Abstract
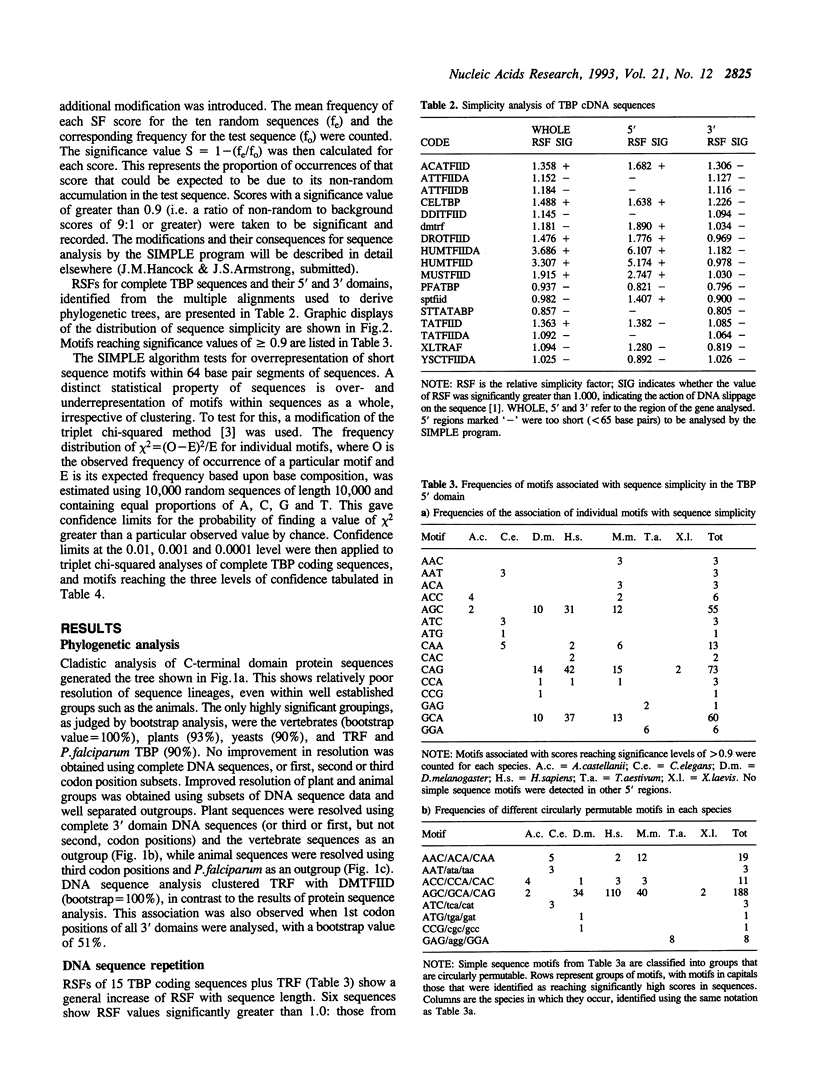
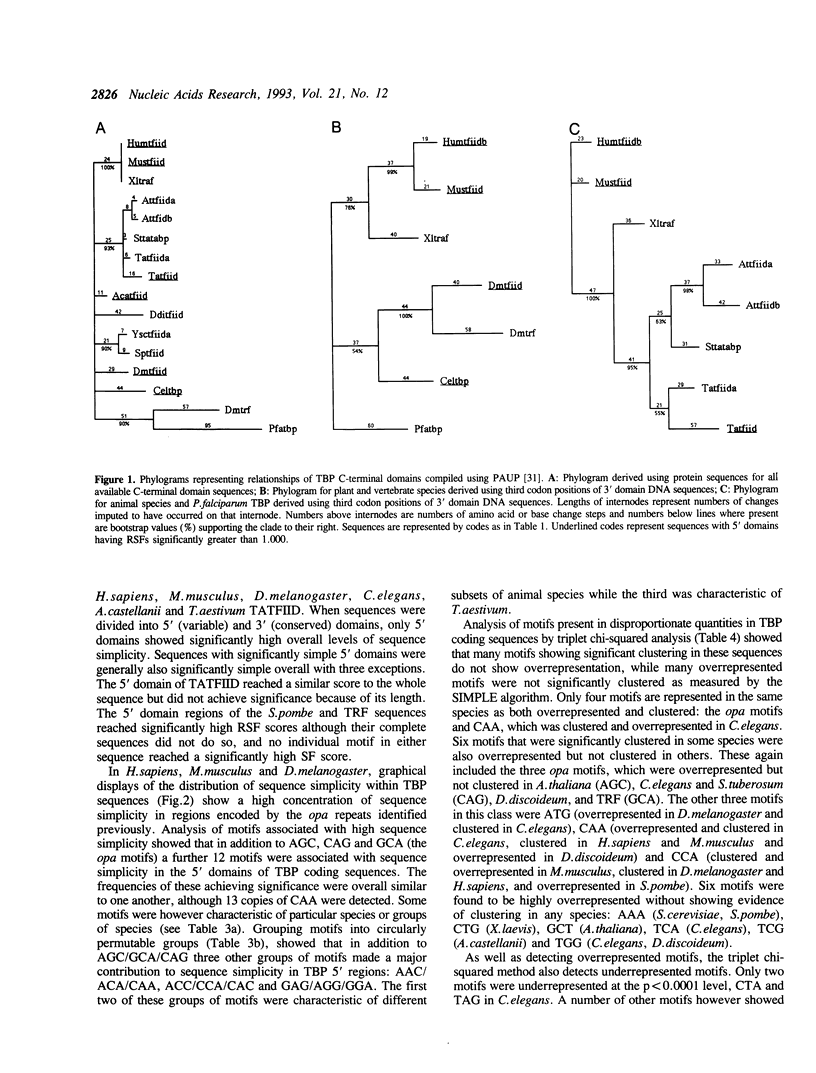
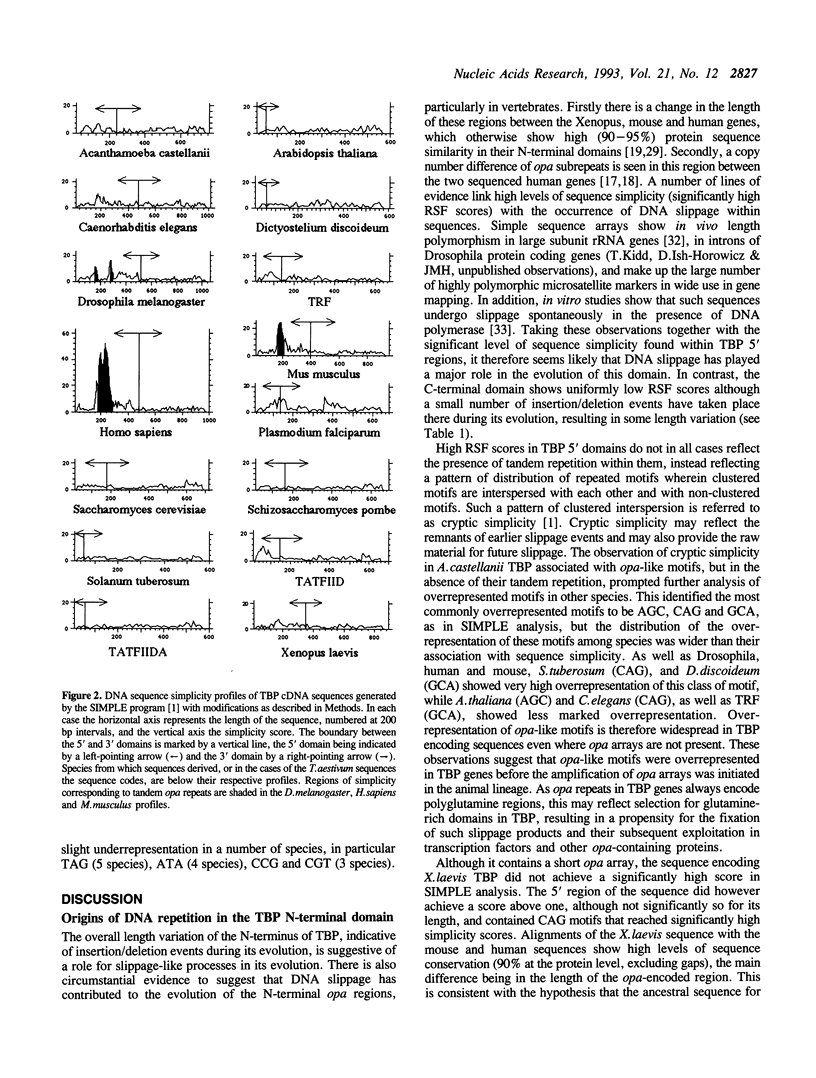
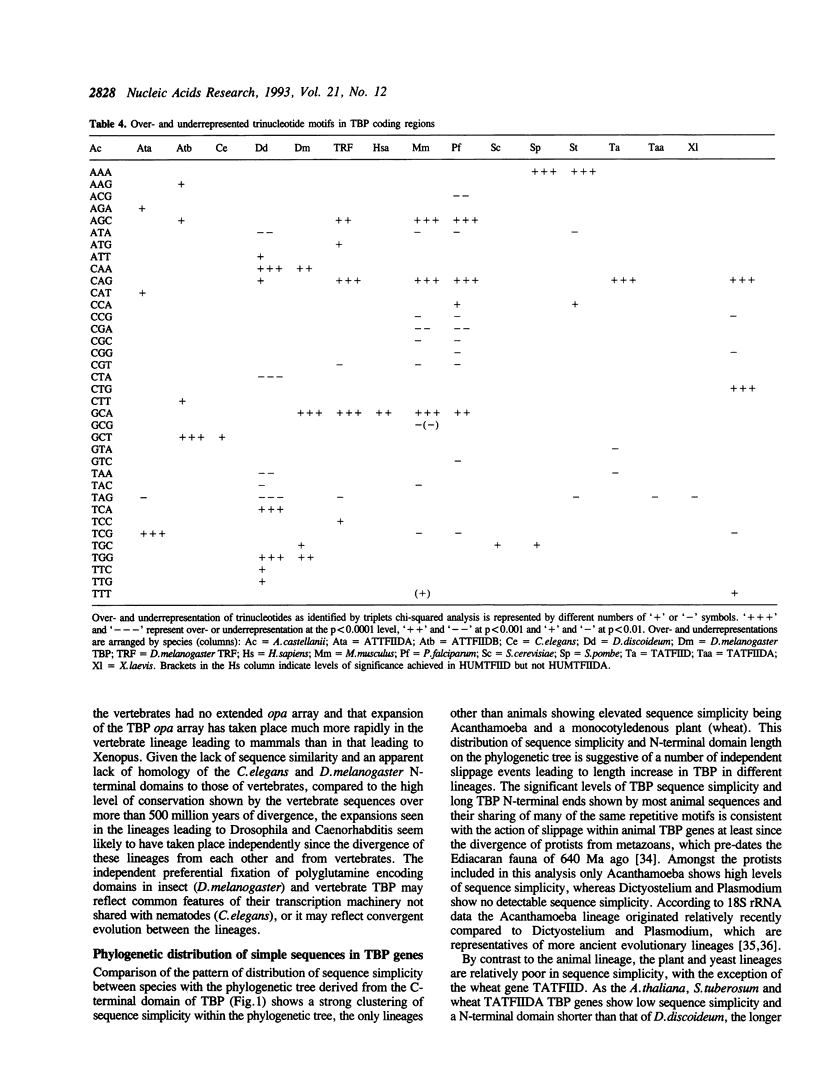
Analysis of TBP gene sequences from a variety of species for clustering of short sequence motifs and for over- and underrepresentation of short sequence motifs suggests involvement of slippage in the recent evolution of the TBP N-terminal domains in metazoans, Acanthamoeba and wheat. AGC, GCA and CAG are overrepresented in TBP genes of other species, suggesting that opa arrays were amplified from motifs overrepresented in ancestral species. The phylogenetic distribution of recently slippage-derived sequences in TBP is similar to that observed in the large subunit ribosomal RNAs, suggesting a propensity for certain evolutionary lineages to incorporate slippage-generated motifs into protein-coding as well as ribosomal RNA genes. Because length increase appears to have taken place independently in lineages leading to vertebrates, insects and nematodes, TBP N-terminal domains in these lineages are not homologous. All gene duplications in the TBP gene family appear to have been recent events despite strong protein sequence similarity between TRF and P. falciparum TBP. The enlargement of the TBP N-terminal domain may have coincided with acquisition of new functions and may have accompanied molecular coevolution with domains of other proteins, resulting in the acquisition of new or more complex mechanisms of transcription regulation.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell S. P., Pikaard C. S., Reeder R. H., Tjian R. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell. 1989 Nov 3;59(3):489–497. doi: 10.1016/0092-8674(89)90032-9. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Cress W. D., Cress A., Triezenberg S. J., Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990 Jun 29;61(7):1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- Cavallini B., Faus I., Matthes H., Chipoulet J. M., Winsor B., Egly J. M., Chambon P. Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud P., Glaessner M. F. The ediacarian period and syste: metazoa inherit the Earth. Science. 1982 Aug 27;217(4562):783–792. doi: 10.1126/science.217.4562.783. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992 May 15;69(4):685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Costa R., Peixoto A. A., Thackeray J. R., Dalgleish R., Kyriacou C. P. Length polymorphism in the threonine-glycine-encoding repeat region of the period gene in Drosophila. J Mol Evol. 1991 Mar;32(3):238–246. doi: 10.1007/BF02342746. [DOI] [PubMed] [Google Scholar]
- Crowley T. E., Hoey T., Liu J. K., Jan Y. N., Jan L. Y., Tjian R. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature. 1993 Feb 11;361(6412):557–561. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djian P., Green H. Vectorial expansion of the involucrin gene and the relatedness of the hominoids. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8447–8451. doi: 10.1073/pnas.86.21.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. A., Flavell R. B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984 Oct;38(3):622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Burke W. D. The silkmoth late chorion locus. II. Gradients of gene conversion in two paired multigene families. J Mol Biol. 1986 Aug 5;190(3):357–366. doi: 10.1016/0022-2836(86)90007-0. [DOI] [PubMed] [Google Scholar]
- Fikes J. D., Becker D. M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990 Jul 19;346(6281):291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- Gasch A., Hoffmann A., Horikoshi M., Roeder R. G., Chua N. H. Arabidopsis thaliana contains two genes for TFIID. Nature. 1990 Jul 26;346(6282):390–394. doi: 10.1038/346390a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J. Roles of TFIID in transcriptional initiation by RNA polymerase II. Cell. 1991 Sep 20;66(6):1067–1070. doi: 10.1016/0092-8674(91)90027-v. [DOI] [PubMed] [Google Scholar]
- Gunderson J. H., McCutchan T. F., Sogin M. L. Sequence of the small subunit ribosomal RNA gene expressed in the bloodstream stages of Plasmodium berghei: evolutionary implications. J Protozool. 1986 Nov;33(4):525–529. doi: 10.1111/j.1550-7408.1986.tb05656.x. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989 Sep 22;58(6):1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- Hancock J. M., Dover G. A. 'Compensatory slippage' in the evolution of ribosomal RNA genes. Nucleic Acids Res. 1990 Oct 25;18(20):5949–5954. doi: 10.1093/nar/18.20.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. M., Dover G. A. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol Biol Evol. 1988 Jul;5(4):377–391. doi: 10.1093/oxfordjournals.molbev.a040505. [DOI] [PubMed] [Google Scholar]
- Hancock J. M., Tautz D., Dover G. A. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol Biol Evol. 1988 Jul;5(4):393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Fujita H., Hasegawa S., Roeder R. G., Horikoshi M. Conserved structural motifs within the N-terminal domain of TFIID tau from Xenopus, mouse and human. Nucleic Acids Res. 1992 Jul 25;20(14):3788–3788. doi: 10.1093/nar/20.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel A. R., Hancock J. M., Dover G. A. Evolution of the cetacean mitochondrial D-loop region. Mol Biol Evol. 1991 Jul;8(4):475–493. doi: 10.1093/oxfordjournals.molbev.a040662. [DOI] [PubMed] [Google Scholar]
- Hoey T., Dynlacht B. D., Peterson M. G., Pugh B. F., Tjian R. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell. 1990 Jun 29;61(7):1179–1186. doi: 10.1016/0092-8674(90)90682-5. [DOI] [PubMed] [Google Scholar]
- Hoffman A., Sinn E., Yamamoto T., Wang J., Roy A., Horikoshi M., Roeder R. G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature. 1990 Jul 26;346(6282):387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Horikoshi M., Wang C. K., Schroeder S., Weil P. A., Roeder R. G. Cloning of the Schizosaccharomyces pombe TFIID gene reveals a strong conservation of functional domains present in Saccharomyces cerevisiae TFIID. Genes Dev. 1990 Jul;4(7):1141–1148. doi: 10.1101/gad.4.7.1141. [DOI] [PubMed] [Google Scholar]
- Holdsworth M. J., Grierson C., Schuch W., Bevan M. DNA-binding properties of cloned TATA-binding protein from potato tubers. Plant Mol Biol. 1992 Jun;19(3):455–464. doi: 10.1007/BF00023393. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Kao C. C., Lieberman P. M., Schmidt M. C., Zhou Q., Pei R., Berk A. J. Cloning of a transcriptionally active human TATA binding factor. Science. 1990 Jun 29;248(4963):1646–1650. doi: 10.1126/science.2194289. [DOI] [PubMed] [Google Scholar]
- Kawata T., Minami M., Tamura T., Sumita K., Iwabuchi M. Isolation and characterization of a cDNA clone encoding the TATA box-binding protein (TFIID) from wheat. Plant Mol Biol. 1992 Aug;19(5):867–872. doi: 10.1007/BF00027083. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990 Jun 29;61(7):1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- Kelly R., Gibbs M., Collick A., Jeffreys A. J. Spontaneous mutation at the hypervariable mouse minisatellite locus Ms6-hm: flanking DNA sequence and analysis of germline and early somatic mutation events. Proc Biol Sci. 1991 Sep 23;245(1314):235–245. doi: 10.1098/rspb.1991.0115. [DOI] [PubMed] [Google Scholar]
- McAndrew M. B., Read M., Sims P. F., Hyde J. E. Characterisation of the gene encoding an unusually divergent TATA-binding protein (TBP) from the extremely A+T-rich human malaria parasite Plasmodium falciparum. Gene. 1993 Feb 28;124(2):165–171. doi: 10.1016/0378-1119(93)90390-o. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Muhich M. L., Iida C. T., Horikoshi M., Roeder R. G., Parker C. S. cDNA clone encoding Drosophila transcription factor TFIID. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9148–9152. doi: 10.1073/pnas.87.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newfeld S. J., Smoller D. A., Yedvobnick B. Interspecific comparison of the unusually repetitive Drosophila locus mastermind. J Mol Evol. 1991 May;32(5):415–420. doi: 10.1007/BF02101281. [DOI] [PubMed] [Google Scholar]
- Patel N. H., Ball E. E., Goodman C. S. Changing role of even-skipped during the evolution of insect pattern formation. Nature. 1992 May 28;357(6376):339–342. doi: 10.1038/357339a0. [DOI] [PubMed] [Google Scholar]
- Paulsson G., Lendahl U., Galli J., Ericsson C., Wieslander L. The Balbiani ring 3 gene in Chironomus tentans has a diverged repetitive structure split by many introns. J Mol Biol. 1990 Jan 20;211(2):331–349. doi: 10.1016/0022-2836(90)90355-P. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Ruiz Linares A., Hancock J. M., Dover G. A. Secondary structure constraints on the evolution of Drosophila 28 S ribosomal RNA expansion segments. J Mol Biol. 1991 Jun 5;219(3):381–390. doi: 10.1016/0022-2836(91)90178-9. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992 Jan 25;20(2):211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. C., Kao C. C., Pei R., Berk A. J. Yeast TATA-box transcription factor gene. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L., Elwood H. J., Gunderson J. H. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Sumita K., Fujino I., Aoyama A., Horikoshi M., Hoffmann A., Roeder R. G., Muramatsu M., Mikoshiba K. Striking homology of the 'variable' N-terminal as well as the 'conserved core' domains of the mouse and human TATA-factors (TFIID). Nucleic Acids Res. 1991 Jul 25;19(14):3861–3865. doi: 10.1093/nar/19.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Tautz C., Webb D., Dover G. A. Evolutionary divergence of promoters and spacers in the rDNA family of four Drosophila species. Implications for molecular coevolution in multigene families. J Mol Biol. 1987 Jun 5;195(3):525–542. doi: 10.1016/0022-2836(87)90181-1. [DOI] [PubMed] [Google Scholar]
- Tautz D., Trick M., Dover G. A. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986 Aug 14;322(6080):652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Treier M., Pfeifle C., Tautz D. Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J. 1989 May;8(5):1517–1525. doi: 10.1002/j.1460-2075.1989.tb03536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- Wong J. M., Liu F., Bateman E. Cloning and expression of the Acanthamoeba castellanii gene encoding transcription factor TFIID. Gene. 1992 Aug 1;117(1):91–97. doi: 10.1016/0378-1119(92)90494-a. [DOI] [PubMed] [Google Scholar]
- Zhou Q. A., Schmidt M. C., Berk A. J. Requirement for acidic amino acid residues immediately N-terminal to the conserved domain of Saccharomyces cerevisiae TFIID. EMBO J. 1991 Jul;10(7):1843–1852. doi: 10.1002/j.1460-2075.1991.tb07710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


