Abstract
There has been a rapid increase in the incidence of diabetes as well the associated vascular complications. Both genetic and environmental factors have been implicated in these pathologies. Increasing evidence suggests that epigenetic factors play a key role in the complex interplay between genes and the environment. Actions of major pathological mediators of diabetes and its complications such as hyperglycaemia, oxidant stress, and inflammatory factors can lead to dysregulated epigenetic mechanisms that affect chromatin structure and gene expression. Furthermore, persistence of this altered state of the epigenome may be the underlying mechanism contributing to a ‘metabolic memory’ that results in chronic inflammation and vascular dysfunction in diabetes even after achieving glycaemic control. Further examination of epigenetic mechanisms by also taking advantage of recently developed next-generation sequencing technologies can provide novel insights into the pathology of diabetes and its complications and lead to the discovery of much needed new drug targets for these diseases. In this review, we highlight the role of epigenetics in diabetes and its vascular complications, and recent technological advances that have significantly accelerated the field.
Keywords: Epigenetics, Diabetes, Vascular complications, MicroRNA, Next-generation sequencing
1. Introduction
Changes in dietary habits and lifestyles associated with rapid economic growth have dramatically increased the incidence of diabetes, obesity, and related vascular complications. This has created a global epidemic of these metabolic disorders and a major health-care concern. Both type 1 diabetes (T1D) and type 2 diabetes (T2D) are associated with hyperglycaemia, oxidant stress and inflammation, and significantly increased risk for macrovascular complications such as atherosclerosis and stroke and microvascular complications such as diabetic nephropathy (DN), neuropathy, and retinopathy.1–6 Several biochemical mechanisms and genetic factors have been implicated in the pathology of diabetes and its complications. However, it is evident that due to the influences of gene–environmental interactions, epigenetic mechanisms such as chromatin histone modifications and DNA methylation regulate at least a part of these pathological mechanisms. Therefore, further understanding of these chromatin-based molecular mechanisms may lead to the development of better therapeutic strategies. In this review, we highlight the role of epigenetic mechanisms in diabetes and its vascular complications, and recent technological advances that have helped accelerate the field.
2. Cellular mechanisms of diabetic vascular complications and metabolic memory
Diabetes and diabetogenic agents such as high glucose (HG), advanced glycation end-products (AGEs), angiotensin II (Ang II), transforming growth factor-β (TGF-β), and oxidized lipids have adverse effects in major target cells involved in vascular dysfunction including endothelial cells (ECs), vascular smooth muscle cells (VSMCs), monocytes, and renal mesangial cells (MCs).2–4,7–12 Several signal transduction mechanisms including oxidant stress, activation of receptor for AGEs (RAGE), protein kinase C (PKC), tyrosine kinases, mitogen-activated protein kinases (MAPKs), and transcription factor nuclear factor-κB (NF-κB) have been implicated in these events.4,6,12–15 However, evidence shows that current therapies based on these mechanisms are not fully efficacious in preventing complications, suggesting the need for the identification of novel therapeutic targets. In particular, it has been noted that some individuals with diabetes experience a continued progression of vascular complications even after glycaemic control subsequent to a period of prior hyperglycaemic exposure, a phenomenon termed ‘metabolic memory'. This has been demonstrated in multiple clinical trials such as the Diabetes Control and Complications Trial (DCCT) and follow-up Epidemiology of Diabetic Complications and Interventions Trial (EDIC) in T1D subjects, and later in other clinical trials with T2D patients. These studies demonstrated that intensive glycaemic control could reduce the progression of diabetic complications but could not prevent them, lower fasting blood glucose levels at the time of diagnosis correlated with delayed vascular complications, and early intervention to control hyperglycaemia could lead to better outcomes for macrovascular complications.16–21 Furthermore, episodes of post-prandial hyperglycaemia may also increase the risk for vascular complications.22
Experimental models in cell culture and animals have revealed a similar metabolic memory phenomenon. Earlier studies showed that retinal complications persisted even after the reversal of hyperglycaemia in dogs.23 Islet transplantation in diabetic rats, 6 weeks after the onset of diabetes, led to reduced rates of retinopathy, but islet transplantation 12 weeks post-diabetes could not prevent retinopathy.24 Studies with streptozotocin (STZ)-injected T1D rats showed that the reinstitution of glycaemic control after a short period of hyperglycaemia had protective effects in the eyes, including reduction in parameters of oxidant stress and inflammation. However, reinstitution of glycaemic control after prolonged diabetes failed to show similar protection.25,26
ECs cultured in HG displayed a sustained increase in the expression of key extracellular and pro-fibrotic genes27 and persistently increased oxidant stress,28 despite subsequent glucose normalization. Furthermore, transient exposure of ECs to HG for 16 h resulted in sustained increases in the expression of NF-κB p65, oxidant stress, and inflammatory genes up to 6 days after glucose normalization.29,30 In another model, VSMCs cultured from the arteries of obese T2D db/db mice exhibited a sustained increase in inflammatory gene expression, NF-κB activation, migration, oxidant stress, and adhesion to monocytes relative to VSMCs cultured from non-diabetic genetic control db/+ mice.15,31,32
Thus, both in vivo and in vitro studies show that the deleterious effects of prior hyperglycaemic exposure have long-lasting effects on target organs even after subsequent glycaemic control underscoring the beneficial effects of intensive glycaemic control in diabetes. They also suggest that the oxidant stress triggered by HG, AGEs, lipids, and other related factors may be a key mediator. Furthermore, increasing evidence now supports epigenetic mechanisms as important components in metabolic memory and the pathology of diabetic complications.
3. Epigenetics and gene transcription
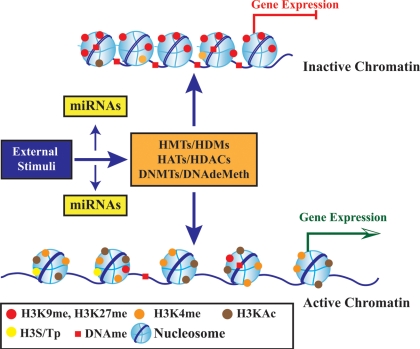
Chromatin function plays a critical role in the regulation of transcription in mammalian cells. Dynamic switching between ‘active' and ‘inactive' states of chromatin in response to extracellular and intrinsic signals is a central mechanism involved in gene regulation33 and is termed as ‘epigenetic mechanism’, because it does not alter the underlying DNA sequence. Chromatin is a highly organized structure composed of chromosomal DNA arranged into repeating units of nucleosome core particles in the nucleus. Nucleosomes are made up of 147 bp DNA wrapped around a histone octamer unit consisting of dimers of core histone proteins H2A, H2B, H3, and H4, held together by an H1 linker.34 DNA methylation is one of the most stable epigenetic modifications and traditionally regarded as the major mediator of epigenetic regulation. However, this view has been modified now to include post-translational modifications (PTMs) of nucleosomal histones and the more recently discovered small non-coding RNAs or microRNAs (miRNAs) and large intergenic non-coding RNAs as additional epigenetic components.35,36 Epigenetic mechanisms regulate both short-term (non-heritable) and long-term (heritable) effects and thus have a significant impact on diverse biological processes.37 Epigenetic changes genome-wide are now collectively referred to as the ‘Epigenome’ and intense research efforts using state-of-the-art ultra-high-throughput profiling technologies have yielded unprecedented insights into the mammalian epigenome (Figure 1).35,38,39
Figure 1.
Schematic diagram showing the role of epigenetic mechanisms in transcription regulation. Chromatin consists of nucleosomes which are made up of DNA wrapped around histone octamers containing dimers of core histone H2A, H2B, H3, and H4. Histone modifications such as H3 lysine-9 methylation (H3K9me) or H3 lysine-27 methylation (H3K27me) are generally repressive marks, whereas H3 lysine-9/14 acetylation (H3KAc) and H3 lysine-4 methylation (H3K4me) are generally activation marks. H3Kme is regulated by histone methyltransferases/demethylases (HMTs/HDMs) and H3KAc is regulated by HATs/HDACs. Histone modifications coupled with DNA methylation at CpG islands by DNMTs/DNA demethylases (DNAdeMeth) determine the active or inactive state of chromatin leading to gene expression or repression, respectively. This dynamic state of the chromatin is subject to alteration by external stimuli via the regulation of epigenetic machinery and miRNAs leading to gene expression and pathophysiological phenotypes.
DNA methylation is exerted by DNA methyltransferases (DNMTs) at the 5′ position of cytosine residues in CpG dinucleotides by transferring methyl groups from S-adenosyl methionine. Hypermethylation of promoter CpG islands generally results in transcription repression.40 DNA methylation in somatic cells was once considered to be generally irreversible, but recent studies have demonstrated evidence for DNA demethylation by both passive and active mechanisms. However, the mechanism of DNA demethylation and the identity of DNA demethylases require further verification.41 DNA methylation is important in many cellular processes including the silencing of repetitive elements, X-inactivation, imprinting, and development. Long-term silencing of tumour suppressors genes via hypermethylation of promoter CpG islands is a well-established mechanism in the development of cancer.35,40
Chromatin histone PTMs such as lysine acetylation, and methylation of lysine and arginine are the other well-studied epigenetic modifications. They act in concert with other PTMs including phosphorylation, ubiquitination, and sumoylation to fine tune gene expression by controlling chromatin access to transcription factors to the cognate cis-elements at promoter and enhancer regions.33,42–44 Combinatorial actions of these modifications form a ‘histone code’ that dictates the ‘repressed’ or ‘active’ states of chromatin.45
Histone H3 lysine acetylation (H3KAc) such as H3K9Ac, H3K14Ac, and H3K27Ac is generally associated with active gene promoters. Histone acetyltransferases (HATs) mediate H3KAc and histone deacetylases (HDACs) remove it.42 Histone H3 arginine methylation (H3Rme) is mediated by protein arginine methyltransferases (PRMT) such as co-activator-associated arginine methyltransferase 1 (CARM1) and generally activates gene expression.42,43 Histone lysine methyltransferases (HMTs) mediate histone lysine methylation (HKme), which can be associated with either active or repressive gene expression depending on the lysine modified. Furthermore, HMTs can mediate mono- (me1), di- (me2), or trimethylation (me3) of specific lysine residues to add an extra layer of regulation.42,44 H3K9me2, -me3 and H3K27me2, -me3 are generally repressive marks, whereas H3K4me is generally an active mark. H3Kme is relatively stable and evidence shows that it can be epigenetically transferred.46 The recent discovery of histone lysine demethylases (HDMs) that remove methylation marks from specific lysine residues demonstrated the dynamic nature of HKme.47 Emerging evidence shows an important regulatory role of HMTs and HDMs in diverse physiological processes and disease conditions.36,47 Interestingly, some of these enzymes can also modify non-histone proteins including p53 and NF-κB,43,48 further re-enforcing their growing importance in cellular processes.
In addition to genetic pre-disposition, environmental factors and nutrient states may alter epigenetic states to play critical roles in normal development as well as susceptibility to diseases including diabetes and related complications.20,36,49–51 Diabetes can further exacerbate the effects of environmental and other risk factors to accelerate vascular complications. Increased understanding of chronic effects of the diabetic milieu on the dynamic regulation of the epigenome could offer mechanistic clues to the metabolic memory phenomenon and provide unique opportunities to develop novel therapeutic approaches.
4. DNA methylation: relation to diabetes and vascular complications
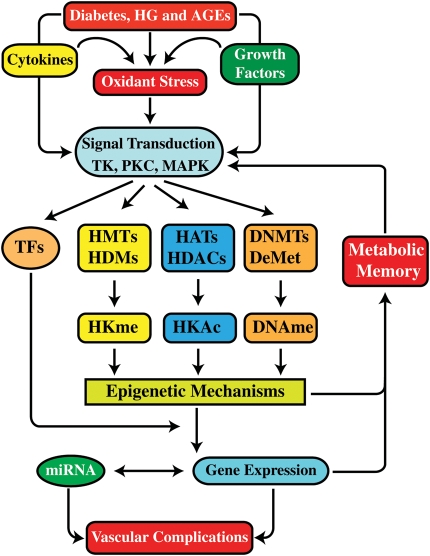
Only limited studies have examined the role of DNA methylation in the pathogenesis of diabetes and vascular diseases. Previous studies showed that the regulation of Agouti gene expression by DNA methylation plays an important role in the development of obesity and diabetes in mice.52 Studies using the intrauterine growth retardation model demonstrated that the islet dysfunction and development of diabetes in rats is associated with epigenetic silencing via promoter DNA methylation of Pdx1, a key transcription factor that regulates β-cell differentiation and insulin gene expression.53 Peroxisome proliferator-activated receptor-γ co-activator 1α (PGC-1α) regulates insulin production in pancreatic β-cells. Studies with T2D animals showed that DNA hypermethylation at the promoter of its gene PPARGC1A reduces PGC-1α expression and inhibits insulin production.54 Interestingly, PPARGC1A promoter was also hypermethylated in skeletal muscles from T2D patients, but at non-CpG nucleotides. DNA methylation in myotubes was induced by TNF-α and free fatty acid palmitate in a DNMT3B-dependent manner, whereas both insulin and glucose had no effect.55 Studies in renal cells exposed to HG and in renal tissues from STZ-induced T1D rats did not show significant differences in DNA methylation at candidate gene promoters.56,57 However, significant differential DNA methylation was observed at 19 gene promoters in genomic DNA from T1D patients with DN compared with patients without DN.58 Notably, one of the hypermethylated genes was UNC13B, which was linked with DN in genetic studies.58 These findings suggest a link between DNA methylation, diabetes, and its complications (Figure 2).
Figure 2.
Schematic diagram showing the role of epigenetic mechanisms in diabetes and metabolic memory implicated in accelerated vascular complications. Diabetes and the associated hyperglycaemia (HG) and AGEs can lead to production of pro-inflammatory mediators such as cytokines and growth factors. Together, they activate multiple signal transduction pathways including oxidant stress, tyrosine kinases (TK), PKC, and MAPKs leading to activation of transcription factors (TFs) such as NF-κB, and dysregulation of epigenetic mechanisms including HKme, histone lysine acetylation (HKAc), and DNA methylation (DNAme) via the action of corresponding methyltransferases, demethylases, acetylases, and deacetylases. In addition, miRNAs can further fine tune the expression of key players involved in these pathways. The net outcome of these events is the loss of repressive chromatin marks and gain of activation marks, leading to the formation of open chromatin state at the promoters of pathological genes allowing increased access to transcription factors. Persistence of this altered state of the epigenome through unknown mechanisms can lead to ‘metabolic memory’ linked with chronic inflammation and other cellular defects associated with micro- and macrovascular complications. HMTs, histone methyltransferases; HDMs, histone demethylases; HATs, histone acetyltransferases; HDACs, histone deacetylases; DNMTs, DNA methyltransferases; DeMet, DNA demethylases.
The role of DNA methylation in the pathogenesis of cardiovascular diseases (CVDs) is not completely understood. Atherosclerosis was associated with global hypomethylation in SMCs of atherosclerotic lesions from humans, and animal models such as high-fat diet-fed ApoE null mice and balloon-injured rabbits.59–61 Furthermore, altered DNA methylation of several candidate genes linked with atherosclerosis was identified in both VSMCs and ECs, and in mouse models. These include hypoxia-inducible factor-1α, c-fos, p53 and oestrogen receptor, growth factors, arachidonic acid-metabolizing enzymes (15-lipoxygenase), vasodilator endothelial nitric oxide synthase, and matrix metalloproteinases.62,63 Alterations in genomic DNA methylation were also demonstrated in leucocytes derived from ApoE null mice preceding the development of atherosclerosis.64 Other CVD risk factors such as hyperhomocysteinaemia, hypercholesterolaemia, and inflammation have also been implicated in DNA methylation changes associated with atherosclerosis.64–66 Altered global DNA methylation was noted in peripheral blood monocytes of patients with increased risk for CVDs.67 Risk for CVDs and diabetes increases with age, and ageing is associated with hypomethylation of genomic DNA.68 Interestingly, the formation of AGEs increases over age and this process is accelerated in diabetes. AGEs and their receptor RAGE have been implicated in inducing oxidant stress, inflammation, and vascular complications.7,9,14 However, whether AGEs can alter DNA methylation profiles in diabetes is still unclear. Thus, several risk factors associated with diabetes and CVDs could potentially alter DNA methylation. With the availability of improved technologies for surveying the DNA methylome, it is likely that important information will soon become available from several clinical cohorts.
5. Histone PTMs: relation to diabetes and vascular complications
The role of histone PTMs has been extensively studied in cancer.35,36 However, much less is known in diabetes and its complications.20,21 Histone PTMs have been implicated in β-cell function and insulin production. Under HG conditions, islet-specific transcription factor Pdx1 was shown to stimulate insulin expression by recruiting co-activators p300 and the HMT SET7/9, which increased histone acetylation and H3K4me2, respectively, and the formation of open chromatin at the insulin promoter. In contrast, under low-glucose conditions, Pdx1 could recruit co-repressors HDAC1/2, leading to inhibition of insulin gene expression. Furthermore, Pdx1 also mediated β-cell-specific expression of SET7/9, which could regulate genes involved in glucose-induced insulin secretion.69,70 Evidence also showed the regulatory roles of H3K27me3, H3K4me3, and Polycomb group of proteins such as Bmi-1, the H3K27me3 transferase Ezh2, its demethylase JMJD3, and the H3K4me3 transferase MLL in the expression of tumour suppressor p16INK4a in β-cell proliferation and regeneration.71 Histone PTMs (H3K4me2 and H3K9me2), H3K4 demethylase lysine-specific demethylase 1 (LSD1), and an H3K9me2 methyltransferase SET domain bifurcated 1 (SETDB1) were implicated in adipogenesis.72 Interestingly, mice deficient in the H3K9me2 demethylase Jhdm2a (JMJD1a) developed obesity and hyperlipidaemia.73 Several transcription regulators involved in islet differentiation are also regulated by acetylation, suggesting a role for HATs/HDACs in diabetes.74 SIRT1, a member of Sirtuin family with HDAC activity, has been shown to modulate energy metabolism and inflammation. SIRT1 overexpression or activation by resveratrol could improve insulin resistance and SIRT1 activators are being developed for diabetes treatment.75 However, the role of other HDACs and the potential use of HDAC inhibitors in diabetes are not very clear.74,75
Inflammation plays a key role in diabetes and its vascular complications. The role of NF-κB in mediating inflammatory gene expression is well established.76 Diabetic conditions can promote inflammatory gene expression via NF-κB activation and enhance monocyte binding to ECs and VSMCs, and subsequent monocyte to macrophage differentiation.15,31,77–83 A number of studies have now explored epigenetic mechanisms in inflammatory gene expression in vascular cells and monocytes. Gene induction by pro-inflammatory agents was associated with increased histone lysine acetylation in ECs and VSMCs, whereas inhibition of HDACs increased inflammatory gene expression.84–88 Increased inflammatory gene expression required collaboration of transcription factors such as NF-κB and cAMP response element-binding protein with HATs including p300/CBP, steroid receptor co-activator-1, and pCAF.84–89 Oxidized LDL-induced chemokine expression was associated with H3KAc and phosphorylation, and recruitment of HATs along with NF-κB in ECs, and these were reversed by pre-treatment with statins.90 Recent studies showed alterations in histone modification patterns, along with changes in expression of the corresponding HMTs, in VSMCs and ECs from aortas of adult mice exposed to hypercholesterolaemia.91 Studies in monocytes showed that H3K9/14Ac and HATs CBP/p300, H3R17me and its methyltransferase CARM1, play key roles in inflammatory gene expression.92,93 HDACs also played key roles in lipopolysaccharide (LPS)-induced inflammatory gene expression in monocytes and macrophages. However, HDAC inhibitors surprisingly suppressed a subset of inflammatory genes but increased pro-atherogenic genes, suggesting that further studies are needed to clarify their role in atherosclerosis and inflammation.94,95 H3Kme also plays a key role in inflammatory gene expression in monocytes. Levels of the repressive mark H3K9me3 were reduced at early time points but restored to control levels at later time periods in monocytes stimulated with pro-inflammatory agents, suggesting a role of H3K9me3 in negative feedback mechanisms associated with inducible inflammatory gene expression.96 Similar results were obtained in VSMCs.32 LPS-induced inflammatory gene expression in macrophages was associated with reduced H3K27me3 and increased expression of an H3K27me3 demethylase JMJD3, which was suggested to fine tune LPS-induced responses.97,98 TNF-α-induced expression of a subset of inflammatory genes in monocytes increased promoter H3K4me and occupancy of SET7/9.99 Thus, epigenetic mechanisms can regulate inflammatory gene expression and CVD susceptibility even under non-diabetic states. These mechanisms may be further accentuated by diabetic conditions and contribute to accelerated complications and metabolic memory (Figure 2).
Inflammatory gene expression induced by diabetic stimuli like HG and a RAGE ligand S100B was associated with increased H3K9/14Ac along with increased recruitment of NF-κB and HATs CBP/p300 at inflammatory gene promoters in THP-1 monocytes. In vivo relevance was demonstrated by noting increased histone lysine acetylation at these promoters in monocytes obtained from T1D and T2D patients.92 Elevated inflammatory gene expression was associated with increased H3K4me and SET7/9 recruitment in TNF-α treated monocytes and in macrophages from T1D mice.99 Furthermore, genome-wide location studies using chromatin immunoprecipitation (ChIP) coupled with microarray analysis (ChIP-on-chip) revealed significant changes in H3K4me2 and H3K9me2 patterns at key gene regions in HG-treated THP-1 monocytes, with relevant changes being observed in primary monocytes from diabetes patients.100 In addition, ChIP-on-chip profiling in blood lymphocytes from T1D patients vs. controls demonstrated significantly increased H3K9me2 levels at a subset of genes, analysis of which linked them to key autoimmune and inflammatory pathways often associated with the development of T1D and its complications.101 These genome-wide studies reveal the utility of epigenomic approaches and suggest that while a reasonably stable methylation pattern is maintained in normal individuals in a cell-type-specific manner and this pattern can be disrupted under disease states.
6. Epigenetic mechanisms in metabolic memory
Recent studies using cell culture and animal models mimicking metabolic memory have identified the role of epigenetic histone modifications such as H3KMe in vascular cells. ChIP assays using cultured db/db VSMCs that exhibit enhanced pro-inflammatory responses demonstrated significantly reduced levels of the repressive H3K9me3 mark at inflammatory gene promoters relative to db/+ even after several passages in vitro.32 Furthermore, db/db VSMCs were hyper-responsive to pro-inflammatory stimuli such as TNF-α with increased inflammatory gene expression, persistently reduced levels of repressive H3K9me3, and occupancy of the H3K9me3 methyltransferase Suv39h1 at inflammatory gene promoters, suggesting defective feedback or repressive mechanisms in db/db cells. Diabetic db/db VSMCs also exhibited reduced levels of Suv39h1, and reconstitution of Suv39h1 reversed the pro-inflammatory phenotype of db/db cells. Human VSMCs treated with HG also exhibited elevated inflammatory genes and reduced H3K9me3 at their promoters.32 In another model using ECs exposed to HG, metabolic memory was associated with enhanced p65 (NF-κB) expression and increased H3K4me1 and SET7/9 HMT recruitment at the p65 promoter, with oxidant stress and reactive dicarbonyls such as methylglyoxal being implicated.29 Additional studies also revealed persistently reduced H3K9me2 and H3K9me3 levels and increased recruitment of LSD1 at the p65 promoter in ECs exposed to HG.30 In a rat model of diabetic retinopathy mimicking metabolic memory, poor glucose control for prolonged periods followed by euglycaemia could not prevent progression of retinopathy or histone acetylation in these retinas, suggesting a role for epigenetic mechanisms in metabolic memory of microvascular complications.102 Taken together, these findings suggest that diabetic stimuli can trigger changes in the chromatin that can have long-lasting effects on the expression of target genes and support a key role for epigenetic histone PTMs in metabolic memory. It is likely that various other histone PTMs, DNA methylation, and related chromatin factors could also be identified as effectors of diabetes-induced vascular complications (Figure 2). Since diabetes is a multifactorial disease involving other factors besides hyperglycaemia, the net outcome may be from their combined effects in several target cells and these still remain to be identified.
Reversal of epigenetic mechanisms or ‘epigenetic therapy’ can be effective in pathological conditions as shown in the treatment of certain cancers.103 An anti-inflammatory agent and HAT inhibitor curcumin ameliorated HG-induced inflammatory gene expression and histone acetylation at their promoters as well as changes in HAT and HDAC activities in human monocytes.104 There has been much interest in the development of HDAC or HMT inhibitors for CVDs and diabetic complications.49,74,75 Given the well-known roles of oxidant stress, growth factors, AGEs, PKC, and extracellular matrix proteins in diabetic complications, it would be worth determining whether inhibitors of these factors can also interfere with downstream epigenetic mechanisms. Recent studies demonstrated that in MCs, epigenetic HKme and recruitment of HMT SET7 play key roles in HG- and TGF-β-mediated fibrotic gene expression and these effects were significantly blocked by a TGF-β neutralizing antibody.105 Overall, these studies suggest that approaches aimed at reversal of epigenetic mechanisms could be beneficial in ameliorating diabetes-associated complications.
7. miRNAs in vascular complications
The 22-nucleotide small non-coding miRNAs play important roles in diverse biological processes and disease conditions such as cancer, diabetes, DN, cardiogenesis, angiogenesis, and vascular development by post-transcriptional mechanisms.106–112 Interaction of mature miRNAs with specific binding sites in the 3′ untranslated regions of target mRNAs in the RNA-induced silencing complex leads to either mRNA degradation or inhibition of translation.106 Each miRNA can regulate multiple targets including signal transduction components, transcription and epigenetic factors, and provide another level of epigenetic mechanism to fine tune gene regulation in response to environmental stimuli.106 Evidence shows that miRNAs can affect the function of both ECs and VSMCs relevant to vascular diseases.109–112 miRNAs are implicated in phenotypic switching, proliferation, migration, and neointimal thickening in VSMCs, as well as capillary formation, migration, senescence, expression of adhesion molecules, and angiogenic growth and transcription factors in ECs.109,113–117 In monocytes and macrophages, miRNAs regulate inflammation, response to oxidized lipids, oxidative stress, immune function, cholesterol homeostasis, and differentiation.109,118–122
Several studies have implicated miRNAs in diabetes pathogenesis.110,123–126 However, the role of miRNAs in diabetes vascular complications is less studied. RAGE ligands increased pro-inflammatory gene COX-2 by post-transcriptional mechanisms via down-regulation of miR-16 in THP-1 monocytes.127 HG increased miR-1/miR-206 via SRF and MEFK1/2 and promoted apoptosis in cardiomyocytes.128 miRNAs have been implicated in the regulation of key renal genes related to DN. miR-192 and miR-216a were shown to regulate collagen 1 (α2) (Col1a2) and E-cadherin gene expression in renal cells.108,129–132 Furthermore, miR-216a and miR-217 were up-regulated in glomeruli from diabetic mice, and by TGF-β in MCs and could increase hypertrophy.130 miR-93 was down-regulated by HG leading to up-regulation of its target VEGF-A in DN models.133
Recent studies showed that miR-125b levels were up-regulated in cultured diabetic db/db VSMCs. Furthermore, miR-125b could down-regulate Suv39h1 protein, increase inflammatory gene expression, reduce H3K9me3 at their promoters, and enhance VSMC–monocyte binding.134 These results present evidence of miRNA-based mechanisms in epigenetic regulation of inflammatory gene expression in diabetes and potential contribution to metabolic memory of vascular complications. Overall, it is clear that miRNAs may contribute to multiple complications of diabetes and can therefore be evaluated as potential therapeutic targets.
8. Emerging technologies for epigenomics research
Candidate gene-based approaches have provided evidence of epigenetic mechanisms in diabetes and vascular complications. However, the full impact of epigenome alterations in disease development can be better understood by taking advantage of genome-wide approaches such as ChIP-on-chip and massively parallel next-generation sequencing (NGS) methods.135,136 NGS technologies have revolutionized the efforts to map the human genome and epigenome by allowing sequencing of millions of short DNA fragments in a single run and providing large volumes of accurate sequence data in a more robust fashion. In these methods, DNA fragments are directly sequenced and counts of sequence reads used as a direct quantitative measure of the chromatin modification or gene expression.135 Several platforms are available that use proprietary technologies to generate a library of short DNA fragments linked with adapters and capture them on to beads or cross-link to solid supports or deposit them into single wells, where DNA sequencing is performed.136 The commonly used platforms include Illumina's Genome Analyzers, Roche's 454 sequencing system, Applied Biosystem's SOLiDTM, Helicos Biosciences tSMSTM, and Pacific Biosciences SMRTR.136 These NGS technologies are now used in all aspects of genome-wide profiling including gene expression (RNA-Seq), miRNA sequencing, transcription factor binding and histone PTMs (ChIP-Seq), and DNA methylation, thus providing an integrated genomic platform.135–141 Traditionally, DNA methylation is studied by methods involving bisulfite conversion of genomic DNA, digestion with methylation sensitive restriction enzymes, and also affinity-based methods to enrich methylated DNA using antibodies to methyl-binding proteins or to methylcytosine in assays such as methylated-CpG island recovery assay (MIRA) or methylated DNA immunoprecipitation (Me-DIP).142–144 For genome-wide profiling, DNA obtained from these methods is hybridized to DNA microarrays142 or sequenced by NGS approaches. Such NGS methods helped reveal cell-type-specific differences in DNA methylomes between embryonic stem cells, pluripotent stem cells, and fully differentiated cells.136,145,146 DNA methylation profiling revealed 19 CpG sites associated with the risk of DN in T1D patients.58 Methods to analyse genome-wide DNA methylation patterns in clinical samples are being developed.147 There is no doubt that these emerging technologies will significantly increase our knowledge of DNA methylation in disease states.
Histone PTMs and transcription factor binding in vivo have been studied using ChIP technique in which formaldehyde cross-linked DNA–protein complexes are immunoprecipitated with specific antibodies against histone PTMs, transcription factors, or other chromatin factors of interest. Immunoprecipitated DNA is amplified using genomic DNA primers specific to promoters of genes being examined.148,149 Utility of ChIP assays has been augmented by ChIP-on-chip technique in which ChIP-enriched DNA is hybridized to various microarrays including high-resolution promoter tiling arrays or whole-genome arrays.140,149,150 These approaches have increased our understanding of genome-wide histone PTMs at regulatory regions, enhancers, cell-specific differences, and bivalent promoters in stem cells.38,151–154 ChIP-on-chip was also used to identify diabetes and HG-specific histone PTM signatures in monocytes and lymphocytes.100,101 ChIP-linked to NGS (ChIP-Seq) has now significantly improved the scope of genome-wide location studies of histone PTMs.139–141 ChIP-Seq was used to profile several different histone PTMs in T cells and characterize the histone PTM patterns specific to enhancers, miRNA promoters, and chromatin states of pluripotent and lineage-committed cells.155–158
The major challenge of NGS technologies lies in the analyses of the large volumes of data generated, complex bioinformatics, gene ontology, and data analysis tools required to reach biologically meaningful conclusions. Several software tools are currently available and many more are constantly emerging. Integration of data obtained from multiple techniques such as RNA-Seq, DNA-methylation, and ChIP-Seq will yield a wealth of information to address several unanswered questions such as how interactions between various components of the epigenome, including histone PTMs, DNA methylation, and miRNAs can lead to gene activation or repression under pathophysiological conditions.141 These integrative approaches will be highly useful to identify novel epigenetic and transcriptional regulation mechanisms across several tissues and cells relevant to diabetes and vascular complications. Another potentially useful application of NGS could be in personal genomics and epigenomics to catalogue variations between individuals and in different disease states to provide improved and customized therapies, including combinations of conventional drugs with those targeting epigenetic factors. The Human Epigenome Project is expected to yield key information in this connection.36,38,159 Technological breakthroughs will permit large-scale epigenomic analyses of various disease states and offer opportunities to develop novel therapies to combat diabetes and associated vascular complications.
Conflict of interest: none declared.
Funding
The authors are supported by grants from the National Institutes of Health (NIDDK and NHLBI), the Juvenile Diabetes Research Foundation, and the American Diabetes Association.
References
- 1.Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- 2.He Z, King GL. Microvascular complications of diabetes. Endocrinol Metab Clin North Am. 2004;33:215–238. doi: 10.1016/j.ecl.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Ziyadeh FN, Sharma K. Overview: combating diabetic nephropathy. J Am Soc Nephrol. 2003;14:1355–1357. doi: 10.1097/01.asn.0000065608.37756.58. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan R, Nadler JL. Lipid inflammatory mediators in diabetic vascular disease. Arterioscler Thromb Vasc Biol. 2004;24:1542–1548. doi: 10.1161/01.ATV.0000133606.69732.4c. [DOI] [PubMed] [Google Scholar]
- 5.King GL. The role of inflammatory cytokines in diabetes and its complications. J Periodontol. 2008;79:1527–1534. doi: 10.1902/jop.2008.080246. [DOI] [PubMed] [Google Scholar]
- 6.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 7.Yan SF, Ramasamy R, Schmidt AM. Mechanisms of disease: advanced glycation end-products and their receptor in inflammation and diabetes complications. Nat Clin Pract. 2008;4:285–293. doi: 10.1038/ncpendmet0786. [DOI] [PubMed] [Google Scholar]
- 8.Marrero MB, Fulton D, Stepp D, Stern DM. Angiotensin II-induced signaling pathways in diabetes. Curr Diabetes Rev. 2005;1:197–202. doi: 10.2174/1573399054022802. [DOI] [PubMed] [Google Scholar]
- 9.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 10.Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-beta as a key mediator. Diabetes. 1995;44:1139–1146. doi: 10.2337/diab.44.10.1139. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan R, Cai Q. Monocyte retention in the pathology of atherosclerosis. Future Cardiol. 2005;1:331–340. doi: 10.1517/14796678.1.3.331. [DOI] [PubMed] [Google Scholar]
- 12.Averill MM, Bornfeldt KE. Lipids versus glucose in inflammation and the pathogenesis of macrovascular disease in diabetes. Curr Diab Rep. 2009;9:18–25. doi: 10.1007/s11892-009-0005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geraldes P, King GL. Activation of protein kinase C isoforms and its impact on diabetic complications. Circ Res. 2010;106:1319–1331. doi: 10.1161/CIRCRESAHA.110.217117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AM, Yan SD, Wautier JL, Stern D. Activation of receptor for advanced glycation end products: a mechanism for chronic vascular dysfunction in diabetic vasculopathy and atherosclerosis. Circ Res. 1999;84:489–497. doi: 10.1161/01.res.84.5.489. [DOI] [PubMed] [Google Scholar]
- 15.Reddy MA, Li SL, Sahar S, Kim YS, Xu ZG, Lanting L, et al. Key role of Src kinase in S100B-induced activation of the receptor for advanced glycation end products in vascular smooth muscle cells. J Biol Chem. 2006;281:13685–13693. doi: 10.1074/jbc.M511425200. [DOI] [PubMed] [Google Scholar]
- 16.Writing Team DCCT/EDIC Research Group. Effect of intensive therapy on the microvascular complications of type 1 diabetes mellitus. JAMA. 2002;287:2563–2569. doi: 10.1001/jama.287.19.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colagiuri S, Cull CA, Holman RR. Are lower fasting plasma glucose levels at diagnosis of type 2 diabetes associated with improved outcomes? U.K. prospective diabetes study 61. Diabetes Care. 2002;25:1410–1417. doi: 10.2337/diacare.25.8.1410. [DOI] [PubMed] [Google Scholar]
- 18.Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med. 2005;353:2643–2653. doi: 10.1056/NEJMoa052187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceriello A. Hypothesis: the ‘metabolic memory', the new challenge of diabetes. Diabetes Res Clin Pract. 2009;86(Suppl. 1):S2–S6. doi: 10.1016/S0168-8227(09)70002-6. [DOI] [PubMed] [Google Scholar]
- 20.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol. 2010;299:F14–F25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol. 2010;6:665–675. doi: 10.1038/nrendo.2010.188. [DOI] [PubMed] [Google Scholar]
- 22.Ceriello A, Esposito K, Piconi L, Ihnat MA, Thorpe JE, Testa R, et al. Oscillating glucose is more deleterious to endothelial function and oxidative stress than mean glucose in normal and type 2 diabetic patients. Diabetes. 2008;57:1349–1354. doi: 10.2337/db08-0063. [DOI] [PubMed] [Google Scholar]
- 23.Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycemic control. Diabetes. 1987;36:808–812. doi: 10.2337/diab.36.7.808. [DOI] [PubMed] [Google Scholar]
- 24.Hammes HP, Klinzing I, Wiegand S, Bretzel RG, Cohen AM, Federlin K. Islet transplantation inhibits diabetic retinopathy in the sucrose-fed diabetic Cohen rat. Invest Ophthalmol Vis Sci. 1993;34:2092–2096. [PubMed] [Google Scholar]
- 25.Kowluru RA. Effect of reinstitution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes. 2003;52:818–823. doi: 10.2337/diabetes.52.3.818. [DOI] [PubMed] [Google Scholar]
- 26.Chan PS, Kanwar M, Kowluru RA. Resistance of retinal inflammatory mediators to suppress after reinstitution of good glycemic control: novel mechanism for metabolic memory. J Diabetes Complications. 2010;24:55–63. doi: 10.1016/j.jdiacomp.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy S, Sala R, Cagliero E, Lorenzi M. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci USA. 1990;87:404–408. doi: 10.1073/pnas.87.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ihnat MA, Thorpe JE, Kamat CD, Szabo C, Green DE, Warnke LA, et al. Reactive oxygen species mediate a cellular ‘memory' of high glucose stress signalling. Diabetologia. 2007;50:1523–1531. doi: 10.1007/s00125-007-0684-2. [DOI] [PubMed] [Google Scholar]
- 29.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li SL, Reddy MA, Cai Q, Meng L, Yuan H, Lanting L, et al. Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes. 2006;55:2611–2619. doi: 10.2337/db06-0164. [DOI] [PubMed] [Google Scholar]
- 32.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 35.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 37.Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 39.Maunakea AK, Chepelev I, Zhao K, Bruneau B. Epigenome mapping in normal and disease States. Circ Res. 2010;107:327–339. doi: 10.1161/CIRCRESAHA.110.222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 41.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- 44.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 46.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 48.Pradhan S, Chin HG, Esteve PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pons D, de Vries FR, van den Elsen PJ, Heijmans BT, Quax PH, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30:266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 50.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinney SE, Simmons RA. Epigenetic mechanisms in the development of type 2 diabetes. Trends Endocrinol Metab. 2010;21:223–229. doi: 10.1016/j.tem.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 53.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling C, Del Guerra S, Lupi R, Ronn T, Granhall C, Luthman H, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Brennan EP, Ehrich M, Brazil DP, Crean JK, Murphy M, Sadlier DM, et al. DNA methylation profiling in cell models of diabetic nephropathy. Epigenetics. 2010;5:396–401. doi: 10.4161/epi.5.5.12077. [DOI] [PubMed] [Google Scholar]
- 57.Williams KT, Garrow TA, Schalinske KL. Type I diabetes leads to tissue-specific DNA hypomethylation in male rats. J Nutr. 2008;138:2064–2069. doi: 10.3945/jn.108.094144. [DOI] [PubMed] [Google Scholar]
- 58.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33–42. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hiltunen MO, Turunen MP, Hakkinen TP, Rutanen J, Hedman M, Makinen K, et al. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 60.Laukkanen MO, Mannermaa S, Hiltunen MO, Aittomaki S, Airenne K, Janne J, et al. Local hypomethylation in atherosclerosis found in rabbit ec-sod gene. Arterioscler Thromb Vasc Biol. 1999;19:2171–2178. doi: 10.1161/01.atv.19.9.2171. [DOI] [PubMed] [Google Scholar]
- 61.Hiltunen MO, Yla-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arterioscler Thromb Vasc Biol. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 62.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 63.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 64.Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–29154. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 65.Zaina S, Lindholm MW, Lund G. Nutrition and aberrant DNA methylation patterns in atherosclerosis: more than just hyperhomocysteinemia? J Nutr. 2005;135:5–8. doi: 10.1093/jn/135.1.5. [DOI] [PubMed] [Google Scholar]
- 66.Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 67.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 69.Chakrabarti SK, Francis J, Ziesmann SM, Garmey JC, Mirmira RG. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic beta cells. J Biol Chem. 2003;278:23617–23623. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- 70.Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–193. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Parrizas M. Histone demethylase LSD1 regulates adipogenesis. J Biol Chem. 2010;285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray SG, De Meyts P. Role of histone and transcription factor acetylation in diabetes pathogenesis. Diabetes Metab Res Rev. 2005;21:416–433. doi: 10.1002/dmrr.559. [DOI] [PubMed] [Google Scholar]
- 75.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 77.Yerneni KK, Bai W, Khan BV, Medford RM, Natarajan R. Hyperglycemia-induced activation of nuclear transcription factor kappaB in vascular smooth muscle cells. Diabetes. 1999;48:855–864. doi: 10.2337/diabetes.48.4.855. [DOI] [PubMed] [Google Scholar]
- 78.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 79.Guha M, Bai W, Nadler JL, Natarajan R. Molecular mechanisms of tumor necrosis factor alpha gene expression in monocytic cells via hyperglycemia-induced oxidant stress-dependent and -independent pathways. J Biol Chem. 2000;275:17728–17739. doi: 10.1074/jbc.275.23.17728. [DOI] [PubMed] [Google Scholar]
- 80.Hatley ME, Srinivasan S, Reilly KB, Bolick DT, Hedrick CC. Increased production of 12/15 lipoxygenase eicosanoids accelerates monocyte/endothelial interactions in diabetic db/db mice. J Biol Chem. 2003;278:25369–25375. doi: 10.1074/jbc.M301175200. [DOI] [PubMed] [Google Scholar]
- 81.Shanmugam N, Reddy MA, Guha M, Natarajan R. High glucose-induced expression of proinflammatory cytokine and chemokine genes in monocytic cells. Diabetes. 2003;52:1256–1264. doi: 10.2337/diabetes.52.5.1256. [DOI] [PubMed] [Google Scholar]
- 82.Meng L, Park J, Cai Q, Lanting L, Reddy MA, Natarajan R. Diabetic conditions promote binding of monocytes to vascular smooth muscle cells and their subsequent differentiation. Am J Physiol Heart Circ Physiol. 2009;298:H736–H745. doi: 10.1152/ajpheart.00935.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 84.Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 85.Edelstein LC, Pan A, Collins T. Chromatin modification and the endothelial-specific activation of the E-selectin gene. J Biol Chem. 2005;280:11192–11202. doi: 10.1074/jbc.M412997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sahar S, Reddy MA, Wong C, Meng L, Wang M, Natarajan R. Cooperation of SRC-1 and p300 with NF-kappaB and CREB in angiotensin II-induced IL-6 expression in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2007;27:1528–1534. doi: 10.1161/ATVBAHA.107.145862. [DOI] [PubMed] [Google Scholar]
- 87.Reddy MA, Sahar S, Villeneuve LM, Lanting L, Natarajan R. Role of Src tyrosine kinase in the atherogenic effects of the 12/15-lipoxygenase pathway in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2009;29:387–393. doi: 10.1161/ATVBAHA.108.179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fish JE, Matouk CC, Rachlis A, Lin S, Tai SC, D'Abreo C, et al. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem. 2005;280:24824–24838. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 89.Rossig L, Li H, Fisslthaler B, Urbich C, Fleming I, Forstermann U, et al. Inhibitors of histone deacetylation downregulate the expression of endothelial nitric oxide synthase and compromise endothelial cell function in vasorelaxation and angiogenesis. Circ Res. 2002;91:837–844. doi: 10.1161/01.res.0000037983.07158.b1. [DOI] [PubMed] [Google Scholar]
- 90.Dje N'Guessan P, Riediger F, Vardarova K, Scharf S, Eitel J, Opitz B, et al. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:380–386. doi: 10.1161/ATVBAHA.108.178319. [DOI] [PubMed] [Google Scholar]
- 91.Alkemade FE, van Vliet P, Henneman P, van Dijk KW, Hierck BP, van Munsteren JC, et al. Prenatal exposure to apoE deficiency and postnatal hypercholesterolemia are associated with altered cell-specific lysine methyltransferase and histone methylation patterns in the vasculature. Am J Pathol. 2010;176:542–548. doi: 10.2353/ajpath.2010.090031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 93.Miao F, Li S, Chavez V, Lanting L, Natarajan R. Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol Endocrinol. 2006;20:1562–1573. doi: 10.1210/me.2005-0365. [DOI] [PubMed] [Google Scholar]
- 94.Bode KA, Schroder K, Hume DA, Ravasi T, Heeg K, Sweet MJ, et al. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122:596–606. doi: 10.1111/j.1365-2567.2007.02678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Halili MA, Andrews MR, Labzin LI, Schroder K, Matthias G, Cao C, et al. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–1114. doi: 10.1189/jlb.0509363. [DOI] [PubMed] [Google Scholar]
- 96.Saccani S, Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002;16:2219–2224. doi: 10.1101/gad.232502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 98.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem. 2007;282:13854–13863. doi: 10.1074/jbc.M609446200. [DOI] [PubMed] [Google Scholar]
- 101.Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 2008;57:3189–3198. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110:1306–1313. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 104.Yun JM, Jialal I, Devaraj S. Epigenetic regulation of high glucose-induced proinflammatory cytokine production in monocytes by curcumin. J Nutr Biochem. 2010 doi: 10.1016/j.jnutbio.2010.03.014. ; doi:10.1016/j.jnutbio.2010.03.014. Published online ahead of print 21 July 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010;21:2069–2080. doi: 10.1681/ASN.2010060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kato M, Natarajan R. microRNA cascade in diabetic kidney disease: big impact initiated by a small RNA. Cell Cycle. 2009;8:3613–3614. doi: 10.4161/cc.8.22.9816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 110.Muhonen P, Holthofer H. Epigenetic and microRNA-mediated regulation in diabetes. Nephrol Dial Transplant. 2009;24:1088–1096. doi: 10.1093/ndt/gfn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 120.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 121.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 122.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 124.Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mi QS, He HZ, Dong Z, Isales C, Zhou L. microRNA deficiency in pancreatic islet cells exacerbates streptozotocin-induced murine autoimmune diabetes. Cell Cycle. 2010;9:3127–3129. doi: 10.4161/cc.9.15.12596. [DOI] [PubMed] [Google Scholar]
- 126.Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2:9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J Biol Chem. 2008;283:36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, et al. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010;584:3592–3600. doi: 10.1016/j.febslet.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 129.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi J, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang B, Herman-Edelstein M, Koh P, Burns W, Jandeleit-Dahm K, Watson A, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794–1802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kato M, Wang L, Putta S, Wang M, Yuan H, Sun G, et al. Posttranscriptional upregulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem. 2010;285:34004–34015. doi: 10.1074/jbc.M110.165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice leads to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59:2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ansorge WJ. Next-generation DNA sequencing techniques. Nat Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 136.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 137.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 138.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 143.Rauch TA, Pfeifer GP. The MIRA method for DNA methylation analysis. Methods Mol Biol. 2009;507:65–75. doi: 10.1007/978-1-59745-522-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sorensen AL, Collas P. Immunoprecipitation of methylated DNA. Methods Mol Biol. 2009;567:249–262. doi: 10.1007/978-1-60327-414-2_16. [DOI] [PubMed] [Google Scholar]
- 145.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Turner FB, Cheung WL, Cheung P. Chromatin immunoprecipitation assay for mammalian tissues. Methods Mol Biol. 2006;325:261–272. doi: 10.1385/1-59745-005-7:261. [DOI] [PubMed] [Google Scholar]
- 149.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 150.Huebert DJ, Kamal M, O'Donovan A, Bernstein BE. Genome-wide analysis of histone modifications by ChIP-on-chip. Methods. 2006;40:365–369. doi: 10.1016/j.ymeth.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 151.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miao F, Wu X, Zhang L, Riggs AD, Natarajan R. Histone methylation patterns are cell-type specific in human monocytes and lymphocytes and well maintained at core genes. J Immunol. 2008;180:2264–2269. doi: 10.4049/jimmunol.180.4.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 155.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 156.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Human Epigenome Task Force. Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]