP1 mutation which does not visibly affect nuclear import may reveal additional functions of the Ran GTPase system. Exp Cell Res. 1998;244:171–183. doi: 10.1006/excr.1998.4174. [DOI] [PubMed] [Google Scholar]
- Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Lührmann R, Görlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Powers MA, Lund E, Forbes D, Dahlberg JE. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc Natl Acad Sci USA. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum SR, Blobel G. A distinct parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker K, Macara IG. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol Cell Biol. 2000;20:3510–3521. doi: 10.1128/mcb.20.10.3510-3521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Macara IG. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J Biol Chem. 1995;270:14405–14411. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Carey KL, Macara IG. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff FR. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Solsbacher J. Transport between the cytoplasm and the nucleus. Protoplasma. 1999;209:166–172. [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt EC. Mtr10p functions as a nuclear import receptor for the mRNA binding protein Npl3p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996a;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Jensen ON, Podtelejnikov AV, Neubauer G, Shevchenko A, Mortensen P, Mann M. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem Soc Trans. 1996b;24:893–896. doi: 10.1042/bst0240893. [DOI] [PubMed] [Google Scholar]
- Shulga N, James P, Craig EA, Goldfarb DS. A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J Biol Chem. 1999;274:16501–16507. doi: 10.1074/jbc.274.23.16501. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Bischoff FR, Schlenstedt G. Cse1p is involved in export of yeast importin α from the nucleus. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Vogel F, Schlenstedt G. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol Cell Biol. 2000;20:8468–8479. doi: 10.1128/mcb.20.22.8468-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Taura T, Schlenstedt G, Silver PA. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- Taura T, Krebber H, Silver PA. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Arndt A, Kutay U, Görlich D, Wittinghofer A. Structural view of the Ran-importin β interaction at 2.3 Å resolution. Cell. 1999a;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999b;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Yan C, Lee LH, Davis LI. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]