Abstract
Alternative splicing of fibroblast growth factor receptor 2 (FGFR2) occurs in a cell-type-specific manner with the mutually exclusive use of exon IIIb or exon IIIc. Specific inclusion of exon IIIb is observed in epithelial cells, whereas exon IIIc inclusion is seen in mesenchymal cells. Epithelium-specific activation of exon IIIb and repression of exon IIIc are coordinately regulated by intronic activating sequence 2 (IAS2) and intronic splicing activator and repressor (ISAR) elements in FGFR2 pre-mRNA. Previously, it has been suggested that IAS2 and a 20-nucleotide core sequence of ISAR form a stem structure that allows for the proper regulation of FGFR2 alternative splicing. Replacement of IAS2 and the ISAR core with random sequences capable of stem formation resulted in the proper activation of exon IIIb and repression of exon IIIc in epithelial cells. Given the high degree of phylogenetic conservation of the IAS2-ISAR core structure and the fact that unrelated stem-forming sequences could functionally substitute for IAS2 and ISAR elements, we postulated that the stem structure facilitated the approximation of intronic control elements. Indeed, deletion of the entire stem-loop region and juxtaposition of sequences immediately upstream of IAS2 with sequences immediately downstream of the ISAR core maintained proper cell-type-specific inclusion of exon IIIb. These data demonstrate that IAS2 and the ISAR core are dispensable for the cell-type-specific activation of exon IIIb; thus, the major, if not the sole, role of the IAS2-ISAR stem in exon IIIb activation is to approximate sequences upstream of IAS2 with sequences downstream of the ISAR core. The downstream sequence is very likely a highly conserved GCAUG element, which we show was required for efficient exon IIIb activation.
Fibroblast growth factor receptor 2 (FGFR2) contains a single transmembrane domain, an intracellular tyrosine kinase domain, and an extracellular fibroblast growth factor (FGF) binding domain, which is composed of immunoglobulin (Ig)-like domains II and III. Alternative splicing of FGFR2 transcripts produces two variants of the Ig-III domain with different carboxy-terminal halves, which lead to distinct ligand binding specificity. The two forms of the Ig-III domain are derived from the tissue-specific inclusion of either exon IIIb or exon IIIc (36, 44). FGFR2(IIIb) primarily binds FGF10 and FGF7 and is the isoform of choice in epithelial cells, whereas FGFR2(IIIc) binds FGF2 with high affinity and is predominantly expressed in mesenchyme (36, 51). Proper cell-type-specific expression of each isoform is essential for maintaining FGF/FGFR2 signaling, which governs epithelial-mesenchymal interactions required for organogenesis in mouse embryos (17, 22). Mutations that alter the ligand specificity of FGFR2(IIIc) or those that lead to inappropriate expression of exon IIIb in mesenchyme have been linked to several developmental syndromes in humans (22, 43, 52). The physiological importance of regulating FGFR2 isoform choice is highlighted further by studies that show a switch from FGFR2(IIIb) to FGFR2(IIIc) during the progression of prostate carcinomas (6, 51).
The mutually exclusive incorporation of exon IIIb or exon IIIc is regulated by the complex interplay of cis-acting elements in FGFR2 pre-mRNA and trans-acting factors, some of which appear to be cell type specific. To study the mechanism of regulation, we employed two cell lines derived from Dunning rat prostate tumors. The DT3 cell line is a well-differentiated carcinoma and expresses FGFR2(IIIb) exclusively, whereas the AT3 cell line is poorly differentiated and solely expresses FGFR2(IIIc) (51). Skipping of exon IIIb in AT3 cells is facilitated by the presence of weak splice sites flanking this exon, an exonic silencing sequence in exon IIIb, and two intronic silencing elements. The upstream intronic splicing silencer (UISS) and the downstream intronic splicing silencer (DISS), which resides within the intronic control element (ICE), flank exon IIIb (8, 13, 20, 50). The exonic silencing sequence functions to recruit hnRNP A1 to exon IIIb, thereby repressing its inclusion (16), while UISS and ICE antagonize exon IIIb definition by binding the polypyrimidine tract binding protein (PTB) and other factors yet to be characterized (8, 49, 50). The silencing of exon IIIb is countered in epithelial cells by the action of several cis-acting elements. Intronic activating sequence 1 (IAS1), which is located downstream of exon IIIb, serves as a binding site for the splicing factor TIA-1. The binding of TIA-1 to IAS1 has been demonstrated to activate the weak 5′ splice site of exon IIIb as well as weak splice sites of other exons (15). IAS2 is located in the middle of the DISS within the ICE, while the intronic splicing activator and repressor (ISAR, also known as IAS3) is located over 700 nucleotides downstream of IAS2. Both of these cell-type-specific elements serve to activate the inclusion of exon IIIb (7, 13, 14). IAS2 and ISAR also function to repress exon IIIc inclusion in a cell-type-specific manner (7, 28, 50a). It is believed that IAS2 and a portion of ISAR, hereafter referred to as ISAR core, activate exon IIIb inclusion by creating a stem structure that disrupts the silencing activity of UISS and ICE (14, 28, 40, 49).
It has been shown that sequences within IAS2 and ISAR and their potential to form a stem have been highly conserved for over 600 million years (38), suggesting the importance of structure and perhaps sequence in the activation of exon IIIb inclusion and the repression of exon IIIc inclusion. Recently, it has been shown that similar stem structures are predicted to form in FGFR1 in several species (40). In addition, it has been shown that other sequences capable of stem formation can functionally substitute for IAS2 and ISAR (40).
In this study, we demonstrate that some, but not all, unrelated stem-forming sequences can functionally substitute for IAS2 and ISAR core. We also find that the 735 nucleotides separating IAS2 and ISAR core serve no function in the regulation of FGFR2 exon choice. Most importantly, we determine that the major function, if not the only function, of the IAS2-ISAR core stem in exon IIIb activation is to approximate intronic sequences upstream of IAS2 to sequences downstream of ISAR core. Splicing precursors where these sequences have been juxtaposed are capable of recapitulating epithelial cell-type-specific exon IIIb inclusion independent of IAS2 and ISAR core. Finally, we identified a GCAUG element downstream of ISAR core that is important for cell-type-specific exon IIIb inclusion and exon IIIc repression.
MATERIALS AND METHODS
Plasmid construction.
The plasmid DNA constructs used in this study were made by using standard cloning techniques described previously (7). The previously described minigenes pI-11 and pI-11 FS (7) are hereafter referred to as pI12 and pI12DE-WT (pI12 double exon wild type). To create minigenes containing only exon IIIb, PCR amplification of parental DE constructs with primers INT3BF2 and INT2R2 (7) was followed by digestion with SpeI and XhoI. The PCR products were cloned back into the XbaI and XhoI sites of pI12. To create minigenes containing only exon IIIc, parental DE constructs were digested with XbaI and XhoI and cloned into pI12. The pI12DE-Rep, pI12DE-Blue, pI12DE-Blue(c), and pI12DE-PyPu splicing constructs were created by digesting pI12DE-ΔIAS2 (50a) with ClaI and performing nondirectional annealed oligonucleotide cloning with the following oligonucleotides: IAS2 Rep F, IAS2 Rep R, IAS2 Blue F, IAS2 Blue R, IAS2 PyPu F, and IAS2 PyPu R. To create pI12DE-Blue Blue, pI12DE-Blue Blue(c), pI12DE-Blue(c) Blue, pI12DE-Blue(c) Blue(c), pI12DE-PyPu, and pI12DE-PyPuΔBulge, a portion of the plasmids containing substitutions in IAS2 were subcloned into pBluescript (Stratagene) by using XbaI and XhoI. These plasmids were then digested with NdeI and NsiI to remove ISAR, and annealed oligonucleotide cloning was performed to replace ISAR core within the wild-type ISAR sequence with the following oligonucleotides: ISAR Blue F, ISAR Blue R, ISAR Blue(c) F, ISAR Blue(c) R, ISAR PyPu F, ISAR PyPu R, ISAR PyPu ΔBulge F, and ISAR PyPu ΔBulge R. The FGFR2 sequence cloned into pBluescript was then subcloned back into pI12DE-WT digested with XbaI and XhoI. The XbaI-XhoI fragment of pI12DE-WT was subcloned into pBluescript (Stratagene) and then subjected to QuikChange mutagenesis (Stratagene) to create the following minigenes: pI12DE-ΔU, pI12DE-ΔG, pI12DE-+G, pI12DE-ΔBulge, pI12DE-C5, pI12DE-C10, pI12DE-C15, pI12DE-C20, pI12DE-C25, pI12DE-C30, pI12DE-C11, pI12DE-C13, pI12DE-C14, and pI12DE-C16. To create pI12DE-ΔLP, PCR amplification of pI12DE-WT with the primer sets INT3BF2 with IAS2-cla-R and INT3CR (7) with HinP1I-ISAR-F was followed by digestion with SpeI/ClaI and XhoI/HinP1I, respectively. The PCR products were sequentially cloned into pI12 digested with XbaI and ClaI and then with ClaI and XhoI. pI12DE-ΔSTLP was cloned by PCR amplification of pI12DE-WT with the primer sets INT3BF2 with del-stem-loop-R and del-stem-loop-F with INT3CR (7), followed by digestion with SpeI/ClaI and BsmBI/XhoI, respectively. The PCR products were sequentially cloned into pI12 digested with XbaI/ClaI and then BsmBI/XhoI. pI12DE-ΔSTLP C15 was cloned by PCR amplification of pI12DE-WT with INT3BF2 (7) and del-stem-loop-R and PCR amplification of pI12DE-C15 with del-stem-loop-C15-F and INT3CR (7), followed by digestion with SpeI/ClaI and BsmBI/XhoI. The PCR products were sequentially cloned into pI12 digested with XbaI/ClaI and then with BsmBI/XhoI. pI12DE-ΔSTLP C10-18 was cloned in the same manner as pI12DE-ΔSTLP C15 except del-stem-loop-C10-18-F and INT3CR (7) were used for PCR amplification from pI12DE-C15. pI12DE-C10-18 was cloned by PCR amplification of pI12DE-C15 with Intron2F and Core 10-18R, followed by digestion with BsiWI and XbaI. The PCR product was cloned into pI12DE-ΔSTLP C10-18 digested with XbaI and BsiWI. All constructs utilizing PCR amplification were sequenced for verification. All oligonucleotide sequences will be provided upon request.
Cell culture and transfection.
AT3 and DT3 cells were maintained in Dulbecco's modified Eagle medium (low glucose) supplemented with 10% fetal bovine serum (HyClone). Stable transfections were performed as previously described (7), with one exception; after trypsinization, the cells were reseeded in 25-cm2 flasks containing 500 μg of Geneticin (Gibco)/ml for selection of stable cell populations.
RNA isolation and RT-PCR assay of transfected minigenes.
Cellular RNA for reverse transcription (RT)-PCR and the Invader RNA assay was isolated by using the method of Chomczynski and Sacchi (10) or Trizol (Invitrogen). RT-PCRs using T7 and SP6 primers were performed as previously described (7). PCR products were either loaded directly onto 5% nondenaturing acrylamide gels or added to restriction digests with either AvaI or HincII (New England Biolabs). Analysis and quantification of PCR products from double-exon digests was performed as previously described (8). Analysis and quantification of PCR products from single-exon constructs (IIIb or IIIc) was performed in the following manner. Phosphorimager quantification of single-inclusion (U-IIIb-D or U-IIIc-D) and skipped bands (U-D) was performed with ImageQuant. A ratio of included to skipped products was calculated by dividing the moles of single inclusions by the sum of the moles of single inclusions and the moles of the skipped product.
Invader RNA assay.
The Invader RNA assay (Third Wave Technologies, Madison, Wis.) was carried out essentially as described by Eis et al. (18) except that 1 μM probe was used for all probe sets and 0.25 μM Invader oligonucleotides were used for the IIIb-IIIc and U-IIIc probe sets as described in Wagner et al. (50a). To analyze all splice variants for double-exon minigenes, the Invader RNA assays were run in the biplex format by using the probe set combinations IIIb-D/U-D and IIIb-IIIc/U-IIIc as previously described (50a). To analyze splice variants for single-exon transfected minigenes, the Invader RNA assays were run in the biplex format by using the probe set combinations IIIb-D/U-D and IIIc-D/U-D. Different concentrations of each RNA splice variant were used to calculate a standard graph comparing attomoles of RNA to fluorescence. From the fluorescence readings, absolute levels of each splice variant were calculated (see the legends to Fig. 1 and 5).
FIG. 1.
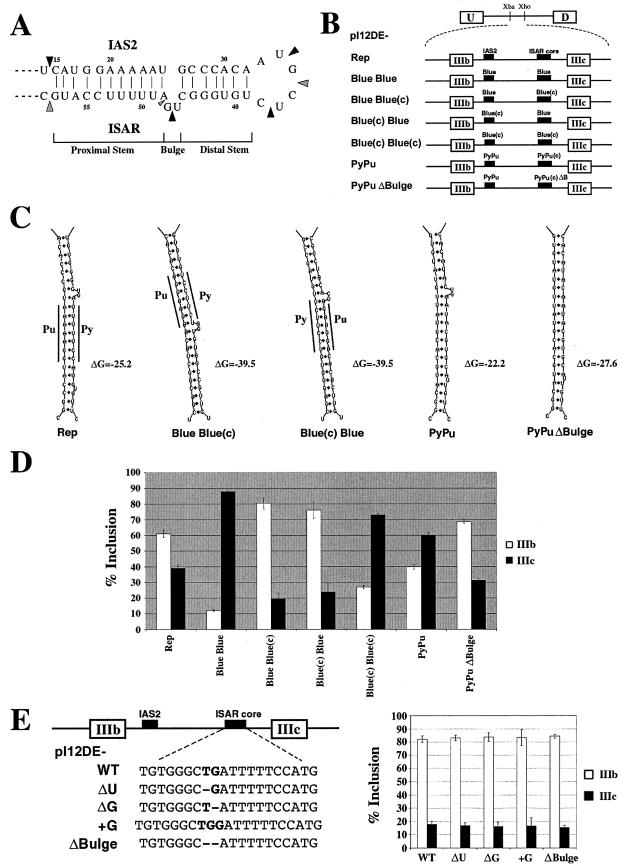
Stem formation dictates exon IIIb inclusion. (A) The structure of an in vitro synthesized 84-nucleotide chimeric RNA molecule containing the rat IAS2 and ISAR core sequences separated by a 6-nucleotide loop was probed with RNase A and RNase T1. Strong RNase A cleavage sites are indicated by large black arrowheads, and strong RNase T1 cleavage sites are indicated by large gray arrowheads. A small gray arrowhead indicates the weak RNase T1 cleavage site. (B) Minigenes used to test the sequence specificity of stem formation for exon IIIb activation in epithelial cells. pI12DE-Rep, pI12DE-Blue Blue(c), pI12DE-Blue(c) Blue, pI12DE-PyPu, and pI12DE-PyPuΔBulge are capable of stem formation, while pI12DE-Blue Blue and pI12DE-Blue(c) Blue(c) are not capable of stem formation. IAS2 and ISAR core or the sequences that replace them are represented as black boxes. (C) Sequence composition of the stems formed by the corresponding minigenes in panel B. Pu and Py demarcate the stretches of purines and pyrimidines in the stem structures. ΔG indicates the predicted Gibbs free energy value for each stem in kcal/mole as calculated by mFold (34). (D) Minigenes that are capable of stem formation recover activation of exon IIIb to various degrees (see Discussion). The percentage of exon inclusion (% IIIb inclusion = 100 × no. of U-IIIb-D transcripts/[no. of U-IIIb-D transcripts + no. of U-IIIc-D transcripts]; % IIIc inclusion = 100 × no. of U-IIIc-D transcripts/[no. of U-IIIb-D transcripts + no. of U-IIIc-D transcripts]) for the minigenes in panel B that were stably transfected into DT3 cells was determined by Invader RNA assay. (E) The two-nucleotide bulge in the stem structure is not necessary for IIIb inclusion. The left panel shows the minigenes used to test the effects of bulge mutations on exon IIIb inclusion. The right panel shows the quantification of RT-PCR analysis of stably transfected minigenes in DT3 cells.
FIG. 5.
A GCAUG element is critical for activation of exon IIIb in minigenes lacking IAS2-ISAR stem structure. (A) Schematic of minigene constructs used in panels B and C. The mutated nucleotides are indicated in bold print. IAS2 and ISAR core are represented as black boxes; ISAR core resides within the full ISAR element (represented as a gray box). The nucleotides in the gray box are within ISAR. (B) The percent inclusion among single-inclusion transcripts (U-IIIb-D and U-IIIc-D) for minigenes in panel A, which were stably transfected into DT3 cells, was determined by Invader RNA assay (e.g., % U-IIIb-D = 100 × no. of U-IIIb-D transcripts/[no. of U-IIIb-D transcripts + no. of U-IIIc-D transcripts]). (C) Quantification of all spliced products for minigenes in panel A, which were stably transfected in DT3 cells, was determined by Invader RNA assay (e.g., % U-IIIb-D = 100 × no. of U-IIIb-D transcripts/[no. of U-D transcripts + no. of U-IIIb-D transcripts + no. of U-IIIc-D transcripts + no. of U-IIIb-IIIc-D transcripts]).
RNA structure probing.
An 84-nucleotide chimeric RNA that included the Rattus norvegicus IAS2 and ISAR core sequences (bold) separated by an artificial 6-nucleotide loop (underlined) (5′-GGGAGAAGAGAAUUCAUGGAAAAAUGCCCACAAUGCUCUGUGGGCUGAUUUUUCCAUGCUAGAGUCGACCUGCAGGCAUGCAUA-3′) was synthesized by using T7 RNA polymerase as described previously (11). Structure probing by limiting digestion with RNase A and RNase T1 followed by primer extension with a 5′-end radiolabeled oligonucleotide (5′-TGCATGCCTGCAGGTC-3′) was performed as described by Mistry et al. (38).
RESULTS
Non-sequence-specific RNA structure mediates proper splicing regulation in DT3 cells.
Compensatory mutations in IAS2 and ISAR core elements, which partially rescue function, and phylogenetic data strongly suggest that a stem-like structure forms between IAS2 and ISAR core (14, 28, 38, 40). In order to provide physical evidence for the predicted structure, we synthesized a chimeric RNA molecule containing the rat IAS2 and ISAR core sequences joined by a six-nucleotide loop and probed its structure in vitro (11). After the RNA was partially digested by RNase T1, which cleaves after single-stranded guanosine residues, or RNase A, which cleaves preferentially after single-stranded uridine residues, we mapped cleavage sites with primer extension. Residues in the predicted stem-forming IAS2 and ISAR core sequences were protected from digestion, whereas residues in the predicted GU bulge and the loop were cleaved (Fig. 1A). Both structure probing and phylogeny suggest a stem structure divided into three parts: a proximal stem, a two-nucleotide bulge, and a distal stem (Fig. 1A).
The primary sequence of the proximal stem has been conserved from mammals to Strongylocentrotus purpuratus; of particular interest is the stretch of adenosines and uridines in IAS2 and ISAR, respectively (38). Also conserved is a predicted two-nucleotide bulge that was shown above to form in the rat stem structure. Although the presence of the bulge has been maintained throughout evolution, the nucleotide content of the bulge has varied, as has the primary sequence of the distal portion of the stem. These observations led us to question whether both the sequence of the proximal stem and the overall structure were critical for the proper regulation of FGFR2. In order to test the importance of sequence composition, we made several minigene constructs, as shown in Fig. 1B. Two of these minigenes [pI12DE-Blue Blue(c) and pI12DE-Blue(c) Blue] have the capacity to form a stem containing a two-nucleotide bulge, while the other two minigenes [pI12DE-Blue Blue and pI12DE-Blue(c) Blue(c)] cannot (Fig. 1B and C). These minigenes were transfected into DT3 and AT3 cells, and RNA from stably transfected cell populations was analyzed by RT-PCR or the Invader RNA assay (18, 50a). The transfections into AT3 cells were performed to control for any effect on the silencing of exon IIIb, which is mediated in part by silencers flanking IAS2. Alterations of IAS2 and ISAR were not expected to affect IIIc inclusion in AT3 cells, and indeed, we observed nearly exclusive use of exon IIIc with each minigene tested (data not shown). In DT3 cells transfected with pI12DE-Blue Blue(c) and pI12DE-Blue(c) Blue, we observed 80 and 76% exon IIIb inclusion, respectively (Fig. 1D). These heterologous stems allowed for even greater exon IIIb inclusion than did pI12DE-Rep (80 and 76% compared to 61%), which contains the authentic IAS2 and ISAR core sequences (Fig. 1C and D). The constructs that could not form a stem lost the ability to effectively include exon IIIb. To determine if the different stem structures were separately involved in activating exon IIIb and repressing exon IIIc, we made minigenes that contained only exon IIIb or exon IIIc as the internal exon (7). These single-exon constructs revealed that stem formation in epithelial cells activated exon IIIb inclusion and repressed exon IIIc inclusion (data not shown). These data and the recent report of Muh et al. (40) suggested that the replacement of IAS2 and ISAR core with presumably unrelated complementary sequences could elicit cell-type-specific regulation of exon IIIb inclusion.
We noticed, however, that all functional stems, including those reported in Muh et al. (40), contained stretches of purines in one strand and complementary pyrimidines on the other (Fig. 1C). Indeed, a stretch of five adenosines and two guanosines in IAS2 and a stretch of five uridines and two cytidines are highly conserved in vertebrates (38). To test whether a stretch of purines and a complementary track of pyrimidines were important, we generated pI12DE-PyPu, which contains alternating pyrimidines and purines in the stem structure (Fig. 1B and C). When this construct was transfected into AT3 cells, almost exclusive use of exon IIIc was observed (data not shown). When this construct was transfected into DT3 cells, pI12DE-PyPu included exon IIIb 40% of the time, which is lower than the 61% IIIb inclusion observed with pI12DE-Rep (Fig. 1D). This result suggested that the stretches of purines and pyrimidines, while not critical, may influence the proper regulation of FGFR2 alternative splicing (see below). Combining the results discussed above and the recent findings of Muh et al. (40), we concluded that many different stems can substitute for IAS2 and ISAR core; however, clearly not all stems regulate exon choice equally.
Deletion of the two-nucleotide bulge has no effect on FGFR2 splicing regulation.
Bulges in RNA secondary structures tend to function by providing unique recognition sites for protein interactions, either by creating a specific RNA topography for protein binding in an otherwise helical structure or by kinking the RNA backbone, allowing access for protein recognition (24). In the case of the human immunodeficiency virus type 1 (HIV-1) trans-activation response element RNA stem structure, a tri-nucleotide bulge serves as a docking site for the binding of Tat. Mutating the bulge destroys the ability of the RNA to bind Tat (45, 46).
Given that both the IAS2 and ISAR core in vitro structure probing and phylogenetic data indicated the presence of a two-nucleotide bulge, we wanted to determine if mutating the bulge would affect exon choice. To that end, we created pI12DE-PyPuΔBulge, which contains the same stem as pI12DE-PyPu with a deletion of the bulge (Fig. 1B and C). When this minigene was transfected into AT3 cells, exon IIIc was included almost exclusively (data not shown). Interestingly, when pI12DE-PyPuΔBulge was transfected into DT3 cells, it gave 69% IIIb inclusion, which is higher than the 40 and 61% IIIb inclusion observed with pI12 PyPu and pI12 Rep, respectively (Fig. 1D). This result demonstrates that the presence of a two-nucleotide bulge is not necessary for IIIb activation; indeed, the presence of this bulge decreased exon IIIb inclusion in the context of the PyPu stem. Moreover, given that pI12DE-PyPuΔBulge lacks any stretches of pyrimidines and purines, these data argue against any role for the conserved sequence composition in the proximal stem of the IAS2-ISAR core structure in exon IIIb activation.
In order to analyze the function of the GU bulge in the authentic stem formed between IAS2 and ISAR core (Fig. 1A), we created pI12DE-ΔU, which contains a deletion of the U, pI12DE-ΔG, which contains a deletion of the G, pI12DE-+G, which has an extra G in the bulge, and pI12DE-ΔBulge, which has the bulge entirely deleted (Fig. 1E). These minigenes were transfected into AT3 and DT3 cells, and RNA from stable cell populations was pooled and analyzed by RT-PCR and the Invader RNA assay. All of the minigenes that were transfected into AT3 cells resulted in the nearly exclusive use of exon IIIc (data not shown). None of the minigenes tested in DT3 cells resulted in a significant decrease in exon IIIb inclusion (Fig. 1E). All the constructs tested included exon IIIb in greater than 80% of all single-inclusion splicing events. These results were confirmed with the Invader RNA assay (data not shown), demonstrating that the bulge has no apparent function in the proper regulation of exon choice.
A six-nucleotide loop is sufficient to mediate proper regulation of FGFR2 splicing.
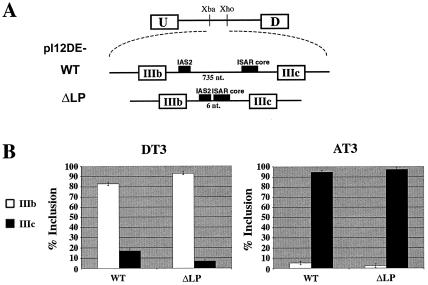
The IAS2-ISAR core structure contains a 735-nucleotide loop. If loop sequences were to contribute to function, deletion of the loop would be predicted to impact splicing regulation. A previously reported loop deletion construct, which resulted in a 126-base pair loop, was shown to have no effect on FGFR2 exon choice (7). Moreover, it was recently shown that a deletion of the majority of the loop sequence between IAS2 and ISAR core resulted in the proper epithelium-specific inclusion of exon IIIb (40). However, in this study the minimal spacer region between IAS2 and ISAR was not determined. To test the importance of these sequences, we created a minigene with a six-nucleotide loop (Fig. 2A). The six-nucleotide loop minigene (pI12DE-ΔLP) had a deletion of the entire loop region between IAS2 and ISAR core with the exception of two nucleotides downstream of IAS2, three nucleotides of unrelated spacer sequence, and one nucleotide upstream of ISAR core. Transcripts from this minigene included exon IIIc almost exclusively in AT3 cells and included exon IIIb predominantly in DT3 cells (Fig. 2B). Remarkably, pI12DE-ΔLP was more tightly regulated than the wild-type minigene, as exon IIIb was included in 92% of all single-exon inclusion events in DT3 cells compared to 83% for pI12DE-WT, and exon IIIc was included 98.5% of the time in AT3 cells compared to 93% for pI12DE-WT (Fig. 2B). Analysis of all possible splicing events in DT3 cells further suggested tight regulation of pI12DE-ΔLP, as double inclusion (transcripts that include both IIIb and IIIc) decreased from 28% for pI12DE-WT to 18% for pI12DE-ΔLP (data not shown). This suggests that the decreased loop length allowed for greater repression of exon IIIc in epithelial cells, perhaps because smaller loops may facilitate stem formation. These results demonstrated that the loop was not essential for proper splicing regulation in either cell line.
FIG. 2.
Loop sequences are not necessary for cell-type specific exon inclusion. (A) Minigenes used to test the importance of loop sequence on cell-type-specific exon inclusion. pI12DE-WT contains 735 nucleotides between IAS2 and ISAR, whereas pI12DE-ΔLP contains 6 nucleotides between IAS2 and ISAR. IAS2 and ISAR core are indicated as black boxes. (B) Quantification of RT-PCR analysis of stably transfected minigenes in DT3 and AT3 cells.
Stem formation functions to approximate sequences upstream of IAS2 and downstream of ISAR core.
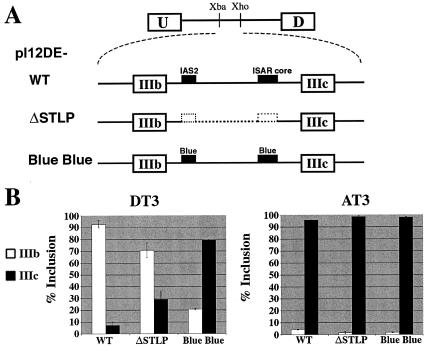
The data presented above and data previously published by Muh et al. (40) show that both the IAS2-ISAR core structure and the loop sequences between IAS2 and ISAR core can be replaced with heterologous stem-loop structures. How does this non-sequence-specific structure mediate cell-type-restricted activation of exon IIIb and repression of exon IIIc? Two general scenarios can provide an answer to this question. In the first scenario, the RNA duplex must be recognized by cell-type-specific trans-acting factors that control exon choice. These trans-acting factors would bind the stem without sequence specificity, which would be provided by neighboring cis elements. In a second scenario, the RNA duplex need not be recognized but rather functions by approximation of sequences. Given that many and diverse stems can substitute for the IAS2-ISAR core structure, we favored approximation as the operative mechanism. In order to distinguish between the above scenarios, we created a minigene that deleted IAS2, the loop sequence, and ISAR core (pI12DE-ΔSTLP in Fig. 3A). The deletion in ΔSTLP transcripts was designed to mimic the approximation of sequences caused by the IAS2-ISAR core stem. RT-PCR and the Invader RNA assay revealed that ΔSTLP transcripts predominantly included exon IIIb in DT3 cells (70%) and, almost exclusively, exon IIIc in AT3 cells (Fig. 3B and data not shown). Cell-type-specific exon choice was achieved even though this construct lacked IAS2, ISAR core, or any RNA stem. The fact that ΔSTLP transcripts included IIIc predominantly in AT3 cells strongly suggests that exon IIIb was not globally activated by intron shortening or by unintended disruption of intronic silencer elements upstream of the deletion. These results ruled out the first scenario, in which double-stranded RNA binding trans-acting factors are required for exon IIIb activation, and demonstrated that approximating sequences upstream of IAS2 to sequences downstream of ISAR core was sufficient for this activation.
FIG. 3.
Approximation of sequences upstream of IAS2 to sequences downstream of ISAR core allows for the proper cell-type-specific exon inclusion. (A) Minigenes used to test the importance of approximating sequences upstream of IAS2 with sequences downstream of ISAR core. pI12DE-WT is described in the legend to Fig. 2A. The pI12DE-ΔSTLP minigene contains a deletion of the entire IAS2-ISAR core stem-loop structure from the 5′ end of IAS2 to the 3′ end of ISAR core, which mimics the predicted outcome of IAS2-ISAR stem formation. The pI12DE-Blue Blue minigene, which is not capable of stem formation, is described in the legend to Fig. 1D. IAS2 and ISAR core are indicated as black boxes. (B) Quantification of RT-PCR analysis of stably transfected minigenes in DT3 and AT3 cells.
Although approximation was sufficient for exon IIIb activation in DT3 cells, it was not sufficient for efficient exon IIIc repression. We noted that double-inclusion transcripts, which contain both IIIb and IIIc, increased from 20% of all spliced products for pI12DE-WT to 45% for pI12DE-ΔSTLP (data not shown; see Fig. 5C and text below). These data suggest that although exon IIIb inclusion was not significantly affected by the deletion of the stem-loop structure, this structure was required for efficient exon IIIc repression in epithelial cells.
Sequences downstream of the ISAR core are important for exon IIIb inclusion in DT3 cells.
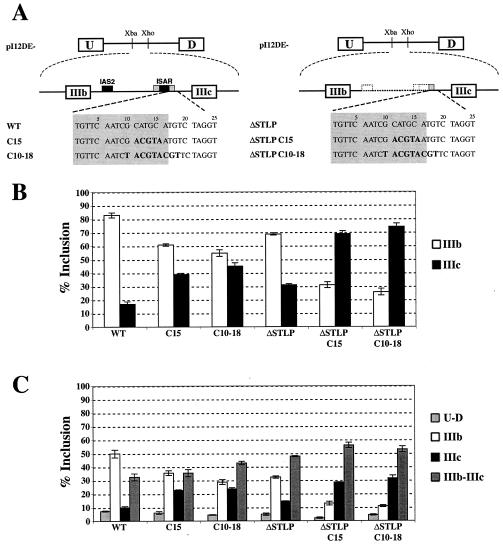
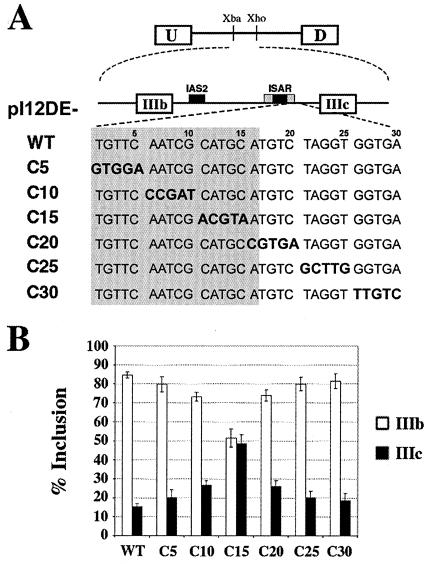
Downstream of ISAR core we had observed high conservation of sequence from echinoderms to mammals (38). Understanding that strong sequence conservation within introns can be a good predictor of important cis elements, we performed a mutational analysis of the sequence downstream of ISAR core. Initially, we created six minigenes, which contained five-nucleotide substitutions sequentially positioned downstream of ISAR core (Fig. 4A). These minigenes were transfected into DT3 cells, and the isolated RNA from stable cell populations was analyzed by RT-PCR. The C5, C25, and C30 mutations (in nucleotides 1 to 5, 21 to 25, and 26 to 30 downstream of ISAR core) had no effect on exon IIIb inclusion compared to that for wild type, and the C10 and C20 mutations (in nucleotides 6 to 10 and 16 to 20) slightly decreased exon IIIb inclusion compared to that for wild type. When nucleotides 11 to 15 were mutated in the C15 construct, exon IIIb levels dropped from 85 to 51% (Fig. 4B), suggesting that these nucleotides play a key role in exon IIIb inclusion and IIIc repression in DT3 cells. To determine if these nucleotides were separately involved in activating exon IIIb and repressing exon IIIc, we created single-exon IIIb and single-exon IIIc minigenes and transfected them into the DT3 cell line. This analysis demonstrated that the primary effect of mutating these nucleotides is to lower IIIb inclusion from 60 to 36%; moreover, the C15 mutation also allowed a slight increase of from 21 to 32% in exon IIIc inclusion in DT3 cells (data not shown). These results suggest that an important cis element is located 11 to 15 nucleotides downstream of ISAR core and that its effect is more critical for exon IIIb inclusion than for exon IIIc repression. This region contains the sequence GCAUGCAUG, which has two overlapping GCAUG motifs, and the C15 mutation is the only one to destroy both GCAUG copies. These GCAUG motifs could be the regulatory elements downstream of ISAR core that are approximated by the IAS2-ISAR core RNA duplex.
FIG. 4.
A GCAUG element immediately downstream of ISAR core plays a role in exon IIIb inclusion. (A) Minigenes with five-nucleotide mutations downstream of ISAR core. The five-base-pair substitution mutations are indicated with bold print. IAS2 and ISAR core are represented as black boxes; ISAR core resides within the full ISAR element (represented as a gray box). The nucleotides in the gray box are within ISAR. (B) Quantification of RT-PCR analysis of stably transfected minigenes in DT3 cells.
While the GCAUG element plays a role in both activation of exon IIIb and repression of exon IIIc, the IAS2-ISAR core structure is required only for exon IIIc repression.
We demonstrated above that the major function of the IAS2-ISAR stem for exon IIIb activation is to approximate sequences upstream of IAS2 with those downstream of ISAR core, and we also showed that a GCAUG sequence element downstream of ISAR core is required for activation of exon IIIb and repression of exon IIIc. We also presented data that suggested a role for a secondary structure in repression of exon IIIc. We decided to evaluate this role and to test the importance of the GCAUG element in the absence of this structure. In addition to pI12DE-C15 (described above), we created pI12DE-C10-18, which completely mutates the GCAUG overlapping repeats by substituting nine nucleotides in place of the GCAUGCAUG sequence downstream of ISAR core (Fig. 5A). We also created pI12DE-ΔSTLP C15 and pI12DE-ΔSTLP C10-18, which contained the C15 and C10-18 mutations in minigenes lacking the IAS2-ISAR stem-loop sequences (Fig. 5A).
The six minigenes shown in Fig. 5A were transfected into DT3 cells, and RNA isolated from stable transfections was analyzed by RT-PCR (data not shown) and by the Invader RNA assay (Fig. 5B and C). As expected, the C15 and C10-18 mutations in the context of the pI12DE-WT minigene led to a significant decrease in exon IIIb inclusion. This decrease in exon IIIb inclusion was observed both by measuring the percentage of single-inclusion transcripts that included IIIb (100 × no. of U-IIIb-D transcripts/[no. of U-IIIb-D transcripts + no. of U-IIIc-D transcripts]) (Fig. 5B) and by measuring the percentage of all transcripts that included IIIb (100 × [no. of U-IIIb-D transcripts + no. of U-IIIb-IIIc-D transcripts]/[no. of U-D transcripts + no. of U-IIIb-D transcripts + no. of U-IIIc-D transcripts + no. of U-IIIb-IIIc-D transcripts]) (Fig. 5C). As noted above with RT-PCR analysis, the mutations of the GCAUG element also led to increased exon IIIc inclusion, suggesting that this element was required for efficient repression of exon IIIc in DT3 cells. Analysis of the deletion of IAS2, the loop region, and ISAR core (ΔSTLP) by the Invader RNA assay (Fig. 5B and C) confirmed the results of our RT-PCR analysis (Fig. 3). Specifically, the overall inclusion of exon IIIb (the sum of the no. of U-IIIb-D and the no. of U-IIIb-IIIc-D transcripts) was not affected in ΔSTLP relative to that for WT (Fig. 5C), and all of the decrease in IIIb single-inclusion transcripts observed in Fig. 5B could be accounted for by an increase in double-inclusion transcripts. The data, however, indicate that the ΔSTLP transcripts are not capable of efficient exon IIIc repression. The C15 and C10-18 mutations in the context of ΔSTLP transcripts led to precisely the expected results: a decrease in exon IIIb inclusion due to the disruption of the GCAUG element and a dramatic increase in overall exon IIIc inclusion due to the combined losses of the GCAUG element and the IAS2-ISAR core structure.
DISCUSSION
Proper exon IIIb or exon IIIc choice in FGFR2 transcripts depends on base pairing between IAS2 and ISAR core sequences (14), as revealed by compensatory mutations (14, 28) and phylogenetic analysis (38, 40). The latter revealed remarkable conservation of the IAS2-ISAR core secondary structure between echinoderms and mammals, whose common ancestor was last extant 600 million years ago (38). Perhaps equally impressive was the conservation of sequence noted for the proximal stem of the IAS2-ISAR core structure (38) (Fig. 1A). Although at first glance this could be taken as evidence for the importance of the primary sequence, the specific nucleotides conserved, GAAAAAU(U) in IAS2 and AUUUUU(U) in ISAR core, hint otherwise. The conserved tracts, A5 in IAS2 and U5 in ISAR core, could be the result of evolutionary drift secondary to the lower mutability of A and T versus that of G and C, as has been observed with human pseudogenes (21). Indeed, the results obtained with the PyPu ΔBulge transcript (above), which although fully functional does not have a tract of purines base paired to a tract of pyrimidines, demonstrate that structure and not sequence was important. Yet not all stem structures provided equal regulation. Even though we cannot categorically explain these differences between RNA structures, their relative stability, as calculated from the predicted ΔG (Fig. 1C), correlated well with activation of exon IIIb inclusion (see discussion of the findings of Brown et al., below). Thus, we conclude, in agreement with Muh et al. (40), that base pairing between IAS2 and ISAR core, rather than the unique sequences of these elements, is critical for regulation of FGFR2 alternative splicing.
Secondary structures have been proposed to act as cis elements, and their action can be exerted in multiple ways. RNA duplexes can be recognized, usually in a sequence-independent fashion, by double-stranded RNA binding proteins (19, 47). Specificity can be provided by direct or indirect interactions with neighboring cis-acting elements or single-stranded regions within the structures (30). RNA duplexes can also either occlude or favorably display other cis-acting elements (2, 3, 41). Finally, RNA duplexes can approximate otherwise distant cis elements. In this manuscript, we show that the major, if not the sole, function of the IAS2-ISAR core structure in exon IIIb activation is to approximate elements upstream of IAS2 to elements downstream of the ISAR core. To properly mimic the propinquity mediated by the IAS2-ISAR core structure, we deleted all the sequences from the 5′ end of IAS2 to the 3′ end of ISAR core, inclusively, in the plasmid of ΔSTLP. ΔSTLP transcripts included exon IIIb predominantly in DT3 cells and exon IIIc almost exclusively in AT3, which indicated correct regulation in the absence of IAS2, ISAR core, and the structure they form. Therefore, we conclude that the IAS2-ISAR core structure works by bringing together other elements. Note that a previous approximation that did not precisely mimic IAS2-ISAR core stem formation did not promote exon IIIb inclusion in epithelial cells (14). Approximation of cis elements by RNA secondary structures has precedent in pre-mRNA processing. The elaborate RexRE structure promotes polyadenylation of human T-cell leukemia virus type 1transcripts by approximating two required elements, the canonical AAUAAA hexanucleotide and the downstream G/U element, which in the primary sequence lie hundreds of nucleotides away (1). Brown et al. (4) showed that many RNA secondary structures can substitute for the RexRE to activate polyadenylation in this context and suggested that the strength of the base pairing interaction and the spatial distance between the approximated elements were critical for activity. RNA duplexes have also been shown to enhance splice site pairing in single intron transcripts (9, 31, 42) and exon inclusion in two intron transcripts in yeast (25). In these cases, the proximity of sequences presumably enhances the interactions between splicing complexes assembled at the ends of introns. Here, we report a case of duplex-mediated approximation of intronic control elements in FGFR2 transcripts, a first for cell-type-specific regulatory sequences.
Even though several questions remain about how approximation leads to exon IIIb activation in epithelial cells, parsimony suggests that the IAS2-ISAR core stem exists in both epithelium and mesenchyme, but only in the former are the approximated sequences recognized by epithelium-specific factors. The identity of the approximated sequences has not been unambiguously ascertained; however, data shown above and prior work suggest likely candidates. In this manuscript, we show that exon IIIb activation requires nucleotides 11 to 15 downstream of ISAR core. These nucleotides are found within the sequence (+10) GCAUGCAUG (+18), which contains two repeats of the previously characterized intronic activating sequence, GCAUG (5, 12, 23, 26, 29, 32, 39). This cis element has been shown to interact with several trans-acting factors and is believed to counteract the action of splicing repressors such as PTB (27, 33, 37). Thus, it is likely that the GCAUG-containing element is the relevant sequence downstream of ISAR core. Our previous work suggests that the pertinent sequence upstream of IAS2 is the intronic splicing silencer (ISS) immediately upstream of IAS2 (50). The juxtaposition of the GCAUG element to the ISS, which binds PTB, must be required to counteract the silencer and mediate inclusion of exon IIIb in epithelial cells.
Although our data indicate that the cell-type-specific factor(s) that control(s) exon IIIb inclusion need not interact with IAS2-ISAR core sequences, note that approximation does not restore repression of exon IIIc in epithelial cells. Careful quantification of exon IIIc (single-inclusion) and exons IIIb and IIIc (double-inclusion) transcript levels by the Invader RNA assay demonstrated reproducibly higher levels of exon IIIc inclusion in ΔSTLP transcripts than in WT transcripts. While it is possible that the topology of the ΔSTLP transcripts does not fully reproduce the approximation created by the IAS2-ISAR core structure, it is also conceivable that the IAS2-ISAR core structure plays a direct role in repressing exon IIIc in epithelial cells. One could envision an exon IIIc repressor that interacts with the IAS2-ISAR core and also with adjacent single-stranded RNA cis elements. This model might be similar to the interactions observed between the Sm protein heterohexamer and multiple sites within U snRNAs (35).
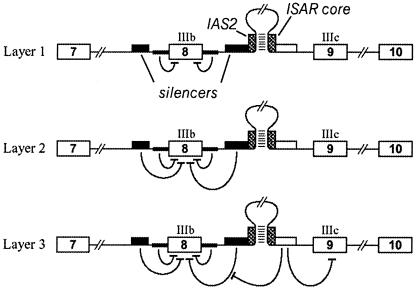
The tissue-specific use of exon IIIb appears to be regulated by the superimposition of three layers of control (Fig. 6). The first layer is determined by the weak splice sites bordering exon IIIb (e.g., the 5′ splice site is GUAACA instead of the canonical GURAGU) and leads to poor exon definition. The second layer is provided by dual ISSs, upstream and downstream of exon IIIb, which mediate their action via PTB and other unidentified repressors. Whereas exon IIIb silencing is dominant in fibroblasts, all the required factors for silencing are also present in epithelial cells. In these cells, however, a cell-type-specific third layer that requires the IAS2-ISAR core structure activates exon IIIb. We propose that exon IIIb activation is carried out by approximation of the GCAUG element to the neighborhood of the ISS. Only in epithelial cells is this element active, very likely because of the differential expression of factors that recognize the GCAUG element. The coordinated repression of exon IIIc requires an overlapping but not identical set of elements and factors.
FIG. 6.
Layers of control in the alternative splicing of FGFR2 transcripts. The schematic shows three layers of control that determine the choice between inclusion of exon IIIb and exon IIIc. A primary transcript from the FGFR2 gene is shown containing the IAS2-ISAR core structure within intron 8. Layer 1 shows the negative effect of weak splice sites on the definition of exon IIIb. Layer 2 represents the influence of silencer elements that regulate exon IIIb inclusion. These elements operate in both mesenchymal and epithelial cells. Layer 3 depicts epithelial-specific activity that promotes inclusion of IIIb and repression of IIIc. We propose that approximation of sequences downstream of the ISAR core to the vicinity of intronic silencers counters these elements and promotes inclusion of exon IIIb.
It is not surprising that tissue-specific regulation of alternative splicing requires multiple layers, each integrating the action of multiple factors. Much has already been discussed about the importance of combinatorial regulation as a way of providing diversity of functional states with a small number of regulators (48). The existence of tiers of regulation is also to be expected, since alternative splicing systems have developed by sequential evolution and are not established de novo (38). In the case of FGFR2 transcripts, the top tier of control is determined by the approximation of cis elements via a stable RNA duplex.
Acknowledgments
We thank Robert Brazas and Robin Wharton for critical reading of the manuscript and members of the Garcia-Blanco laboratory for many helpful discussions. We also thank Annette Kennett for her assistance in the preparation of the manuscript.
This research was supported by PHS grants to M.A.G.-B. (GM63090). E.J.W. and A.P.B. acknowledge the support of a Department of Defense predoctoral fellowship, and A.P.B. also acknowledges the support of a PHS training grant.
REFERENCES
- 1.Ahmed, Y. F., G. M. Gilmartin, S. M. Hanly, J. R. Nevins, and W. C. Greene. 1991. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell 64:727-737. [DOI] [PubMed] [Google Scholar]
- 2.Blanchette, M., and B. Chabot. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 3:405-419. [PMC free article] [PubMed] [Google Scholar]
- 3.Bratt, E., and M. Ohman. 2003. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA 9:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, P. H., L. S. Tiley, and B. R. Cullen. 1991. Effect of RNA secondary structure on polyadenylation site selection. Genes Dev. 5:1277-1284. [DOI] [PubMed] [Google Scholar]
- 5.Carlo, T., D. A. Sterner, and S. M. Berget. 1996. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA 2:342-353. [PMC free article] [PubMed] [Google Scholar]
- 6.Carstens, R. P., J. V. Eaton, H. R. Krigman, P. J. Walther, and M. A. Garcia-Blanco. 1997. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene 15:3059-3065. [DOI] [PubMed] [Google Scholar]
- 7.Carstens, R. P., W. L. McKeehan, and M. A. Garcia-Blanco. 1998. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol. Cell. Biol. 18:2205-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstens, R. P., E. J. Wagner, and M. A. Garcia-Blanco. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7388-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chebli, K., R. Gattoni, P. Schmitt, G. Hildwein, and J. Stevenin. 1989. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol. Cell. Biol. 9:4852-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Colvin, R. A., and M. A. Garcia-Blanco. 1992. Unusual structure of the human immunodeficiency virus type 1 trans-activation response element. J. Virol. 66:930-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deguillien, M., S. C. Huang, M. Moriniere, N. Dreumont, E. J. Benz, Jr., and F. Baklouti. 2001. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood 98:3809-3816. [DOI] [PubMed] [Google Scholar]
- 13.Del Gatto, F., and R. Breathnach. 1995. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 15:4825-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Moerlooze, L., B. Spencer-Dene, J. Revest, M. Hajihosseini, I. Rosewell, and C. Dickson. 2000. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127:483-492. [DOI] [PubMed] [Google Scholar]
- 18.Eis, P. S., M. C. Olson, T. Takova, M. L. Curtis, S. M. Olson, T. I. Vener, H. S. Ip, K. L. Vedvik, C. T. Bartholomay, H. T. Allawi, W. P. Ma, J. G. Hall, M. D. Morin, T. H. Rushmore, V. I. Lyamichev, and R. W. Kwiatkowski. 2001. An invasive cleavage assay for direct quantitation of specific RNAs. Nat. Biotechnol. 19:673-676. [DOI] [PubMed] [Google Scholar]
- 19.Fierro-Monti, I., and M. B. Mathews. 2000. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 25:241-246. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, E., F. Del Gatto, P. Champion-Arnaud, M. C. Gesnel, and R. Breathnach. 1993. Control of BEK and K-SAM splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol. Cell. Biol. 13:5461-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graur, D., and W.-H. Li. 2000. Fundamentals of molecular evolution, 2nd ed. Sinauer, Sunderland, Mass.
- 22.Hajihosseini, M. K., S. Wilson, L. De Moerlooze, and C. Dickson. 2001. A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc. Natl. Acad. Sci. USA 98:3855-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedjran, F., J. M. Yeakley, G. S. Huh, R. O. Hynes, and M. G. Rosenfeld. 1997. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl. Acad. Sci. USA 94:12343-12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermann, T., and D. J. Patel. 2000. RNA bulges as architectural and recognition motifs. Struct. Fold Des. 8:R47-R54. [DOI] [PubMed] [Google Scholar]
- 25.Howe, K. J., and M. Ares, Jr. 1997. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl. Acad. Sci. USA 94:12467-12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh, G. S., and R. O. Hynes. 1994. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 8:1561-1574. [DOI] [PubMed] [Google Scholar]
- 27.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, R. B., R. P. Carstens, Y. Luo, and W. L. McKeehan. 2001. 5′- and 3′-terminal nucleotides in the FGFR2 ISAR splicing element core have overlapping roles in exon IIIb activation and exon IIIc repression. Nucleic Acids Res. 29:3557-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamoto, S. 1996. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J. Biol. Chem. 271:17613-17616. [PubMed] [Google Scholar]
- 30.Lehmann, K. A., and B. L. Bass. 1999. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 291:1-13. [DOI] [PubMed] [Google Scholar]
- 31.Libri, D., F. Stutz, T. McCarthy, and M. Rosbash. 1995. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA 1:425-436. [PMC free article] [PubMed] [Google Scholar]
- 32.Lim, L. P., and P. A. Sharp. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 35.McConnell, T. S., R. P. Lokken, and J. A. Steitz. 2003. Assembly of the U1 snRNP involves interactions with the backbone of the terminal stem of U1 snRNA. RNA 9:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miki, T., D. P. Bottaro, T. P. Fleming, C. L. Smith, W. H. Burgess, A. M. Chan, and S. A. Aaronson. 1992. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc. Natl. Acad. Sci. USA 89:246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 38.Mistry, N., W. Harrington, E. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA 9:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muh, S. J., R. H. Hovhannisyan, and R. P. Carstens. 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277:50143-50154. [DOI] [PubMed] [Google Scholar]
- 41.Muro, A. F., M. Caputi, R. Pariyarath, F. Pagani, E. Buratti, and F. E. Baralle. 1999. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 19:2657-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman, A. 1987. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 6:3833-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oldridge, M., E. H. Zackai, D. M. McDonald-McGinn, S. Iseki, G. M. Morriss-Kay, S. R. Twigg, D. Johnson, S. A. Wall, W. Jiang, C. Theda, E. W. Jabs, and A. O. Wilkie. 1999. De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am. J. Hum. Genet. 64:446-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orr-Urtreger, A., M. T. Bedford, T. Burakova, E. Arman, Y. Zimmer, A. Yayon, D. Givol, and P. Lonai. 1993. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev. Biol. 158:475-486. [DOI] [PubMed] [Google Scholar]
- 45.Puglisi, J. D., R. Tan, B. J. Calnan, A. D. Frankel, and J. R. Williamson. 1992. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257:76-80. [DOI] [PubMed] [Google Scholar]
- 46.Roy, S., U. Delling, C. H. Chen, C. A. Rosen, and N. Sonenberg. 1990. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 4:1365-1373. [DOI] [PubMed] [Google Scholar]
- 47.Ryter, J. M., and S. C. Schultz. 1998. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17:7505-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 49.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 50a.Wagner, E. J., M. Curtis, N. Robson, A. P. Baraniak, P. Eis, and M. A. Garcia. RNA, in press. [DOI] [PMC free article] [PubMed]
- 51.Yan, G., Y. Fukabori, G. McBride, S. Nikolaropolous, and W. L. McKeehan. 1993. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol. Cell. Biol. 13:4513-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, K., A. B. Herr, G. Waksman, and D. M. Ornitz. 2000. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc. Natl. Acad. Sci. USA 97:14536-14541. [DOI] [PMC free article] [PubMed] [Google Scholar]