Abstract
Genome-wide studies have revealed that mammalian genomes are pervasively transcribed. This has led to the identification and isolation of novel classes of non-coding RNAs (ncRNAs) that influence gene expression by a variety of mechanisms. Here we review the characteristics and functions of regulatory ncRNAs in chromatin remodelling and at multiple levels of transcriptional and post-transcriptional regulation. We also describe the potential roles of ncRNAs in vascular biology and in mediating epigenetic modifications that might play roles in cardiovascular disease susceptibility. The emerging recognition of the diverse functions of ncRNAs in regulation of gene expression suggests that they may represent new targets for therapeutic intervention.
Keywords: Non-coding RNA, ncRNA, Vascular biology, Epigenetics, Gene regulation
1. Introduction
Recent high-throughput transcriptomic analyses have revealed that eukaryotic genomes transcribe up to 90 % of the genomic DNA.1 Only 1–2% of these transcripts encode for proteins, whereas the vast majority are transcribed as non-coding RNAs (ncRNAs). Evolutionarily, the repertoire of protein-coding genes has remained relatively static, whereas the amount of non-coding sequences has markedly increased along with the complexity of the organism.2 A biological basis for this observation is supported by the growing evidence of the functionality of these transcripts. The fact that most putative ncRNAs are expressed at substantially lower levels than mRNAs further suggests that these RNAs mainly fulfil regulatory functions. Indeed, there is increasing evidence for regulatory roles of ncRNAs during development3–5 and in response to stress and environmental stimuli.6–9 A major goal of contemporary molecular biology is to identify and functionally characterize the full spectrum of ncRNAs with respect to normal physiological functions and roles in human diseases.
ncRNAs can be divided into infrastructural ncRNAs and regulatory ncRNAs. Constitutively expressed infrastructural ncRNAs include ribosomal, transfer, small nuclear, and small nucleolar RNAs. Regulatory ncRNAs can be classified into microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), and long non-coding RNAs (lncRNAs).10 In addition, a novel class of promoter-associated RNAs (PARs) and enhancer RNAs (eRNAs) has been recently described.8,9,11 In this review, we will focus on highlighting the characteristics and biological roles of the regulatory RNAs, with particular emphasis on their roles in vascular biology. The potential therapeutic applications of regulating ncRNA expression will also be discussed.
2. Types of ncRNAs and their biological roles
2.1. MicroRNAs
MiRNAs are evolutionarily conserved, small single-stranded molecules (20–24 nucleotides), that have been postulated to regulate the expression of ∼50% of the genes in a cell at the post-transcriptional level.12 In contrast to other endogenous small RNAs, miRNAs derive from transcripts forming distinctive hairpin structures (Table 1). Processing of the hairpin into the mature miRNA by Drosha and Dicer13,14 allows interaction with Argonaute (Ago) proteins to form RNA-induced silencing complex (RISC).15,16 Strand selection for RISC is dictated by the thermodynamic stabilities of the two duplex ends: the strand having its 5′ terminus at the less stably base-paired end of the duplex is favoured.12 The miRNAs then pair with mRNAs, most favourably to the 3′ untranslated region (UTR), to guide their translational repression or deadenylation and degradation (Table 1).13,15,17,18 Challenging this view, a recent report by Guo et al.19 suggested that destabilization of the target mRNA is the predominant reason (≥84 %) for reduced protein levels by endogenous miRNAs. In addition to its classical roles, miRNAs have also been shown to regulate gene expression through promoter targeting and translational activation.12,20,21 Interestingly, the former is likely to involve epigenetic mechanisms.21
Table 1.
Regulatory ncRNAs produced from eukaryotic genomes and their characteristics and functions
| Type | Long name | Length (nt) | Characteristics | Function |
|---|---|---|---|---|
| miRNA | Micro RNA | 20–24 | Pri-miRNA produced in the nucleus as capped and polyadenylated ssRNA with a imperfectly paired stem-loop structure | Perfect complementarity: Ago2-mediated cleavage of mRNA |
| Processing by Drosha and Dicer lead to a production of mature dsRNA with exact ends | Non-perfect complementarity: Suppression of translation or mRNA degradation (deadenylation, decapping, and exonucleocytic degradation) | |||
| Effector phase occurs primarily in the cytoplasm mediated by Ago proteins | Minor functions in transcriptional silencing and translational activation | |||
| piRNA | PIWI-interacting RNA | 24–31 | Precursor ssRNA, which is modified to contain 3′-terminal 2′-O-methyl | Silencing of transposable elements in the germline |
| Strong preference for uridine at the 5′ end | ||||
| siRNA | Small interfering RNA | 20–24 | Canonical form long, linear, perfectly base-paired dsRNA | Perfect match: endonucleocytic cleavage |
| Processed by Dicer into mature siRNA with heterogenous end composition | Non-perfect match or endonuclease-inactive RISC: translational repression or exonucleocytic degradation | |||
| Effector functions occur primarily in the cytoplasm supported by Ago proteins | Induction of heterochromatin formation | |||
| Silencing of the same locus from which they are derived | ||||
| PAR (aPASR, TSSa-RNA, tiRNA, PROMPT) | Promoter-associated RNA | 16–200 | Weakly expressed ssRNAs | Partly unknown but indications of transcriptional regulation (example interaction with Polycomb group of proteins) |
| Short half-life | ||||
| Bidirectional expression reflecting PolII distribution | ||||
| eRNA | Enhancer RNA | 100–9000 | ssRNA produced bidirectionally from enhancer regions enriched for H3K4me1, PolII and coactivators such as p300 | Mostly unknown but plays a role in transcriptional gene activation |
| Short half-life | ||||
| Evolutionarily conserved sequences | ||||
| Dynamically regulated upon signalling | ||||
| Expression correlates positively with nearby mRNA expression | ||||
| lncRNA | Long non-coding RNA | >200 | Precursor ssRNA | Chromatin remodelling |
| Many lncRNAs are subject to splicing, polyadenylation, and other post-transcriptional modifications | Transcriptional regulation | |||
| Mostly nuclear RNAs but a subset also located in the cytoplasm | Post-transcriptional regulation (splicing, TF localization) | |||
| Not evolutionary conserved with the exception of large intergenic ncRNAs, lincRNAs (H3K4me3-H3K36me3 signature) | Precursors for siRNAs | |||
| Component of nuclear organelles (paraspeckles, nuclear speckles) |
aPASR, promoter-associated small RNA; TSSa-RNA, transcription start site-associated RNA; tiRNA, transcription initiation RNA; PROMTs, promoter upstream transcript.
The first observations establishing the significance of miRNAs in the regulation of vascular biology came from experimental studies disrupting the function of Dicer and Drosha in miRNA biogenesis.22–24 Dicer-deficient mice died early during development due to defects in blood vessel formation.22 Similarly, knockdown of Dicer and Drosha in vitro results in reduction in endothelial cell migration, capillary sprouting, and tube formation.23,24 These studies paved the way for an explosion of new studies exploring the roles of individual miRNAs in angiogenesis and in the pathogenesis of vascular diseases. Indeed, numerous miRNAs have been shown to participate in angiogenic processes, making them interesting therapeutic tools. In this respect, miRNAs can be divided into two groups: pro-angiogenic miRNAs or anti-angiogenic miRNAs. Pro-angiogenic miRNAs include miR-17–92,25 miR-27b, Let-7,23 miR-126,26,27 miR-130a,28 miR-210,29 miR-378,30 and miR-296,31 whereas anti-angiogenic miRNAs include miR-15b, miR-16,32 miR-221/222,33 miR-328,34 miR-92a,35 and miR-214.36 Deregulation of miRNA expression is also implicated in many vascular diseases. For example, the roles of miRNA-1,37,38 -23,39 -133,40 and -20841 in cardiac hypertrophy, miR-142 and -32843 in arrhythmia, miRNA-2944 and -13345 in cardiac fibrosis, and miR-1, -21,46 and miR-15 family47 in cardiac ischaemia have been established.
2.2. Piwi-interacting RNAs
PiRNAs are small ncRNAs of 24–31 nt in size named for their ability to form complexes with Piwi proteins of the Argonaute family.48 PiRNAs have a 2′-O-methyl modification on the nucleotide at the 3′ end and usually a uridine at the 5′ end (Table 1).48 PiRNAs were first discovered in Drosophila as repeat-associated siRNAs (rasiRNA), which show complementarity to a variety of transposable and repetitive elements.49 The primary role of these small RNAs has been shown to be suppression of transposon activity during germ line development.50,51 Single-stranded precursors give rise to antisense (AS) piRNAs, which then recognize and target the cleavage of transposons by associated PIWI-proteins. This generates additional sense piRNAs arising from the target transposon sequence. This ‘ping-pong’ cycle goes on to increase the abundance of piRNAs and transposon silencing.50,51
Unlike Drosophila piRNAs, more that 90% of mammalian piRNAs map uniquely in the genome and cluster to a small number of loci.52–54 However, transposon control also occurs in mammals during spermatogenesis through de novo DNA methylation.55 PiRNAs have been mostly uncovered in the germline but growing evidence suggests that their defensive function extends into somatic cells.56,57 Supporting this, a recent study proposed a role for piRNAs in the regulation of the cell cycle of mesenchymal stem cells.58
2.3. Small interfering RNAs
The canonical siRNA is a linear, perfectly base-paired dsRNA, which is processed by Dicer into 20–24 nt siRNAs that direct silencing when loaded onto RISC. They mediate post-transcriptional silencing similar to miRNA silencing. Compared with miRNAs, guide strand recognition is indistinguishable, but it is still unclear if all siRNA sequences are capable of effectively guiding all RNA silencing functions.59 In addition to post-transcriptional gene silencing (PTGS), siRNAs have also been found to direct sequence-specific transcriptional gene silencing by increasing epigenetic marks characteristic of heterochromatin (Table 1).59,60
SiRNAs were first observed during transgene-induced silencing in petunia61 followed by studies in Caenorhabditis elegans.62,63 Initially, RNA interference (RNAi) was considered to be a natural defence mechanism that used exogenous siRNAs to protect organisms from viruses, but it soon became evident that endogenous siRNAs (endo-siRNAs) also play a role in regulating genome functions. In this respect, transposons and repetitive elements were first discovered as the source of endo-siRNAs, suggesting that they may play a similar role as piRNAs in suppressing transposon activity.64,65
Another group of endo-siRNAs consists of natural AS transcripts (NATs). NATs can be divided into different categories based on their orientation to the protein-coding gene: head-to-head (overlapping 5′ ends), tail-to-tail (overlapping 3′ ends) or fully overlapping.66 An interesting subgroup of NATs is composed of AS-transcribed pseudogenes.67,68 Approximately half of all mammalian protein families include pseudogenes, with greatest enrichment found in ribosomal and housekeeping families of genes.69 A high degree of pseudogenization is also exhibited by genes of the SH3_1 (Src homology 3), homeobox, Gp_dh_N/C (glyceraldehyde 3-phosphate dehydrogenase, NAD-binding domain/C-terminal domain), and collagen families implicated in vascular development, homeostasis, and disease.69
2.4. Long ncRNAs
The majority of the non-protein-coding transcripts belong to the group of lncRNAs, which are arbitrarily considered as >200 nt in length (Table 1).10 However, many of these lncRNAs can also act as primary transcripts for the production of short RNAs, making the categorization of this group of ncRNAs ambiguous. Most lncRNAs are characterized by nuclear localization, low expression, low level of sequence conservation and are composed of both poly A + and poly A− transcripts.70,71 LncRNAs can be classified according to their proximity to protein coding genes placing them into five categories: sense, AS, bidirectional, intronic, and intergenic.10
Recently, a subgroup of lncRNAs, named large intergenic non-coding RNAs (lincRNAs), was described based on distinctive chromatin signature that marks actively transcribed genes.72,73 LincRNAs are marked by trimethylation of lysine 4 of histone H3 (H3K4me3) at their promoter and trimethylation of lysine 36 of histone H3 (H3K36me3) along the transcribed region. In contrast to most lncRNAs, lincRNAs exhibit a high conservation between different species. LincRNAs have been suggested to guide chromatin-modifying complexes to specific genomic loci and this way participate in the establishment of cell type-specific epigenetic states.72,73 The most well-described examples are involved in epigenetic gene silencing, exemplified by the role of X-inactive specific transcript Xist in X-chromosome inactivation and H19 or Air in genomic imprinting.10 The H19 gene encodes a 2.3-kb ncRNA, which is highly expressed during embryogenesis but shut off in most tissues after birth.74 Environmental factor, such as maternal undernutrition, has been shown to regulate the expression of H19 in a sex-specific manner; maternal low-protein diet was shown to cause abnormalities in male but not female mice blastocysts.75 Interestingly, this mechanism could potentially contribute to the different susceptibility of cardiovascular diseases between male and female.76 Apart from embryogenesis, it has been shown to play roles in tumour development by promoting the expression of genes involved in metastasis and angiogenesis.74
The majority of lncRNAs are transcribed as complex networks of overlapping sense and AS transcripts with respect to protein-coding loci.77 In humans, 61% of transcribed regions show evidence of AS transcription suggesting a role for AS ncRNAs in transcriptional regulation of the overlapping mRNA.78 Already a decade ago, it was shown that mRNA stability of hypoxia inducible factor alpha (HIF-1α), a physiological regulator of angiogenesis, is modulated by an AS HIF (aHIF) transcript complementary to the HIF-1α 3′UTR.79,80 Prolonged hypoxia or aHIF overexpression was found to trigger the decay of the HIF-1α mRNA.81 Interestingly, HIF also upregulates aHIF expression through a hypoxia response element present in the promoter region of aHIF thus generating a negative feedback loop.81
Subsequent studies have also discovered an AS mRNA to endothelial nitric-oxide synthase (eNOS), termed sONE, which participates in the regulation of endothelial cell-specific gene expression.82 Moreover, sONE mediates the post-transcriptional down-regulation of eNOS during hypoxia.83 As the down-regulation of eNOS may play a role in the aetiology of vascular diseases such as pulmonary arterial hypertension, ways to interfere with this interaction could bear potential for therapeutic purposes.
Recently, an AS RNA produced from the tyrosine kinase with immunoglobulin-like and EGF-like domains 1 (tie-1) locus was also identified as a transcriptional repressor with potential implications in the control of vascular development.84 The tie-1 AS was shown to form a duplex with tie-1 mRNA leading to down-regulation of gene expression. Overexpression of tie-1 AS lncRNAs resulted in defects in endothelial cell junctions and tube formation. Moreover, the levels of tie-1 AS were found to be 5–10-fold higher in human vascular anomaly samples compared with normal tissue suggesting a role in the aetiology of vascular disease. Whether AS mechanisms regulate a broader class of genes with endothelial-restricted pattern of expression forms an interesting area of future research.
2.5. Enhancer RNAs
Another class of ncRNA that has received much recent attention is found expressed at enhancer regions. The size of eRNAs has been shown to range from 0.1 to 9 kB, with an average size of 800 nt.9,11 This situates most of the eRNAs to the category of lncRNAs but owing to their specific histone methylation signature typical of enhancers, they are discussed separately (Table 1).
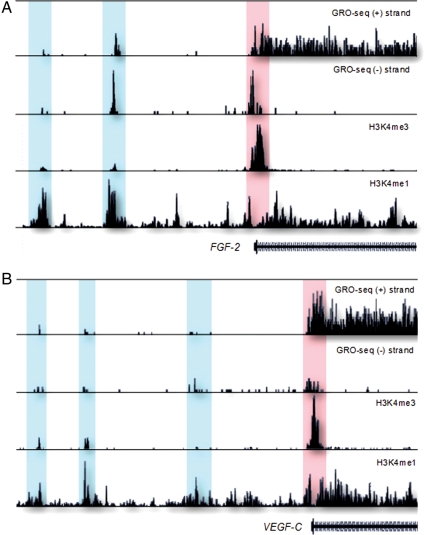
High-throughput sequencing studies of RNA and immune-precipitated chromatin (ChIP) have defined the following characteristics of eRNA transcripts (Table 1). (i) eRNAs are produced from regions defined by high enrichment of monomethylation on lysine 4 of histone 3 (H3K4me1) and low enrichment of H3K4 trimethylation (H3K4me3).8,9 (ii) These regions are enriched for RNA polymerase II (PolII) and transcriptional co-regulators, such as the p300 co-activator. (iii) Transcription of eRNAs initiates from PolII-binding sites and elongates bidirectionally. (iv) DNA sequences encoding eRNAs are evolutionarily conserved. (v) Enhancer-associated RNA transcripts have a short half-life. (vi) RNA transcripts are dynamically regulated upon signalling, and (vii) are positively correlated to levels of nearby mRNA expression.8,9 These latter characteristics have been described in the neuronal cell response to membrane depolarization and in macrophage response to lipopolysaccharide and γ-interferon.8,9 A recent global run-on sequencing (GRO-Seq)85 in human fibroblasts data reveals that eRNAs are also a prominent feature of vascular genes exemplified by fibroblast growth factor 2 (FGF-2) and vascular endothelial growth factor C (VEGF-C) (Figure 1).
Figure 1.
Potential promoter-associated RNAs and enhancer RNAs produced upstream of (A) fibroblast growth factor 2 (FGF-2) and (B) vascular endothelial growth factor C (VEGF-C) genes in human IMR90 cells.85 The promoter-associated RNAs (highlighted in red) colocalize with H3K4me3 histone mark, whereas the eRNAs (highlighted in blue) are revealed by their overlap with H3K4me1 mark.145 Regions upstream of transcription start site (TSS) for FGF-2 and VEGF-C are 50 and 100 kb, respectively. The y-axis indicates the number of sequencing tags.
Although still speculative, several lines of evidence suggest a functional role for eRNAs as transcriptional activators. Reports of enhancer-transcribed RNAs date back to as early as 1992, when Tuan et al.86,87 discovered RNA transcripts at the locus control region (LCR) of the beta-globin locus. The LCR is defined by four erythroid-specific DNAseI hypersensitive sites (HS1–4) and temporally regulates globin genes within the cluster throughout development. But how does the LCR, transcribed 10–50 kb upstream of its target genes, control transcription of the globin gene cluster? In a transient reporter assay, levels of eRNA were decreased by insertion of a transcriptional terminator downstream of the HS2 enhancer. This lead to reduced reporter gene activity driven by the epsilon-globin promoter.88 In another line of experiments, ChIP studies demonstrated PolII recruitment to both HS2 enhancer and adult beta-globin promoter.89 When elongation of PolII was inhibited pharmacologically, PolII enrichment decreased at the beta-globin promoter, but not at the HS2 enhancer.89 This suggests that the recruitment of PolII to promoter, but not to the enhancer, is dependent on RNA synthesis. One may speculate that RNA synthesis from the enhancer is required for promoter PolII recruitment. A recent report further provided evidence of eRNA's functionality by targeting non-coding transcripts using RNAi.11 Depleting eRNA led to a gene-specific decrease in mRNA expression and DNA segments encoding eRNA were sufficient to induce transient reporter activity. Moreover, RNA-mediated enhancer activity appears to be sequence-specific; while keeping the transcription start site (TSS) intact, substituting eRNA with other open reading frames led to decreased enhancer activity.11 This indicated that transcription at the enhancer alone is insufficient for enhancer activity. Collectively, these studies propose a possible transcriptional activation role for eRNA.
Many questions arise as more functional evidence for eRNAs emerge. Kim et al.8 demonstrated that the absence of an intact promoter abolishes eRNA transcription. What is the relationship between promoter and enhancers? Are the low-abundance eRNAs transcriptional noise or are they byproducts of the moving PolII along intervening DNA from enhancers to promoter?90 Do eRNAs maintain an open chromatin state and modulate promoter and enhancer interactions? Do eRNAs serve as platforms for RNA-binding transcription factors and participate in the establishment of cell-type-specific enhancer signature?91,92 Addressing these functional and mechanistic questions should lead to an improved understanding of the role of enhancers in the control of gene expression in different cell systems.
2.6. Promoter-associated RNAs
Similar to the discovery of eRNAs, various genome tiling and high-throughput sequencing methods have unveiled the diverse class of ncRNA linked at promoters. These RNAs can be classified based on their size—ranging from small RNA species of 16–36 to 200 nt (Table 1).70,93–95 Longer >200 nt RNAs have also been described, but it is unclear whether these are precursors of shorter ncRNAs.70,93 They can also be characterized by their location; some are expressed near TSSs, whereas others are expressed from upstream elements of the promoter.96 Furthermore, these RNAs are found expressed in sense and divergent orientation with respect to the TSS. Most of these RNAs are associated with highly expressed genes, while themselves being weakly expressed and exhibiting short half-lives. In fact some of these transcripts were discovered when the RNA degradation machinery was either depleted or functionally deficient.96,97
Increasing number of studies are beginning to connect PARs with transcriptional activation and repression.93,98–100 Studying this group of ncRNAs presents a big challenge since conventional tools like DNA deletion or mutation may interrupt regulatory elements or alternative TSS. Attempts have been made to understand functions of PARs by modulating their levels in cells. Transfection of synthetic RNAs designed to target promoter regions of E-cadherin, vascular endothelial growth factor A (VEGF-A) and p21 increased expression of these genes in human, non-human primates, and rodent cells.101–103 More commonly, however, promoter-targeted siRNAs lead to the repression of the downstream genes, empasizing emerging roles of PARs in transcription.98,103 PARs are also a general feature of vascular genes as significant colocalization of GRO-Seq tags and the hallmark of active promoters H3K4me3 is found for these genes as illustrated for FGF-2 and VEGF-C (Figure 1).
There is a growing body of work unravelling the mechanisms by which PARs participate in the transcriptional regulation. For example, most target genes of the repressive Polycomb group (PcG) protein complex exhibit low levels of the repressive histone mark histone 3 trimethyl lysine 27 (H3K27me3), while being associated with histone marks for transcriptional initiation such as RNA Pol II and H3K4me3. These characteristics suggest that the PcG target gene promoters have adopted a poised state that allows their rapid induction upon cellular responses. Interestingly, short RNAs of 50–200 nt in length originate from the promoters of PcG target genes in primary T cells and embryonic stem cells. Components of the PcG complex bind to stem loop structures of these RNAs and mediate transcriptional repression in cis. These short RNAs are lost upon activation, offering a model in which dissociation of PcG is in the sequence of rapid induction of poised genes.104
3. Mechanisms mediating transcriptional regulation and epigenetics
3.1. Chromatin remodelling
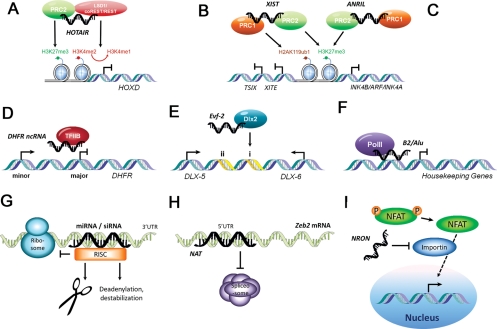
Understanding how ncRNAs regulate transcription has become an area of intense research. LncRNAs have been proposed to regulate transcription by recruiting chromatin-remodelling complexes, which in turn mediate epigenetic changes.10 Epigenetics refers to heritable changes in phenotype and gene expression caused by mechanisms other than the changes in DNA sequences. The repressive PcG is one of the most well-described transcriptional complexes that initiate and maintain epigenetic changes. PcG is characterized as two multiprotein complexes—polycomb repressive complex 1 (PRC1) and 2 (PRC2).105 Components of PRC2 trimethylate H3K27, establishing the silent chromatin state. Components of PRC1 bind H3K27me3 and ubiquitinate lysine 119 on histone 2A. Interestingly, components of PRC1 and PRC2 are also RNA-binding proteins.4,106–108 Locus-specific silencing mediated by PcG might thus be guided by bound lincRNAs. A classic example for this model is X-chromosome inactivation—PcG binds to ncRNA XIST expressed on the targeted X-chromosome and initiates epigenetic silencing by trimethylation of H3K27 in cis (Figure 2B). PcG also mediates transcriptional repression through interaction with histone deacetylases and exerts long-lasting silencing by CpG methylation through interaction with DNA methyltransferase 3 alpha.109,110
Figure 2.
Mechanisms for regulation of epigenetics and gene expression by non-coding RNAs. NcRNAs can function as modulators of epigenetics through (A through C) chromatin remodelling or regulate gene expression at (D through F) transcriptional or (G through I) post-transcriptional level. (A) A 5′ domain of HOTAIR binds polycomb repressive complex 2 (PRC2), whereas a 3′ domain of HOTAIR binds the LSD1/CoREST/REST complex. This allows HOTAIR to coordinate histone H3 lysine 27 methylation and lysine 4 demethylation at the HOXD locus in trans. (B) In cis recruitment of PRC2 by Xist antisense RNA and appearance of H3K27me3 along the inactive X chromosome are among the earliest events in X inactivation. Recruitment of PRC1-mediated H2AK119ub1 parallels the recruitment of PRC2. (C) Similarly, antisense non-coding RNA ANRIL represses the expression from INK4b/ARF/INK4a locus by recruiting and retaining PRC1 and PRC2 complexes in cis. (D) LncRNA transcribed from the minor promoter of dihydrofolate reductase (DHFR) froms a triplex together with the transcription factor TFIIB and the major promoter leading to the dissociation of the preinitiation complex. (E) Enhancer region (i and ii) of Dlx5/6 generates an lncRNA Evf-2 which forms a complex with homeodomain protein Dlx-2 to activate transcription. (F) Transcription of B2 and Alu RNAs is induced upon heat-shock. They inhibit mRNA synthesis by disrupting contacts between RNA polymerase II and promoter DNA. (G) Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are incorporated into RNA-induced silencing complexes (RISCs) that target specific mRNAs for cleavage, translational repression or destabilization depending on the extent of sequence complementarity. (H) Natural antisense transcript (NAT) prevents the binding of the spliceosome to the 5′UTR of the Zeb2 mRNA. This leads to retention in an intron containing internal ribosomal entry site (IRES), which is dispensable for the translation of Zeb2 protein. (I) The nuclear trafficking of nuclear factor of activated T cells (NFAT) is inhibited by the interaction of non-coding repressor of NFAT (NRON) with proteins of the importin-beta superfamily.
Recent studies have further highlighted the mechanistic roles of ncRNAs in PcG-mediated transcriptional regulation. PcG was first described to silence the homeotic (Hox) genes in Drosophila melanogaster during development. HOX antisense intergenic RNA (HOTAIR), a 2.2 kb lincRNA expressed in HOXC cluster, is required for PcG-mediated silencing of the HOXD cluster in trans (Figure 2A).4,111 A recent study demonstrated that the 5′ end of HOTAIR is bound by PRC2, whereas the 3′ end is bound by the LSD1/REST/CoREST complex (Lysine-specific demethylase 1; RE-1 silencing transcription factor; corepressor or REST). LSD1 represses gene expression by removing dimethyl mark from H3K4. In essence, HOTAIR enables tethering of two distinct repressive complexes to chromatin for coupled H3K27 methylation and H3K4 demethylation.112
While HOTAIR represents a model for ncRNA-dependent PcG function, an estimated 20% of lincRNA expressed in human cells are bound by PcG, suggesting that a similar strategy may be more widely applicable.72 Indeed, PcG has been implicated in tumourigenesis by repression of INK4B/ARF/INK4A tumour suppressor genes (inhibitors of CDK4 and CDK6; alternative reading frame of INK4A).113–116 An Antisense Non-coding RNA in the INK4 Locus, or ANRIL, is expressed in this locus and recruits PcG repressive complexes to repress these tumor suppressor genes in cis (Figure 2C).117–119 Polycomb repressive complex 1 component chromobox 7 (CBX7) can bind ANRIL and H3K27me3. Mutation in either binding domains for RNA or H3K27me3 impairs repression of INK4A/ARF/INK4B. Cells harbouring these mutations have reduced proliferative capacity in colony forming assays.119 Intriguingly, ANRIL is the only transcript found to localize at the chromosome region 9q21 associated with cardiovascular disease susceptibility.120 Indeed, a recent study demonstrated that disease-associated genetic variants decrease the efficiency of ANRIL splicing and subsequent production of circular RNA species, thereby influencing PcG-mediated INK4/ARF repression and atherosclerosis susceptibility.121
3.2. Transcriptional regulation
Many lncRNAs have also been demonstrated as negative regulators of transcription. An illustrative example of this is the regulation of dihydrofolate reductase (DHFR).122 The gene encoding DHFR contains a major and a minor promoter, the latter being silenced in quiescent cells. The lncRNAs generated from the minor promoter bind both the major promoter (triplex formation) and the general transcription factor IIB leading to the dissociation of preinitiation complex (Figure 2D).122
Recently, p53 was shown to activate the expression of numerous lincRNAs.123 One such RNA, called lincRNAs-p21 was shown to be essential for the guidance of heterologous nuclear ribonucleoprotein K to the promoters of genes repressed by p53 thus playing an important role in cellular response to apoptotic signals. The exact mechanism by which lincRNA-21 contributes to repression is still unknown but it might act by a similar mechanism as DHFR.123 Interestingly, the specificity of p53-mediated activation of lincRNAs can be further regulated by MEG3 ncRNA, which might modulate the binding of p53 on the promoter of its target genes.124 Knock-out studies in mice have highlighted the important role of MEG3 in the control of vascularization in the brain.125
NcRNAs can also serve as transcriptional coactivators as illustrated by the 3.8 kb polyadenylated Evf2 ncRNA (Figure 2E). Evf2 is transcribed from an ultraconserved region in Dlx5/6 locus, and it forms a complex with the homeodomain-containing protein Dlx2.126 This Dlx2–Evf2 complex functions cooperatively as transcriptional activator of Dlx5/6 expression in an enhancer-specific manner.126 The same group followed up with an in vivo study by generating a mouse model where Evf2 expression is interrupted by insertion of polyadenylation sequences. Interestingly, disruption of Evf2 ncRNA increased Dlx5/6 expression. Reintroduction of Evf2 partially rescued the phenotype; but when a higher amount of Evf2 was reintroduced, expression of Dlx5/6 was further increased as seen in the reporter assay. This result suggests a complex scenario where dosage of Evf2 ncRNA may have different effect on target genes.127
By regulating transcription, ncRNAs can be viewed as sensors of environmental signals, exemplified by two ncRNAs, mouse B2 RNA and human Alu RNA transcribed from short interspersed sequence elements. These ncRNAs have been found to repress mRNA transcription in response to heat shock. They do so by preventing PolII from establishing contacts with the promoter around TATA box during the first step of transcription initiation called closed complex formation (Figure 2F).128 These studies together with other examples of ncRNA transcription factor complexes—like heat-shock RNA-1/heat-shock transcription factor 1,129 ncRNA steroid receptor RNA activator (SRA)/nuclear receptors,130 SRA/master regulator of muscle differentiation MyoD131—illustrate the emerging role of ncRNA in regulating transcriptional responses to external and developmental stimuli through interaction with transcription factors.
3.3. Post-transcriptional regulation
ncRNAs are also implicated in the regulation of post-transcriptional processing, such as splicing, transport, translation, and degradation. The best characterized mechanism is no doubt the PTGS mediated by siRNAs and miRNAs through RNAi pathway (Figure 2G).48 As discussed earlier, both types of RNAs influence the expression of genes by regulating the stability or translation of mRNAs. What separates these two is the fact that siRNAs silence the locus from which they are derived, whereas miRNAs regulate different genes.48
An increasing number of metazoan genes are being found to have naturally occurring AS transcripts.66 AS transcripts overlapping exon–intron boundaries can mask the splicing sites thus enabling alternative splicing. For example, the expression of Zeb2 relies on the splicing of the internal ribosomal entry site (IRES)-containing intron, which is dependent upon the expression of AS transcript (Figure 2H).132 Similar mechanisms have also been described for c-erb2133 and more recently natriuretic peptide precursor.134
Other ncRNAs regulate the transcription by controlling the subcellular localization of transcription factors. One such lncRNA is called non-coding repressor of nuclear factor of activated T cells (NFAT) (NRON), which regulates the nuclear trafficking of NFAT (Figure 2I).135 Upon stimulation, NFAT is dephosphorylated by the calcium-regulated phosphatase calcineurin and localized to the nucleus, where it becomes transcriptionally active. The role of NRON seems to be to prevent the translocation of dephosphorylated NFAT thus modulating its activity. As calcineurin signalling and NFAT activation play a critical role in the development of cardiovascular and skeletal muscle136 and coronary angiogenesis,137 it remains to be seen if this system plays a role in cardiovascular disease processes.
3.4. Other potential mechanisms and perspectives
No study yet delivers a definitive explanation of the role of PARs or the functions of eRNAs.8,9,11,50,64–66,85,94 Are they just a result of spurious transcriptional noise or a result from RNA Pol II molecules failing to elongate? The prevailing view at present suggests a role for these ncRNAs in maintaining the chromatin landscape poised for regulation. Recently, targeting of eRNAs by synthetic siRNAs was shown to decrease the expression of neighbouring protein-coding genes, supporting their functional roles.11 Also, as discussed earlier, transfection of synthetic promoter-associated small RNAs most often leads to a reduction in the expression of the overlapping mRNA promoter.138 Han et al.99 showed that low-copy PARs (extended 5′ UTR) are required for RNA-directed epigenetic gene silencing in human cells. Divergent transcription spanning the promoter could thus be involved in the regulation of mRNA expression. This is supported by another piece of evidence by Morris et al.98 who showed that p21 AS RNA maintains a low level of epigenetic silencing by recruitment of Ago1 and H3K27me3 to the promoter. Subsequently, suppression of AS RNA transcription allows enhanced transcription of the sense/mRNA. According to this model, dysregulation of gene expression in disease conditions could well be due to imbalance in bidirectional transcription. Controlling transcription of the PARs and eRNAs could allow cells to fine tune gene expression, processes which can be foreseen to be harnessed for therapeutic purposes.
In addition, eRNAs could play a structural role in bringing the enhancer areas together with the promoter region by chromatin looping.139 On a genome-wide level, this would suggest a function for eRNAs in maintenance of the three-dimensional conformation of chromosomes by bringing widely separated functional elements into close spatial proximity. Indeed, deep sequencing of chromatin-associated RNAs in human fibroblast cells provided first evidence of the role of ncRNAs in fine-tuning of the chromatin architecture.140 Moreover, recent studies exploiting the latest genomics approach of chromosome conformation capture (3C) demonstrated that sections containing genes co-regulated during the cell cycle and genes containing the same DNA motifs at their promoter regions tend to associate in a statistically significant manner.141 It is tempting to speculate that genes induced by environmental stimuli, also displaying induction of eRNAs9 could associate together through the interplay of ncRNAs and transcriptional co-regulators.
Similarly, we could ask do lncRNAs have master regulatory functions or could their role also be to provide fine-tuning? How do proteins interact with lncRNAs and how does this interaction specify the functional outcome? The non-conserved sequence of lncRNAs could suggest that conserved secondary structure is the key to its functions.10 This is further supported by the fact that many lncRNAs with low sequence similarity are associated with Polycomb proteins.4,118 One can envisage the secondary structure being responsible for interaction with the protein partner but also in the recognition of DNA elements or histone marks. The consequences of lncRNAs binding to a protein partner could then be to modulate its activity, ability to bind other co-regulators, or recognize binding motifs. Illustrative example of such mechanism is the regulation of cyclin D1 (CCND1) by a ncRNA generated from the 5′ regulatory regions of the gene.100 The CCND1 ncRNA is upregulated in response to genotoxic stress, which in turn enables allosteric modulation of the activity of an RNA-binding protein, translocated in liposarcoma (TLS). The modified TLS inhibits the enzymatic activities of CBP/p300 which subsequently represses the CCND1 mRNA expression.
Many questions still remain unanswered but the ever growing evidence strongly points to a central role of ncRNAs in gene regulatory programmes. Shedding light to these intricate and complex roles of ncRNAs will no doubt be a major objective for future investigations.
4. Conclusions
The continual discovery of new regulatory ncRNA species suggests that we are only just beginning to understand their complexity and functions. Nevertheless, it has already become evident that much of their biological and molecular functions are associated with the control of epigenetic pathways, transcription, translation, and turnover.
There is increasing evidence that epigenetic pathways may control vascular endothelial gene expression and modulate cardiovascular disease susceptibility.76 Two recent publications have already described a causative role for epigenetic alterations in the progression of heart failure.142,143 Owing to the extensive roles of ncRNAs in the regulation of gene expression they may well serve as novel diagnostic markers for vascular diseases. For example, the use of miRNAs as biomarkers for cardiovascular disease diagnosis has already been proposed.144 We can expect this to be expanded to the other regulatory ncRNAs when their role in vascular disorders becomes fully established by genome-wide association studies. Furthermore, the potential for therapeutic applications can be imagined. Therapy using small RNAs that target ncRNA transcripts, such as eRNAs or PARs, may represent a new way to treat disease conditions caused by epigenetic changes. The emphasis to fully characterize the mechanism of ncRNA-based gene regulation will no doubt lead into the development of novel therapies for cardiovascular diseases.
Conflict of interest: none declared.
Funding
This work was supported by Leducq Foundation Transatlantic Network of Excellence grant. M.U.K. was supported by Sigrid Jusélius fellowship, Fondation Leducq Career Development award and grants from Academy of Finland, ASLA-Fulbright, Finnish Foundation for Cardiovascular Research, Finnish Cultural Foundation and Orion-Farmos Research Foundation. M.T.Y.L. was supported in part by the UCSD Genetics Training Program through an institutional training grant from the NIH/NIGMS, T32 GM008666, and by the UCSD Medical Scientist Training Program through NIH/NIGMS Training Grant 5 T32 GM007198–37.
References
- 1.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 3.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 16.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 18.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 19.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 23.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 24.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 25.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. [Google Scholar]
- 33.Minami Y, Satoh M, Maesawa C, Takahashi Y, Tabuchi T, Itoh T, et al. Effect of atorvastatin on microRNA 221/222 expression in endothelial progenitor cells obtained from patients with coronary artery disease. Eur J Clin Invest. 2009;39:359–367. doi: 10.1111/j.1365-2362.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, et al. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS ONE. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 36.Chan LS, Yue PY, Mak NK, Wong RN. Role of microRNA-214 in ginsenoside-Rg1-induced angiogenesis. Eur J Pharm Sci. 2009;38:370–377. doi: 10.1016/j.ejps.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 38.Terentyev D, Belevych AE, Terentyeva R, Martin MM, Malana GE, Kuhn DE, et al. miR-1 overexpression enhances Ca(2+) release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56alpha and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514–521. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 41.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 42.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 43.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 44.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. 176p following 178. [DOI] [PubMed] [Google Scholar]
- 46.Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia-reperfusion in mice. FEBS Lett. 2008;582:4137–4142. doi: 10.1016/j.febslet.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 49.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 50.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 51.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 52.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 53.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 54.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 55.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Q, Ma Q, Shehadeh LA, Wilson A, Xia L, Yu H, et al. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem Biophys Res Commun. 2010;396:915–920. doi: 10.1016/j.bbrc.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fire A, Albertson D, Harrison SW, Moerman DG. Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle . Development. 1991;113:503–514. doi: 10.1242/dev.113.2.503. [DOI] [PubMed] [Google Scholar]
- 63.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 66.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 67.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 68.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lam HY, Khurana E, Fang G, Cayting P, Carriero N, Cheung KH, et al. Pseudofam: the pseudogene families database. Nucleic Acids Res. 2009;37:D738–D743. doi: 10.1093/nar/gkn758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 71.Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, et al. Poly A- transcripts expressed in HeLa cells. PLoS One. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 75.Kwong WY, Miller DJ, Ursell E, Wild AE, Wilkins AP, Osmond C, et al. Imprinted gene expression in the rat embryo-fetal axis is altered in response to periconceptional maternal low protein diet. Reproduction. 2006;132:265–277. doi: 10.1530/rep.1.01038. [DOI] [PubMed] [Google Scholar]
- 76.Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol. 2010;7:510–519. doi: 10.1038/nrcardio.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 79.Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/s0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- 80.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 81.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 82.Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279:37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 83.Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, et al. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282:15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 84.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci U S A. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- 89.Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 96.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 97.Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 98.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, et al. RNAa is conserved in mammalian cells. PLoS ONE. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, et al. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism a novel example of epigenetherapy. Circ Res. 2009;105:604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 104.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 106.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 108.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 110.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 111.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jacobs JJL, Kieboom K, Marino S, Depinho RA, Lohuizen MV. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 114.Scott CL, Gil J, Hernando E, Teruya-Feldstein J, Narita M, Martínez D, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci USA. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, Visakorpi T, et al. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- 116.Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 117.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 118.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 121.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 123.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 125.Gordon F, Nutt C, Cheunsuchon P, Nakayama Y, Provencher K, Rice K, et al. Increased expression of angiogenic genes in the brains of mouse MEG3-null embryos. Endocrinology. 2010;151:2443. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 130.Lanz RB, McKenna NJ, Oñate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 131.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 132.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- 134.Annilo T, Kepp K, Laan M. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol. 2009;10:81. doi: 10.1186/1471-2199-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 136.Schulz RA, Yutzey KE. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev Biol. 2004;266:1–16. doi: 10.1016/j.ydbio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 137.Zeini M, Hang CT, Lehrer-Graiwer J, Dao T, Zhou B, Chang CP. Spatial and temporal regulation of coronary vessel formation by calcineurin-NFAT signaling. Development. 2009;136:3335–3345. doi: 10.1242/dev.037903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kartha RV, Subramanian S. MicroRNAs in cardiovascular diseases: biology and potential clinical applications. J Cardiovasc Transl Res. 2010;3:256–270. doi: 10.1007/s12265-010-9172-z. [DOI] [PubMed] [Google Scholar]
- 145.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]