Abstract
Interactions between Cajal bodies (CBs) and replication-dependent histone loci occur more frequently than for other mRNA-encoding genes, but such interactions are not seen with all alleles at a given time. Because CBs contain factors required for transcriptional regulation and 3′ end processing of nonpolyadenylated replication-dependent histone transcripts, we investigated whether interaction with CBs is related to metabolism of these transcripts, known to vary during the cell cycle. Our experiments revealed that a locus containing a cell cycle-independent, replacement histone gene that produces polyadenylated transcripts does not preferentially associate with CBs. Furthermore, modest but significant changes in association levels of CBs with replication-dependent histone loci mimic their cell cycle modulations in transcription and 3′ end processing rates. By simultaneously visualizing replication-dependent histone genes and their nuclear transcripts for the first time, we surprisingly find that the vast majority of loci producing detectable RNA foci do not contact CBs. These studies suggest some link between CB association and unusual features of replication-dependent histone gene expression. However, sustained CB contact is not a requirement for their expression, consistent with our observations of U7 snRNP distributions. The modest correlation to gene expression instead may reflect transient gene signaling or the nucleation of small CBs at gene loci.
INTRODUCTION
The Cajal or coiled body (CB) contains the highest concentration of splicing small nuclear ribonucleoproteins (snRNPs) in the nucleus (Carmo-Fonseca et al., 1992; Huang and Spector, 1992; Matera and Ward, 1993). Pulse-labeling experiments have indicated that CBs do not contain significant levels of DNA or newly synthesized RNA in their interiors, and thus it has been suggested that they probably do not represent sites of RNA synthesis or processing (Monneron and Bernhard, 1969; Callan and Gall, 1991; Raska, 1995; Jordan et al., 1997). However, a very similar structure in amphibian oocytes, the sphere organelle, attaches at its surface to a few specific loci in lampbrush chromosomes, those containing the histone genes (Gall et al., 1981; Callan et al., 1991). Similarly, the CBs of human cells associate specifically and preferentially with histone loci and, in addition, with the U1 and U2 small nuclear RNA (snRNA) loci and the small nucleolar RNA (snoRNA)-encoding U3 gene (Frey and Matera, 1995; Smith et al., 1995; Gao et al., 1997). All of these loci position at the periphery of CBs, not the centers, and although they interact preferentially, none are 100% associated with CBs, suggesting possible transient interactions. A few characteristics set CB-associating genes apart from most other genes that are transcribed by RNA polymerase II. Many are organized in multigene clusters or repeats, and their transcripts all undergo unusual processing (see below). Some evidence suggests a link between U2 expression and CB association levels (Frey et al., 1999), but the functional significance or mechanism for this putative link has yet to be uncovered.
The histone loci that associate with CBs in human cells contain several replication-dependent histone genes that code for the new histones required during S phase (Frey and Matera, 1995). These cell cycle-regulated, intronless histone genes usually produce transcripts with 3′ end stem loops instead of polyA tails (reviewed in Osley, 1991; Dominski and Marzluff, 1999). In contrast, there are replacement histone gene variants that are constitutively expressed and produce polyadenylated mRNA (Wu and Bonner, 1981; Brush et al., 1985; Wells and Kedes, 1985). Whether these have a relationship with CBs has not been previously investigated. The rates of both transcription and 3′ end processing of the replication-dependent genes peak during S phase (reviewed in Osley, 1991; Marzluff, 1992). Two factors required for 3′ end processing of the replication-dependent transcripts, stem-loop binding protein (SLBP) and U7 snRNP, are present in CBs, with U7 being particularly enriched in these structures (Wu and Gall, 1993; Frey and Matera, 1995; Wang et al., 1996c; Abbott et al., 1999). In addition, an S phase-specific colocalization of replication-dependent histone loci and cleavage bodies, which can be spatially associated with CBs, has been observed (Schul et al., 1999). Cleavage bodies are marked by accumulations of some factors required for the cleavage-polyadenylation reaction (Schul et al., 1996), though it is not known whether these factors participate in histone mRNA 3′ end formation, nor whether their S phase overlap with histone loci is simultaneously and spatially associated with CBs. The relationship between replication-dependent histone genes and CBs as a function of cell cycle has not been directly investigated. However, it has been suggested that active histone genes in S phase cells might represent those loci that associate with CBs, and cleavage bodies, for the purpose of regulating gene activity (Schul et al., 1999; Liu et al., 2000).
We have tested three specific predictions of the hypothesis that there is a relationship between replication-dependent histone gene expression and CB association. First, we explored whether the unique aspects of replication-dependent gene expression are required for CB interaction by examining the spatial relationship between CBs and a replacement histone gene. Second, we determined whether the frequency of association between CBs and replication-dependent histone loci changes during different stages of the cell cycle as their expression is modulated. Third, we visualized for the first time individual histone loci and their associated transcripts simultaneously to determine whether the loci actively producing transcripts are the ones contacting CBs. Our findings were examined in light of the distribution of U7 snRNA to further understand the possible role of 3′ end processing in CB–histone gene associations.
MATERIALS AND METHODS
Cell Culture
HeLa S3 cells were grown at 37°C in DMEM-high glucose supplemented with 10% heat-inactivated fetal calf serum and 100 U/ml penicillin and streptomycin (Life Technologies, Gaithersburg, MD). WI-38 fetal human diploid fibroblasts were obtained from American Type Culture Collection (Manassas, VA) and grown at 37°C in Eagle's basal medium (Life Technologies) supplemented with 10% heat-inactivated fetal calf serum and 100 U/ml penicillin and streptomycin. Subconfluent cells grown on coverslips were extracted and fixed before hybridization and/or immunofluorescence protocols as follows (Carter et al., 1991; Xing et al., 1993). Coverslips were rinsed briefly with Hanks' balanced salt solution (Life Technologies) followed by ice-cold cytoskeletal buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8 [Fey et al., 1986]). Cells were then extracted for 2–5 min on ice in cytoskeletal buffer plus 0.5% Triton X-100 and 20 mM vanadyl ribonucleoside complex (Life Technologies) and subsequently fixed in 4% paraformaldehyde in 1× phosphate-buffered saline (PBS), pH 7.4, for 10 min at room temperature. Cells were then stored in 70% ethanol at 4°C.
For bromo-deoxyuridine (BrdU) labeling, cells were grown in fresh, prewarmed media containing 30 μg/ml BrdU (Roche Molecular Biochemicals, Indianapolis, IN) for 15 min and then washed twice with prewarmed media before the extraction and fixation protocol. Mitotic shake-off cells were obtained by washing subconfluent cells with meduim to remove dead cells, allowing cells to recover for 30 min at 37°C, shaking flasks vigorously, collecting dislodged cells from media, and replating cells on coverslips for the indicated times before fixation. Only obvious sister cell pairs were scored from these enriched G1 populations.
Antibodies
Large and small CBs were detected with a 1:200 dilution of rabbit anti-coilin antibody (R288; Andrade et al., 1991), generously supplied by E. Chan (Scripps Research Institute, San Diego, CA) or 1:100 dilutions of mouse monoclonal antibodies 5P11-ρδ and 5P10-π (gift from M. Carmo-Fonseca, University of Lisbon, Lisbon, Portugal; Almeida et al., 1998). A 1:200 dilution of mouse anti-BrdU monoclonal antibody (Partec, Münster, Germany) or a 1:20 dilution of rat anti-BrdU (Harlan Sera-Lab, Harlan Bioproducts for Science, Indianapolis, IN) was used for BrdU immunodetection. Cyclin B was detected with a 1:100 dilution of rabbit antibody H433 (Santa Cruz Biotechnology, Santa Cruz, CA). Primary antibodies were detected with the following, all obtained from Jackson ImmuoResearch (West Grove, PA): fluorescein isothiocyanate (FITC)-goat anti-rabbit IgG, aminomethylcoumarin acetate (AMCA)-donkey anti-rabbit IgG, FITC-donkey anti-mouse IgG, and AMCA donkey anti-mouse IgG. Antibodies were diluted in 4× SSC, 1% bovine serum albumin.
Fluorescent In Situ Hybridization (FISH) Probes
All probes except oligonucleotides were generated by nick translating plasmid DNA in the presence of 120 μM digoxigenin-11-dUTP (Roche) or biotin-16-dUTP (Roche) using the Life Technologies BioNick kit and substituting the buffer with 50 mM Tris-HCl, pH 7.5, 10 mM MgSO4, 50 μg/ml bovine serum albumin, 0.1 mM dithiothreitol, and 60 μM each dATP, dCTP, and dGTP. Probes for the histone gene cluster(s) at 6p21, here called HIST1 based on the orthologous mouse locus HIST1 (Wang et al., 1996a; Albig et al., 1998), were generated from cosmid 6B6 isolated from the telomeric end of the cluster(s) and cosmid F1B9 from the centromeric end, both generously provided by D. Doenecke and W. Albig (Universitat Gottingen, Gottingen, Germany; Albig and Doenecke, 1997; Albig et al., 1997). Cosmid 6B6 contains one copy each of H4, H3, and H1 genes as well as an H3 pseudogene; F1B9 contains one copy each of genes for H1.5, H2B, and H4; two copies of H3 and H2A; and one H2B pseudogene. Probes for the histone gene cluster at 1q21, here referred to as HIST2 according to similarity with the mouse locus HIST2 (Wang et al., 1996b), were generated from either the phage clone λHHG 41, which contains one copy each of the H4 and H3 genes (Sierra et al., 1982), or from the plasmid pFO-003, a subclone of λHHG 41 containing only the H4 gene and flanking sequence (Kroeger et al., 1987). Plasmid pFO-002, also derived from λHHG 41 and containing the same H4 gene in pFO-003 with only 1.8 kb of flanking sequence, was used to detect H4 RNA (Plumb et al., 1983; Pauli et al., 1989). The hybridization signals produced by probe pFO-002 were identical to those of a probe generated from plasmid FO108-T7 (Aziz et al., 1998), which contains only 215 bp of the promoter sequence and most of the coding region of this H4 gene. The H3.3A gene at 1q41 was probed with plasmid W07, which contains the first three exons and introns of that gene and was a gift from D. Wells (University of Houston, Houston, TX; Wells and Kedes, 1985). A 14-kb genomic clone containing the human osteocalcin gene, phBGP, was provided by H. Deluca (University of Wisconsin, Madison, WI; Celeste et al., 1986).
FISH
Hybridizations with nick translated probes were carried out as previously described (Johnson et al., 1991). Briefly, for detection of DNA only, fixed cells were denatured and RNA hydrolyzed in 0.07 N NaOH, 70% ethanol at room temperature for 5 min. Cells were washed twice in 70% ethanol, further denatured in 2× SSC, 70% formamide at 72°C for 2 min, and then dehydrated through a cold ethanol series. Heat-denatured, nick translated probe (50 ng) was applied to denatured slides for hybridization at 37°C overnight and detected with either TRITC (tetramethyl rhodamine)-sheep anti-digoxigenin antibody or FITC-avidin (Roche). For the exclusive detection of RNA, the cell denaturation steps were omitted and vanadyl ribonucleoside complex was added to the hybridization solution at a final concentration of 20 mM. Note that the permeabilization of cells (before fixation, which allows probes to penetrate the nucleus, results in the extraction of some cytoplasmic RNA and that only RNA and not DNA can be detected when cellular DNA is not denatured (Lawrence et al., 1989). To detect nuclear RNA in one color and DNA in another, nondenatured cells were first hybridized to detect only RNA with a digoxigenin-labeled probe, washed as described above, fixed with 4% paraformaldehyde in 1× PBS for 10 min, subjected to NaOH hydrolysis, and then hybridized with a biotinylated probe to detect only DNA (Xing et al., 1995). Hybridization with and detection of biotinylated U7 antisense oligonucleotide (Frey and Matera, 1995), generously supplied by A.G. Matera (Case Western Reserve University, Cleveland, OH), was achieved according to a previously published protocol (Matera et al., 1995).
Immunostaining
Immunofluorescence was typically coupled with the above-mentioned hybridization protocol as follows. For the detection of CBs (Smith et al., 1995), before hybridization fixed cells were stained with anti-coilin antibody for 1.5 h at 37°C and washed successively in 4× SSC, 4× SSC plus 0.1% Triton X-100, and 4× SSC for 10 min each. They were then incubated with appropriate secondary antibody with conjugated fluorochrome, washed as before, and refixed in 4% paraformaldehyde in 1× PBS for 5–10 min before continuing with the hybridization steps. Staining for cyclin B followed the hybridization protocol and an additional fixation step. To detect BrdU incorporated into DNA, cells first were hybridized to probe as described above, except that the dehydration step was omitted and denatured probe was applied to cells quickly rinsed in 2× SSC after heat denaturation. After hybridization and washes, the anti-BrdU antibody was included in the incubation with the probe's secondary antibody. This was followed by 4× SSC washes and detection of anti-BrdU with fluorochrome-conjugated secondary antibody. Some cells were counterstained with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in 1× PBS for 30 s. All samples were mounted in 1 mg/ml phenylenediamine (Sigma, St. Louis, MO), 1× PBS, pH 8.0; 90% glycerol.
Microscopic Analysis
Cells were examined with a Zeiss Axioplan microscope equipped with a filter wheel, triple-bandpass epifluorescence filter (Chroma Technology, Brattleboro, VT), and a 100×, numerical aperature (NA) 1.4 objective (Zeiss, Oberkochen, Germany). Digital images were either acquired by a Photometrics Series 200 charge-coupled device camera and analyzed with a data acquisition system by Hanaway and Associates (Boulder, CO) or acquired with a Photometrics Quantix camera and Metamorph imaging software (Universal Imaging, Media, PA). Digital images shown in figures were adjusted for brightness and contrast with Adobe Photoshop 4.0 such that they reproduce with accuracy and clarity the images as seen through the microscope eyepiece. Associations among CBs, gene signals, and/or RNA foci were usually scored through the microscope eyepiece by at least two investigators. Scoring of some triple-label experiments required the use of digital images. Threshold values were not altered in these cases. Gene/RNA signals were scored as “associated” if they appeared to overlap or contact the edge of CBs with no visible space between the two signals. Based on the limits of resolution of light microscopy, associated objects are within 250 nm of each other.
RESULTS
CBs Preferentially Associate with Replication-dependent Histone Genes but not a Replacement Histone Variant
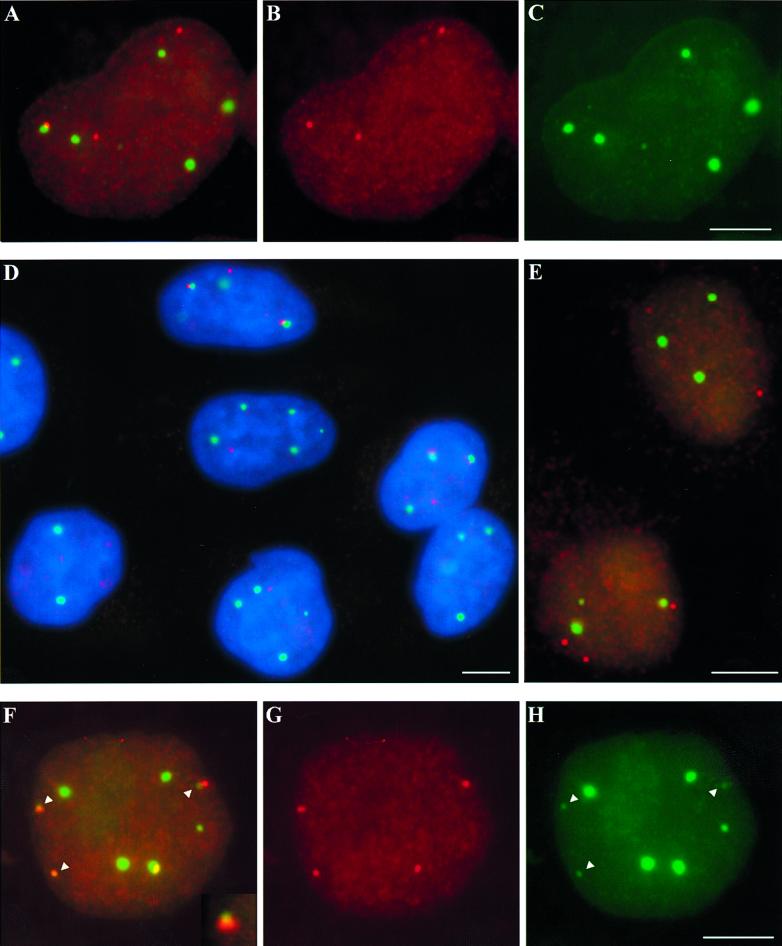
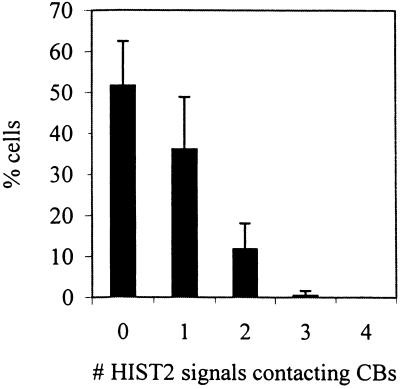
All currently identified replication-dependent histone genes in humans are organized into two multigene clusters, HIST1 and HIST2, which map to chromosomes 6p21.3-p22 and 1q21, respectively (Green et al., 1984; Allen et al., 1991; Albig et al., 1997). This study is based primarily on HIST2, which previous studies reported is more frequently associated with CBs than is HIST1 (Frey and Matera, 1995; Jacobs et al., 1999). HIST2 loci were detected by FISH coupled with indirect immunofluorescence detection of CBs using one of three anti-coilin antibodies (see MATERIALS AND METHODS), p80 coilin being a hallmark protein of CBs (Andrade et al., 1991). Signals from the HIST2 probe never colocalize with closely related HIST1 probe signals when they are cohybridized to interphase nuclei, and the HIST2 probe hybridizes only to locus 1q21 on metaphase chromosomes, verifying its specificity. The HeLa cells used in this study are tetraploid for HIST2, showing four FISH signals when probed for this locus or a linked gene, osteocalcin (Figure 1D, and see below). Similar to previously reported data (Frey and Matera, 1995; Jacobs et al., 1999), we found in unsynchronized HeLa cells that ∼50% of nuclei contain at least one HIST2 locus in contact with a CB (Figure 1, A–D and Table 1). Here we further report that 14% of HIST2 DNA signals are found associated with CBs, and that although most cells with CB-HIST2 associations contain only one associated locus, the numbers of associated loci vary and reach a maximum of three per nucleus (Figure 2).
Figure 1.
CBs preferentially associate with replication-dependent histone loci but not with the histone variant H3.3A. (A-C) In one HeLa nucleus, one HIST2 locus (red, B) contacts the edge of a CB (green, C), whereas two other loci are separate from coilin accumulations. (D) This field of HeLa cells shows a variety of HIST2–CB interactions. Nuclei are counterstained with DAPI (blue), CBs are in green, and HIST2 DNA is red. (E) The polyadenylated, variant histone gene H3.3A (red) rarely associates with CBs (green). (F–H) HIST1 (red, F and G) preferentially interacts with both large and small (arrowheads) CBs (green, F and H). Contacts with small CBs can be either at the periphery (F, inset, and top two arrowheads) or completely overlapping (bottom left arrowhead). CBs were detected with rabbit anti-coilin R288 polyclonal antibody (A, C, D, and E) or mouse anti-coilin 5p11-ρδ monoclonal antibody (F and H). Bars, 5 μm.
Table 1.
Frequencies of associations between CBs and specific genetic loci/transcripts
| Locus | Cytogenetic band | Nuclear target | No. of FISH signals scored | % signals associated with CBsa | % cells with ≥1 associationa |
|---|---|---|---|---|---|
| HIST2 | 1q21 | DNA | 890 | 14 ± 3.8b | 47 ± 9.4 |
| RNA | 556 | 20 ± 5.0 | 44 ± 12c | ||
| Osteocalcin | 1q21 | DNA | 334 | 2.7 ± 0.6 | 8.5 ± 3.5 |
| H3.3A | 1q41 | DNA | 415 | 2.4 ± 2.8 | 5.2 ± 5.0 |
| HIST1 | 6p21.3 | DNA | 768 | 9.7 ± 1.5 | 30 ± 4.3 |
CBs >0.3 μm in diameter were scored.
Standard deviations were calculated based on data from different experiments and scorers.
Cells with visible signals were scored. These cells did not necessarily contain signals from all loci.
Figure 2.
Number of replication-dependent histone loci associated with CBs varies from cell to cell. The histogram shows the percentages of HeLa cells in an asynchronous population that contain a given number of CB-contacting FISH signals of the HIST2 (1q21) locus. About half of the cells has at least one locus associated with a CB, whereas the other half shows no associations. The HeLa cells examined are tetraploid for HIST2 and thus the maximum possible number of contacts per cell is four. Error bars represent SDs among different samples.
Given the small size of the gene signals and small portion of nuclear volume occupied by CBs (<1%), the interactions between CBs and HIST2 loci are unlikely based on random chance. To further demonstrate the specificity of this interaction, we examined an unrelated gene, osteocalcin, located ∼7 Mb from HIST2 in region 1q21 (GenBank accession numbers NT_002807 and NM 003528). We found only 3% of osteocalcin gene signals contacting CBs, with only 9% of cells containing an osteocalcin–CB interaction (Table 1). Even though this gene is linked to the HIST2 locus, this association level is approximately fivefold less than HIST2 and approaches that of a random distribution. A similar difference has been observed between the U2 locus and another locus on chromosome 17 <1 Mb away (Smith et al., 1995).
If the interactions between HIST2 and CBs depend on any of the unique aspects of replication-dependent histone gene expression, then a replacement histone gene would not be expected to preferentially associate with CBs. We tested this hypothesis by examining the replacement histone gene H3.3A, which is constitutively active, contains three introns, and produces polyadenylated mRNA (Wells and Kedes, 1985). Scoring H3.3A relative to CBs indicates that, similar to osteocalcin, this gene does not associate preferentially with CBs (Table 1). These results are consistent with transcriptional regulation or 3′ end processing of replication-dependent histone transcripts playing a role in CB association.
Replication-dependent Histone Gene Associations with CBs Occur throughout Interphase but Are More Frequent during Stages When Genes Are More Highly Expressed
To address whether replication-dependent histone gene activity correlates with CB association, we examined HIST2–CB associations at different stages of the cell cycle. Such an analysis enabled us to test the simple hypothesis that the appearance of HIST2–CB associations in only 50% of unsynchronized cells might be because these cells are the ones in S phase, when HIST2 is most active. Transcription of replication-dependent histone genes occurs throughout interphase but increases by at least two- to fourfold between early G1 and early S phase and decreases in G2 (Heintz et al., 1983; Baumbach et al., 1987; Collart et al., 1991; Harris et al., 1991). Pre-mRNA 3′ end processing rates change in a similar manner (Harris et al., 1991).
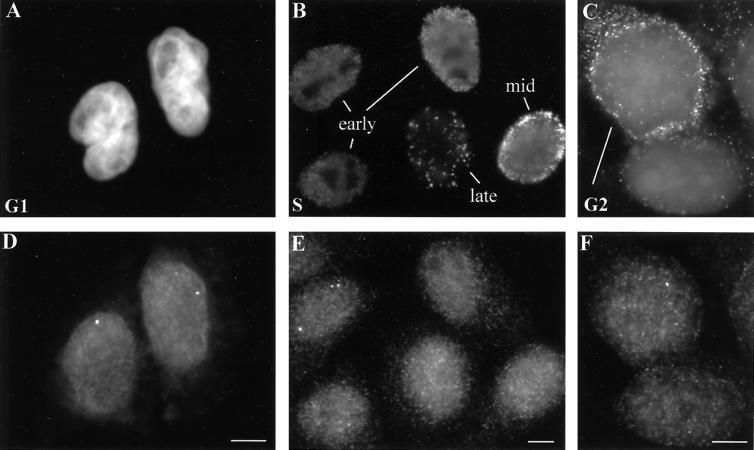
To examine the association level of replication-dependent histone loci with CBs during different stages of interphase, cells in G1, S, and G2 phases were identified in one of three ways. 1) Enriched populations of HeLa cells in G1 were generated by mitotic shake-off, plated, and allowed to grow for 2, 4, or 6 h. G1 cells were identified as those in sister-cell pairs (Figure 3A) and were primarily in G1 and not S phase because fewer than 10% incorporated BrdU when pulse-labeled. In contrast, BrdU incorporation increased to 30% by 8 h after shake-off. 2) Cells in S phase were identified by labeling unsynchronized cells with BrdU for 15 min before fixation. These cells were then subjected to a triple-labeling protocol to detect BrdU-labeled DNA, HIST2 DNA, and coilin. Different patterns of BrdU-labeled replication foci enabled us to determine whether cells were in early, mid, or late S phase (Figure 3B), as has been previously reported (Nakamura et al., 1986; Nakayasu and Berezney, 1989; Kill et al., 1991). Approximately 45% of these unsynchronized cells labeled with BrdU. 3) To identify G2 cells, unsynchronized HeLa cells were triple-labeled for HIST2 DNA, coilin, and cyclin B, which accumulates to detectable levels in the cytoplasm during G2 (Figure 3C) (Bailly et al., 1992).
Figure 3.
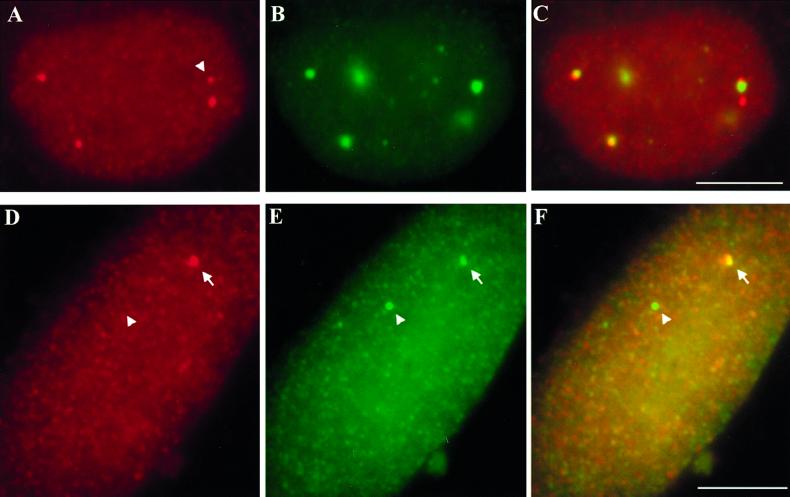
HeLa cells at several different stages of interphase contain RNA foci from replication-dependent histone genes. (A) A pair of mid-G1 daughter cell nuclei, 4 h after mitotic shake-off, is counterstained with DAPI. (B) In an asynchronous population of HeLa cell nuclei pulse-labeled with BrdU, anti-BrdU antibody staining reveals patterns indicative of cells in early, middle, and late S phase. (C) Immunostaining with anti-cyclin B antibody reveals one G2 cell with cytoplasmic cyclin B particularly concentrated around the nuclear periphery (top left) and one cell with only background signal, not in G2 (lower right). (D–F) H4 RNA foci are detected by FISH in the same cells shown in A–C. In these examples, RNA foci are clearly visible in both G1 cells (D), one of the early S phase cells (E), and the G2 nucleus (F). Typically, early S phase cells have the brightest and most numerous RNA foci (E), though RNA has been detected in all stages of S phase. Bars, 5 μm.
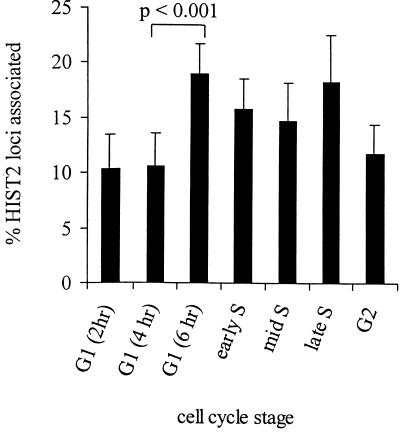
Scoring the associations of HIST2 locus with CBs in cell stage-marked populations indicated that cells with associated loci clearly are not specific to S phase but can be at any stage in interphase. However, we found that the level of association increases modestly (1.8-fold) between mid-G1 and late G1, remains at a higher level through S phase, and then returns to the original level by G2 (Figure 4). Although seemingly small, a Student's t test shows that the increase between mid-G1 (4 h) and late G1 (6 h) is statistically significant (p < 0.001). Moreover, the pattern of change in association frequencies is similar to the reported changes in histone gene expression (Plumb et al., 1983; Baumbach et al., 1987), suggesting that the two are linked. In fact, the increase in CB associations that occurs between mid- and late G1 might actually precede the reported increase in replication-dependent histone gene transcription at the beginning of S phase.
Figure 4.
HIST2 associates with CBs more frequently during late G1 and all of S phase. The histogram shows the percentage of HIST2 DNA FISH signals contacting CBs in HeLa nuclei at different stages of interphase, as determined by mitotic shake-off and DAPI stain, BrdU labeling, or cyclin B staining (Figure 3). G1 cells were examined 2, 4, and 6 h after shake-off as indicated. Error bars represent SDs between different experiments and from different scorers.
Varying Levels of Transcript Are Detected at Individual HIST2 Loci within the Same Nucleus
Although the interactions between replication-dependent histone loci and CBs are significantly more frequent than random expectation, they do not occur for all the loci in a nucleus at the same time, indicating that all loci are not equivalent. This observation led us to ask whether individual homologs of HIST2 might also be different in terms of their expression status, and whether such a difference might be reflected in positioning relative to CBs. If so, this would directly link histone gene expression with CB association. We therefore first investigated whether individual HIST2 loci might be associated with different amounts of transcript in situ.
To detect transcripts from a single histone gene in HIST2, we used a probe for one of the more highly expressed H4 genes (Lichtler et al., 1982). With this probe, we detected small RNA foci in a subset of nuclei (Figures 5, A and D, and 3, D–F). The level of detection varied with different cell preparations, but in some cases >50% of HeLa cells contained nuclear RNA signal. To determine whether the RNA foci correspond to transcripts accumulated at the gene, we used a two-step hybridization protocol to detect separately RNA and DNA in two different colors (see MATERIALS AND METHODS). These hybridizations showed that 99% of the H4 RNA foci overlapped the HIST2 gene signals in both HeLa cells and WI38 diploid fibroblasts (Figure 5, D–F). Moreover, their appearance is responsive to transcription level, as they are brightest and most numerous in early S phase cells (Figure 3E), in accordance with their time of peak transcription (Plumb et al., 1983; Baumbach et al., 1987). However, RNA foci are present in G1 and G2 nuclei (Figure 3, D and F), consistent with previous reports of replication-dependent gene activity (Plumb et al., 1983; Harris et al., 1991).
Figure 5.
Nuclear foci of RNA from replication-dependent histone genes localize to their sites of transcription, not always with CBs. (A–C) Some HIST2 H4 RNA foci (red, A and C) associate with CBs (green, B and C) in HeLa cells and some do not. These RNA foci vary in number and intensity from cell to cell. The cell shown is atypical in that three of the four RNA signals are CB-associated. Note that the weakest RNA focus (arrowhead) contacts a CB, whereas a more intense, nearby focus does not. (D–F) H4 RNA (red, D and F) forms a focus that partially overlaps one HIST2 locus (arrow, green, E and F) in a WI38 diploid fibroblast nucleus. The other homolog of HIST2 has no apparent accumulation of H4 RNA (arrowhead). Bars, 5 μm.
Hybridization to nuclear H4 RNA in several experiments consistently detected different amounts of transcripts accumulated at different loci within the same nucleus. WI38 fibroblasts, though diploid, rarely contain more than one RNA focus (Figure 5D), and HeLa nuclei contain as many as four H4 RNA foci, often with varying intensities within the same nucleus (Figures 5A, arrowhead marks focus smaller than the rest, and 3, D–F). Although the different levels of transcripts detected could result from technical variations, we think this unlikely because RNA signals are compared within the same nucleus. Also, in experiments in which DNA is detected in one color and RNA in another, HIST2 DNA signals have uniform sizes and intensities even though only a subset of these colocalize with RNA foci (Figure 5, D–F). These observations suggest that the intensities of RNA signals reflect real, in vivo differences and, therefore, that individual HIST2 homologs in a given cell either do not synthesize transcripts at the same rate or do not accumulate them to the same extent. This has also been observed for U2 loci (Smith and Lawrence, 2000).
Replication-dependent Histone Transcripts Can Be Detected at Loci Unassociated with CBs
To test whether the HIST2 loci producing transcripts are the ones contacting CBs, as has been previously suggested (Schul et al., 1999; Liu et al., 2000), H4 RNA foci and CBs were detected simultaneously. Data from the scoring of hundreds of cells indicate that a subset of RNA signals contacts CBs (20%), though the majority do not (Table 1). A less typical cell with three of four RNA signals associated with CBs is shown in Figure 5, A–C. These findings suggest that active histone loci need not have sustained contact with CBs to synthesize RNA (see DISCUSSION).
Although our data suggest that CB contact might not be necessary for histone mRNA metabolism, it might still facilitate this process. If so, more highly active loci should be more frequently associated with CBs. We therefore examined whether the size and intensity of individual H4 RNA foci correlate with CB association and found that they do not (Figure 5, A–C, and Table 2). However, this is not necessarily unexpected. Although cells with more and brighter RNA signals are more transcriptionally active in general, individual RNA foci represent steady-state accumulations that can also be affected by RNA processing and transport rates. Importantly, we do find that overall the CB-contacting loci are slightly enriched for those that produce visible RNA foci; 20% of RNA foci, which represent active genes, versus 14% of all HIST2 DNA signals are CB associated (Table 1), consistent with our cell cycle analysis.
Table 2.
Proportion of large and small H4 RNA foci contacting CBs
| H4 RNA focus size | No. of foci scored | % of total focia | % of CB-associated foci | Fold enrichment at CBsb |
|---|---|---|---|---|
| Small | 248 | 33 | 35 | 1.1 |
| Large | 213 | 28 | 24 | 0.9 |
Total foci include all RNA signals in the nuclei scored. These were classified as small, medium, or large based on their relative sizes in multiple cells.
Percentage of CB-associated foci per percentage of total foci.
Histone Gene Activity Does Not Require Continual Association with Highly Concentrated U7 snRNA Sites
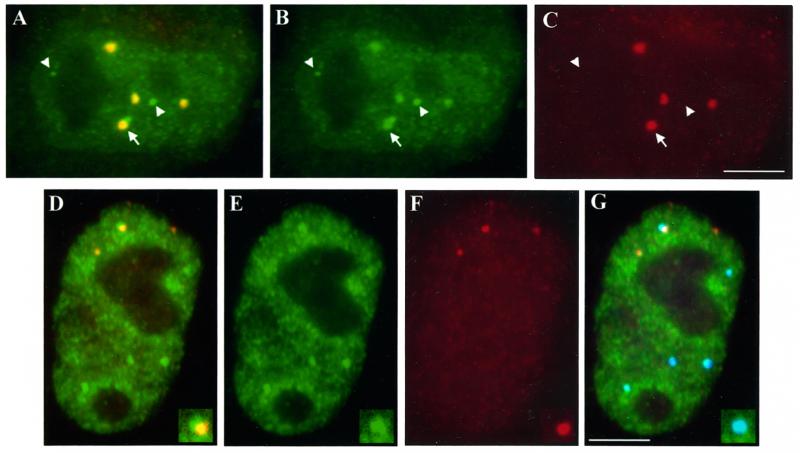
The observation that active replication-dependent histone genes can be separated from CBs raises the question of how their transcripts get processed, because U7 and SLBP have been shown to concentrate in CBs. To address this question, we examined the distribution of U7 in our cultured cell system. We hybridized HeLa cells with an antisense oligonucleotide probe directed against U7 snRNA and then stained them for coilin. Similar to previously reported data (Frey and Matera, 1995), this experiment shows several sites of concentrated U7 oligo signal, many of which colocalize with CBs, as well as a weaker dispersed signal throughout the entire nucleus but excluding the nucleolus (Figure 6, A–C). Of note are several sites of concentrated U7 that do not correspond to CBs and that have not been described previously. Many of these are completely independent of CBs (Figure 6, A–C, arrowheads), but interestingly, some of the brightest are located at the periphery of CBs and appear as “buds” on the larger U7 signal that is coincident with the CB (Figure 6, A–C, arrow). To determine whether the extra U7 sites might have some special relationship to active histone loci, we triple-labeled cells to distinguish H4 RNA, U7, and CBs. This experiment showed that when H4 RNA associates with a concentrated U7 site, it also contacts a CB (Figure 6, D–G); H4 RNA rarely associates with a U7 concentration that is separate from a CB. Thus, although U7 distribution is more complex and widespread than that of CBs, active histone genes still interact selectively with the concentrated U7 sites that correspond to CBs. In addition, we observed that the RNA foci associated with the edges of CBs also overlap the smaller “buds” of U7 with high frequency. This arrangement is reminiscent of the previously described cleavage body and HIST1 DNA during S phase, which was suggested to facilitate the expression of the associated locus (Schul et al., 1999). However, scoring of H4 RNA foci relative to U7 accumulations indicates that this association does not account for the majority (80%) of transcriptionally active loci, which are separate from both CBs and U7 concentrations (Figure 6, D–G).
Figure 6.
U7 snRNA signal is both disperse and concentrated at discrete sites that do not necessarily correspond to CBs or sites of active HIST2 loci. (A–C) A HeLa nucleus hybridized with a U7 antisense oligonucleotide (green, A and B) and stained with anti-coilin antibody R288 (red, A and C) shows that most concentrations of U7 signal coincide with CBs. However, others are completely separate from CBs (arrowheads, note one small accumulation at the nucleolar periphery, left) or localize to the edge of a CB appearing as “buds” from the signal overlapping the CB (arrow). (D–G) Triple labeling to detect U7 snRNA (green, D, E, and G), H4 RNA (red, D, F, and G), and coilin (blue, G) shows one of three active histone loci associated with concentrated U7 signal (D and insets). This association occurs at the periphery of a CB (G and insets) in most HeLa nuclei. Bars, 5 μm.
Related to the above-mentioned experiments, and particularly relevant to concepts mentioned in the DISCUSSION (see below), we detected some CBs that are smaller (≤0.3 μm in diameter) than those usually described (0.5–1 μm in diameter) and are reminiscent of a subset of the concentrated U7 sites that can appear separate from the usual larger CBs (Figures 1H, arrowheads, 1C, and 5B). These are clearly visible when higher concentrations of antibody are used to stain cells, can be detected with two different anti-coilin antibodies (Figure 1, C and H), and have been previously described (Smith et al., 1995; Boudonck et al., 1999; Platani et al., 2000). On average, four very small CBs appear in about two-thirds of the HeLa cells we studied. Double labeling shows that these small CBs contain U7, though some of the smaller U7 sites are still independent of CBs, both large and small (Figure 6, A–C).
It is difficult to determine whether these small coilin concentrations are bona fide CBs. We were unable to resolve whether they contain typical CB components such as the Sm antigen and fibrillarin above “background” levels detected by immunostaining. However, like the typical, larger CBs, we found that the small CBs also associate with HIST2 loci (Table 3). Interestingly, small CBs are twice as frequently associated with HIST1 than they are with HIST2 (Figure 1, F–H, and Table 3). They are also more frequently associated with HIST1 than the larger CBs (Tables 1 and 3). These findings are consistent for both ends of the HIST1 cluster (our unpublished results), which are 2.5 Mb apart and have the potential to localize independently of each other (Lawrence et al., 1990; Smith et al., 1995). HIST1 can localize at the center of the small CBs as well as the edges, unlike with the larger CBs (Figure 1, F–H). The high coincidence of small CBs at HIST1 suggests to us the possibility that new CBs might arise specifically at these loci (see DISCUSSION).
Table 3.
Association frequencies of specific genes and “small” CBs
| Locus | No. of FISH signals scored | % of signals contacting small CBsa |
|---|---|---|
| HIST2 | 644 | 11 ± 3.0 |
| HIST1 | 528 | 26 ± 7.4 |
| Osteocalcin | 241 | 1.0 ± 0 |
Only CBs ∼0.3 μm in diameter were scored.
DISCUSSION
The work presented here provides valuable information regarding the mechanism that mediates the spatial relationship between CBs and specific loci. Our finding that the replacement histone gene H3.3A does not preferentially associate with CBs is consistent with the hypothesis that cell cycle transcriptional regulation and/or U7-mediated 3′ end processing play a role in HIST1– and HIST2–CB interactions. Although the difference between replication-dependent loci and H3.3A might arise coincidentally from their chromosomal locations, we consider this unlikely because we also found that the osteocalcin gene, which is in the same cytogenetic band as HIST2, does not associate with CBs. Rather, the difference is more likely related to divergent 5′ or 3′ regulatory regions of the genes within those loci. This is also suggested by the presence of the 3′ processing factors U7 snRNP and SLBP in CBs (reviewed in Dominski and Marzluff, 1999), and by the very recently reported CB association with cyclin E and cyclin-dependent kinase 2 (CDK2) accumulations, the latter of which occurs specifically during S phase (Liu et al., 2000). Cyclin E plays a role in activating p220/NPAT, a protein that activates replication-dependent histone gene transcription and accumulates at HIST1 and HIST2 loci (Ma et al., 2000; Zhao et al., 2000).
In support of the hypothesis that CB association is related to replication-dependent histone gene expression, our results show that a modest but significant change in CB association levels mirrors the timing of changes in replication-dependent histone gene expression. This finding is further supported by our observations that 20% of HIST2 transcript foci in unsynchronized cells associate with CBs versus 14% of HIST2 gene signals, because the RNA foci are most readily detected in S phase nuclei. Similarly, when U2 expression levels are increased (e.g., by copy number or integration site), CB association levels within that population of cells also increase (Frey et al., 1999), suggesting that a link to expression level may be a common feature of CB-associating loci.
Without further analysis at the level of individual loci, the positive correlation between CB association and gene expression within a cell population might be considered to indicate a straightforward explanation: that loci are more active in RNA production when directly associated with a CB (Schul et al., 1999; Liu et al., 2000). However, our in situ analyses of nuclear RNA foci emanating from the HIST2 locus reveal that this is not the case. Our results clearly indicate that sustained contact with a CB is not required for gene expression, as indicated by the presence of detectable RNA foci at loci unassociated with a CB. This is also the case for U2 loci (Smith and Lawrence, 2000). That CB association is not an absolute requirement for gene expression is indirectly suggested by the observation that some cell types have no visible CBs (Spector et al., 1992; Carmo-Fonseca et al., 1993). Because CBs are often prominent in highly metabolically active cells, it was a priori possible that sustained contact with a CB enriched in specific RNA metabolic factors might instead be required for the enhanced expression of specific genes. However, our results show that this is not the case, even in rapidly dividing cells. Hence, these results argue against mechanisms in which ongoing CB contact is necessary to maintain gene activity, as might be the case if CBs continually supply components necessary for metabolism of that RNA. This is consistent with the widespread distribution of U7 snRNA, which suggests that histone pre-mRNAs can be processed at many sites throughout the nucleus, regardless of the presence of CBs.
If sustained CB contact is not a requirement for histone gene expression, why does this interaction occur, and what purpose might it serve? We envision two broad possibilities. One is that transient interactions might enhance histone gene expression in metabolically active cells. Because CBs have been shown to move about the nucleus (Boudonck et al., 1999; Platani et al., 2000), they may well contact specific genes briefly. Measured speeds of the typical, larger CBs might be fast enough to ensure that each locus is contacted within a given cell cycle or even the short time that its transcripts reside in the nucleus (Heintz et al., 1983; Boudonck et al., 1999; Platani et al., 2000). Furthermore, data from our lab and others indicate that the very small CBs move even faster than the larger ones (Tam and Lawrence, unpublished data; Platani et al., 2000). An average of one or two brief interactions, particularly during late G1 and early S phase, could conceivably be sufficient to maintain the rate at which the HeLa cells studied here divide.
Although sustained contact with active genes would imply that CBs act by providing necessary metabolic factors, transient associations would be more consistent with the possibility that CBs are involved in regulating expression by signaling a change in activity status. This model of CBs acting as transient regulators of replication-dependent histone genes is supported by our observation that HIST2–CB association levels begin to increase in late G1, perhaps in anticipation of gene up-regulation that occurs at the G1/S transition. However, the possibility that transient CB interactions serve to deposit a small amount of necessary factors at specific gene sites cannot be ruled out. Additionally, we think it unlikely that CBs interact transiently with all genes, as might be suggested by their accumulations of many basic factors necessary for RNA metabolism (Gall et al., 1999). In a given cell cycle, the three to four large CBs clearly do not move throughout the nucleus fast enough to contact such a great number of genes, and whether the three to four small CBs could do so is questionable. Even so, CBs associate significantly more frequently with histone and snRNA loci than they do with others, indicating specialized interactions.
The alternative to CBs acting as transient regulators of genes is that the level of histone gene activity impacts the formation of CBs. These, in turn, may be involved in basal nuclear RNA metabolism, such as the maturation of snRNA and snoRNA and the assembly of snRNPs (Gall et al., 1999; Smith and Lawrence, 2000). If CB formation is nucleated by high levels of histone gene expression, it could provide a link between numerous activities that must be balanced with one another and reflect the cell's overall metabolic rate. This process may be impacted by the possible formation of CBs at other loci such as U2, which are more highly expressed and may better compete for CB-forming factors, consistent with the observations that not all loci appear associated with a CB at a given time.
That CBs might be nucleated at histone loci is suggested by several observations. Very small CBs were found in association with HIST1 with high frequency and may represent new CBs that have yet to grow into more mature and larger CBs (Figure 1F). Small CBs have also been observed previously in the vicinity of U2 loci (Smith et al., 1995). In addition, the average number of CBs, both large and small, increases between G1 and S phase and decreases again in G2, following the activity profile of histone genes (our unpublished observations). This is in apparent contrast to previous reports describing G1 as the time when CB numbers peak, though these studies may have used different cell types or different techniques to select cells at specific stages (Andrade et al., 1993; Smith et al., 1995). A recent report suggests that high levels of U7 snRNA can cause the formation of CB-like bodies (Tuma and Roth, 1999). CBs might therefore be nucleated by localized concentrations of U7 that accumulate at histone loci as a result of gene activity. However, we and others have found that not all U7 accumulations are coincident with CBs (Figure 6; Matera, personal communication), suggesting that if U7 is involved in this process, other nonhomogeneously distributed factors also might be necessary.
Our findings have significantly narrowed the potential mechanisms that may underlie the interactions of CBs with replication-dependent histone loci. They point most strongly to two possible models, either that brief interaction with CBs might signal a sustained effect on histone gene expression or, conversely, that histone gene expression might nucleate new CBs. The recent findings that cyclin E and CDK2 associate with CBs in a cell cycle-dependent manner tend to favor a role for the CB in gene regulation (Liu et al., 2000; Ma et al., 2000; Zhao et al., 2000). However, future work, including investigations of CB interactions in living cells, will be needed to further define why histone loci physically associate with CBs.
ACKNOWLEDGMENTS
We thank Kelly Smith, Carol Johnson, Lisa Hall, John McNeil, and Rose Tam for cell scoring, technical assistance, discussion, and helpful comments on this manuscript. We thank T J Last, André van Wijnen, Werner Albig, Detlef Doenecke, Dan Wells, Hector Deluca, Ed Chan, Maria Carmo-Fonseca, and Greg Matera for generously supplying us with reagents. Thanks also to Janine LaSalle for suggesting the use of cyclin B antibody to identify G2 cells. This publication was made possible in part by grants from the Muscular Dystrophy Association and the National Institutes of Health (GM-49254) to J.B.L.; to J.L.S., J.B.Li., G.S.S., and J.B.La. (National Institutes of Health AR-42262); and to L.S.S. (National Institutes of Health fellowship GM-18846). The contents are solely the responsibility of the authors and do not necessarily reflect the official views of the National Institutes of Health or Muscular Dystrophy Association.
REFERENCES
- Abbott J, Marzluff WF, Gall JG. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- Albig W, Drabent B, Burmester N, Bode C, Doenecke D. The hemochromatosis candidate gene HFE (HLA-H) of man and mouse is located in syntenic regions within the histone gene cluster. J Cell Biochem. 1998;69:117–126. doi: 10.1002/(sici)1097-4644(19980501)69:2<117::aid-jcb3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997;40:314–322. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- Allen BS, Stein JL, Stein GS, Ostrer H. Single-copy flanking sequences in human histone gene clusters map to chromosomes 1 and 6. Genomics. 1991;10:486–488. doi: 10.1016/0888-7543(91)90337-e. [DOI] [PubMed] [Google Scholar]
- Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Tan EM, Chan EKL. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz F, van Wijnen AJ, Stein JL, Stein GS. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J Cell Physiol. 1998;177:453–464. doi: 10.1002/(SICI)1097-4652(199812)177:3<453::AID-JCP8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- Baumbach LL, Stein GS, Stein JL. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 1987;26:6178–6187. doi: 10.1021/bi00393a034. [DOI] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush D, Dodgson JB, Choi OR, Stevens PW, Engel JD. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985;5:1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan HG, Gall JG. Association of RNA with the B and C snurposomes of Xenopus oocyte nuclei. Chromosoma. 1991;101:69–82. doi: 10.1007/BF00357056. [DOI] [PubMed] [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste AJ, Rosen V, Buecker JL, Kriz R, Wang EA, Wozney JM. Isolation of the human gene for bone gla protein utilizing mouse and rat cDNA clones. EMBO J. 1986;5:1885–1890. doi: 10.1002/j.1460-2075.1986.tb04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart D, Ramsey-Ewing A, Bortell R, Lian J, Stein J, Stein G. Isolation and characterization of a cDNA from a human histone H2B gene which is reciprocally expressed in relation to replication-dependent H2B histone genes during HL60 cell differentiation. Biochemistry. 1991;30:1610–1617. doi: 10.1021/bi00220a024. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The non-chromatin substructures of the nucleus: the ribonucleoprotein RNP-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera G. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L, Van Antwerpen R, Stein J, Stein G, Tripputi P, Emanuel B, Selden J, Croce C. A major human histone gene cluster on the long arm of chromosome 1. Science. 1984;226:838–840. doi: 10.1126/science.6494913. [DOI] [PubMed] [Google Scholar]
- Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N, Sive HL, Roeder RG. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector D. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell. 1999;10:1653–1663. doi: 10.1091/mbc.10.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: implication for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- Jordan P, Cunha C, Carmo-Fonseca M. The cdk7-cyclin H-MAT1 complex associated with TFIIH is localized in coiled bodies. Mol Biol Cell. 1997;8:1207–1217. doi: 10.1091/mbc.8.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kill IR, Bridger JM, Campbell KH, Maldonado-Codina G, Hutchison CJ. The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts. J Cell Sci. 1991;100:869–876. doi: 10.1242/jcs.100.4.869. [DOI] [PubMed] [Google Scholar]
- Kroeger P, Stewart C, Schaap T, van Wijnen A, Hirshman J, Helms S, Stein G, Stein J. Proximal and distal regulatory elements that influence in vivo expression of a cell cycle-dependent human H4 histone gene. Proc Natl Acad Sci USA. 1987;84:3982–3986. doi: 10.1073/pnas.84.12.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, McNeil JA. Interphase and metaphase resolution of different distances within the human dystrophin gene. Science. 1990;249:928–932. doi: 10.1126/science.2203143. [DOI] [PubMed] [Google Scholar]
- Lichtler AC, Sierra F, Clark S, Wells JR, Stein JL, Stein GS. Multiple H4 histone mRNAs of HeLa cells are encoded in different genes. Nature. 1982;298:195–198. doi: 10.1038/298195a0. [DOI] [PubMed] [Google Scholar]
- Liu J, Hebert MD, Ye Y, Templeton DJ, Kung H, Matera AG. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J Cell Sci. 2000;113:1543–1552. doi: 10.1242/jcs.113.9.1543. [DOI] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Histone 3′ ends: essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Pauli U, Chiu JF, Ditullio P, Kroeger P, Shalhoub V, Rowe T, Stein G, Stein J. Specific interactions of histone H1 and a 45 kilodalton nuclear protein with a putative matrix attachment site in the distal promoter region of a cell cycle-regulated human histone gene. J Cell Physiol. 1989;139:320–328. doi: 10.1002/jcp.1041390214. [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M, Stein J, Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Res. 1983;11:2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J Cell Biochem. 1995;59:11–26. doi: 10.1002/jcb.240590103. [DOI] [PubMed] [Google Scholar]
- Schul W, Groenhout B, Koberna K, Takagaki Y, Jenny A, Manders E, Raska I, van Driel R, de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Der Kraan I, Matera AG, van Driel R, de Jong L. Nuclear domains enriched in RNA 3′-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol Biol Cell. 1999;10:3815–3824. doi: 10.1091/mbc.10.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra F, Lichtler A, Marashi F, Rickles R, Van Dyke T, Clark S, Wells J, Stein G, Stein J. Organization of human histone genes. Proc Natl Acad Sci USA. 1982;79:1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Smith KP, Lawrence JB. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for pre-U2 within Cajal bodies. Mol Biol Cell. 2000;11:2987–2998. doi: 10.1091/mbc.11.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell. 1992;3:555–569. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma RS, Roth MB. Induction of coiled body-like structures in Xenopus oocytes by U7 snRNA. Chromosoma. 1999;108:337–344. doi: 10.1007/s004120050385. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Krasikov T, Frey MR, Wang J, Matera AG, Marzluff WF. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1Mb. Genome Res. 1996a;6:688–701. doi: 10.1101/gr.6.8.688. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Tisovec R, Debry RW, Frey MR, Matera AG, Marzluff WF. Characterization of the 55-kb mouse histone gene cluster on chromosome 3. Genome Res. 1996b;6:702–714. doi: 10.1101/gr.6.8.702. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996c;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wells D, Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequence and encode polyadenylated mRNAs. Proc Natl Acad Sci USA. 1985;82:2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CHH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Bonner WM. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981;27:321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil JA, Lawrence JB. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]