Abstract
Background
Cancers of the urinary bladder are the fifth most commonly diagnosed malignancy in the US. Early clinical diagnosis of bladder cancer remains a major challenge and the development of non-invasive methods for detection and surveillance is desirable for both patients and health care providers.
Approach
In order to identify urinary proteins with potential clinical utility we enriched and profiled the glycoprotein component of urine samples using a dual-lectin affinity chromatography and LC-MS/MS platform.
Results
From a primary sample set obtained from 54 cancer patients and 46 controls a total of 265 distinct glycoproteins were identified with high confidence, and changes in glycoprotein abundance between groups were quantified by a label-free spectral counting method. Validation of candidate biomarker alpha-1-antitrypsin (A1AT) for disease association was performed on an independent set of 70 samples (35 cancer cases) using an ELISA. Increased levels of urinary alpha-1-antitrypsin (A1AT) glycoprotein were indicative of the presence of bladder cancer (p value < 0.0001) and augmented voided urine cytology results. A1AT detection classified bladder cancer patients with a sensitivity of 74% and specificity of 80%.
Summary
The described strategy can enable higher resolution profiling of the proteome in biological fluids by reducing complexity. Application of glycoprotein enrichment provided novel candidates for further investigation as biomarkers for the non-invasive detection of bladder cancer.
Keywords: bladder cancer, glycoprotein profiling, diagnostic profile, A1AT
1. Introduction
With an estimated 70,980 newly diagnosed cases and 14,330 associated deaths in 2009, urinary bladder cancer is the second most common genitourinary malignant disease in the U.S. and among the five most common malignancies worldwide (1). Transitional cell carcinoma (TCC), the most prevalent subtype, accounts for 90% of bladder cancers, while two other forms, squamous cell carcinomas and adenocarcinomas, constitute 5% and 2% respectively (2). When detected early, the 5-year survival rate is approximately 94%, thus timely intervention dramatically increases patient outcome. At presentation, more than 80% of bladder tumors are non-muscle invasive papillary tumors (Ta or T1) and these superficial lesions are treated conservatively by transurethral resection. However, more than 70% of patients with these lesions will have disease recurrence during the first two years. If left untreated these initially superficial lesions can progress to being muscle-invasive, thus patients are under continued surveillance by expensive and invasive cystoscopic examination for early detection of new tumor developments.
Voided urine cytology (VUC) remains the method of choice for the noninvasive detection of bladder cancer. This method microscopically examines the morphology of the cells of the bladder lining which are collected from the urine. The method is subjective and is open to considerable inter-observer variation, and so accuracy is a problem, especially for low-grade and low-stage tumors. Furthermore, results are not available rapidly, it is prone to inter-observer variation, and it is relatively expensive. However, the diagnostic accuracy of voided urine cytology (VUC) is limited (3, 4). While VUC has specificities up to 98% (5), its utility as a primary detection tool is reduced because of sensitivities ranging from 40% to 76% (6). In a recent paper, we found the sensitivity of urinary cytology to be 33% (7). The major problem with urine cytology is the fact that results are operator dependent, meaning that the skill of the cytologist can result in variable results across institutions (8). This variation in accuracy is reflected in the fact that voided urine cytology is rarely the single test performed during a clinical workup to investigate potential bladder cancers (9).
A number of molecular tests have been developed to detect putative tumor-associated molecules that are released into the urine from malignant lesions. Tests include the bladder tumor antigen (BTA) (10), nuclear matrix protein 22 (NMP-22) (11, 12), BLCA-4, cytokeratin (13), psoriasin (S100A7) (14), fibronectin (FG)/degradation products (FDPs) (15, 16), zinc-alpha2-glycoprotein (17). However, these markers have limited sensitivity, so alone they are not yet sufficient to replace the gold standards of urine cytology or cystoscopy (18, 19). The search for more sensitive, non-invasive urine tests for more reliable detection and surveillance is warranted.
Proteomic profiling approaches provide an unbiased and systematic survey of the proteome, and technologies continue to be developed to address problems of biological variation and proteome complexity. In our own work we have focused on analyzing the glycoprotein component of the proteome. As much as half of the mammalian proteome is composed of glycoproteins that are involved in essential cellular and biological functions, and many of the biomarkers currently used for cancer detection in body fluids are glycoproteins, including PSA (20), and CA-125 (21). Analytical tools for glycoprotein separation have been developed based upon chemical reaction, gel-binding and lectin affinity chromatography. The latter has become a powerful tool for the purification and concentration of glycoproteins. Lectins constitute a group of proteins with unique affinities for carbohydrate structures. Widely used lectins include concanavalin A (ConA) and wheat germ agglutinin (WGA). ConA recognizes alpha-linked mannose, which is very common in N-linked glycans, and WGA selectively binds to N-acetyl-glucosamine (GlcNAc) groups and sialic acid residues.
The aim of this study was to design and apply a comprehensive strategy for the identification of glycoprotein biomarkers in urine for potential utility in bladder cancer detection. We employed dual-lectin affinity (ConA and WGA) chromatography to isolate a wide range of N-linked glycoproteins from urine samples, and incorporated a LC-MS/MS based label-free shotgun method to identify glycoproteins that were differentially expressed according to disease status. One of the most discriminatory novel candidate biomarkers (A1AT) was validated in an independent sample cohort using ELISA. The strategy identified specific glycoprotein biomarkers with potential utility in non-invasive bladder cancer detection.
2. Material and Methods
2.1 Clinical sampling and processing
Under IRB approval and informed consent, urothelial samples and associated clinical information were prospectively collected from individuals with no previous history of urothelia carcinoma. Patients were undergoing complete hematuria workup including office cystoscopy and upper tract imaging by computed tomography of the abdomen and pelvis (without and with intravenous contrast). Urine cytology was performed by standard Papanicolaou staining. All slides were evaluated by a cytopathologist. Urothelial carcinoma grading and staging were performed according to the World Health Organization criteria. Two different clinical cohorts were analyzed in this study. The first group (experimental) consisted of 46 subjects with either a negative evaluation (i.e., normal imaging of upper tract and normal cystoscopy) or asymptomatic donors, and 54 subjects with a visible bladder tumor detected by upper tract imaging and/or cystoscopy, and which was later proven by evaluation of a biopsy to be urothelial carcinoma. The second group (validation) comprised 35 subjects with a negative evaluation and 35 cases of confirmed bladder cancer. A summary of clinical data is given in Table 1. Each sample consisted of 30–50 mL of midstream urine collected in a sterile cup, stored immediately at 4°C, and processed for storage within one hour of collection. Specimens with clearly visible hematuria were excluded from analysis. Urinary creatinine, blood, leukocytes, nitrite, glucose, pH, ketone and bilirubin were measured for all samples prior to processing using MULTISTIX PRO Reagent Strips (Bayer HealthCare, Elkhart, IN). Cells and debris were removed from each urine sample by centrifugation at 500 g and 5000 g for 5 min and 10 min at 4 °C respectively and supernatant was stored at −80°C until processing for proteomic analysis. For chromatography, supernatant was subjected to total protein precipitation by adding four times the sample volumes of cold acetone (−20 °C). We found by testing replicate samples that cold acetone gave a yield of ~90% protein recovery. The sample was left at −20 °C overnight followed by centrifugation at 12000 g for 15 min at 4 °C. The supernatant was removed, and the centrifuge tube was left open in the fume hood to remove the remaining solvent. The pellet was resuspended in binding buffer (20 mM Tris, 0.15 M NaCl, 1 mM MnCl2, and 1 mM CaCl2, pH 7.4) and vortexed vigorously to completely dissolve the pellet. The protein concentration in the sample was determined by the Bradford protein assay (Bio-Rad, Hercules, CA). Throughout the study, all samples were processed and analyzed individually, no pooling of samples was performed.
Table 1.
Demographic and clinicopathologic characteristics of discovery (Phase I) and validation study cohorts
| Phase I | Validation | |||
|---|---|---|---|---|
| Noncancer (%)* | Cancer (%) | Noncancer (%)* | Cancer (%) | |
| N=46 | N=54 | N=35 | N=35 | |
| Median Age (range, y) | 56 (20–77) | 71 (48–89) | 58 (19–85) | 67 (20–87) |
| Male/Female ratio | 35:11 | 47:7 | 28:13 | 34:7 |
| Race | ||||
| White | 35 (76) | 53 (98) | 34 (83) | 41 (100) |
| African American | 6 (13) | 1 (2) | 3 (7) | 0 (4) |
| Other | 5 (11) | 0 (0) | 4 (10) | 0 (0) |
| Positive/suspicious cytology | 0 (0) | 16 (30) | 0 (0) | 11 (31) |
| Clinical stage | ||||
| Tis | n/a | 4 (7) | n/a | 0 (0) |
| Ta | n/a | 9 (17) | n/a | 14 (34) |
| T1 | n/a | 11* (20) | n/a | 9 (22)^ |
| T2 | n/a | 22(41) | n/a | 17 (41)^ |
| T3 | n/a | 2 (4) | n/a | 1 (2) |
| T4 | 3 (6) | 1 (2) | ||
| Tx | 1 (2) | 0 | ||
| Grade | ||||
| 1/2 | n/a | 9 (17) | n/a | 10 (24) |
| 3 | n/a | 45 (83) | n/a | 31 (76) |
2.2 Con A and WGA lectin affinity chromatography
A 700 µL resin slurry of agarose-bound concanavalin A (Con A) and 800 µL resin slurry of wheat germ agglutinin (WGA) (Vector Laboratories, Burlingame, CA) was packed into a 2 mL disposable centrifuge column (Thermo Scientific, Rockford, IL). The resin was first washed three times by binding buffer. 450 µg diluted protein sample (1:4) was then added to the column and incubated for 30 min. The flow-through was discarded. The non specific binding was removed by washing resin four times with 1ml of binding buffer each time. 1ml of elution buffer (20 mM Tris, 0.5 M NaCl, 1 mM MnCl2, 1 mM CaCl2, 0.3 M methyl-α-D-mannopyroside and 0.3 M N-acetyl-D-glucosamine, pH 7.0) was added and mixed for 10 min. The eluate was collected and the elution step was repeated once.
2.3 Tryptic Digestion / PNGase F Treatment
The protein sample eluates were concentrated using a Microcon YM-10 column (Millipore Corp., Bradford, MA) according to the manufacturer’s protocol. Approximately 10 µL of sample was obtained from the filter device. A 1µL portion of 100 mM DTT (Sigma) was added and the resulting mixture was incubated at 45 °C for 30 min. A 1 µL portion of trypsin (Sigma) was added afterwards at 37 °C. The overnight digestion and reduction reaction was terminated by adding 0.1µL of TFA to the digest. The tryptic digestion mixture was completely dried using a SpeedVac concentrator (Labconco Corp., Kansas City, MO) operated at 45 °C. The digest was reconstituted with 20 µL of 50 mM ammonium bicarbonate and 5 unit of PNGase F (Sigma) was added. The deglycosylation reaction was incubated for 24 h in a water bath set at 37 °C. The reaction was stopped by incubating the digest mixture at 75 °C for 20 min. The sample was dried by SpeedVac concentrator (22).
2.4 Nano-RPLC-ESI MS
A Paradigm MG4 micropump (Michrom Biosciences Inc., Auburn, CA) was used to deliver the mobile phase. The pump flow rate was split to achieve a column flow rate of 300 nL/min. The separation column (0.1mm×150mm, C18 AQ particles, 5 µm, 120 Å) was from Michrom BioResources (Auburn, CA, USA). The resolved peptides were analyzed on a linear ion trap mass spectrometer with a nano-LC-ESI source (LTQ, Thermo Finnigan, San Jose, CA). A mobile phase system of two solvents was used, where solvents A and B were composed of 0.1% formic acid and 2% acetonitrile or water in HPLC-grade water and acetonitrile, respectively. A 60 min acetonitrile/water gradient method was used, starting with 5% acetonitrile which was ramped to 40% in 55 min and to 95% in another 5min. The LTQ instrument was operated in positive ion mode. The capillary transfer tube was set at 200 °C, the ESI spray voltage at 2.5 kV, and the capillary voltage at 30 V. Ion activation was achieved by utilizing helium at a normalized collision energy of 35%. The data acquisition and generation of peak list files were automatically done by Xcaliber software (2.1.0.1139). For each cycle of one full mass scan (range of m/z 400–2000), the three most intense ions in the spectrum were selected for tandem MS analysis, unless they appeared in the dynamic or mass exclusion lists.
2.5 Database Queries and Manual Validation
All MS/MS spectra were analyzed using the SEQUEST algorithm, version 27 incorporated in Bioworks software, version 3.1 SR1 (Thermo Finnigan). Peptide fragment lists were generated and submitted to the Swiss-Prot database. The search parameters were as follows: [1] protein database, Uniprot/Swiss-Prot Release version 54.6 of 17-Dec-2007, downloaded from NCBI; [2] allowing two missed cleavages; [3] variable modification, oxidation of M; N+1, the +1 Da was allowed owing to hydrolysis of the amide of the asparagine side chain to release the asparagine-linked oligosaccharides from glycopeptides; [4] peptide ion mass tolerance 1.50 Da; [5] fragment ion mass tolerance 0.0 Da; [6] peptide charges +1, +2, and +3. To further validate data obtained from SEQUEST, Trans-Proteomic Pipeline (TPP) software was used where the threshold score was set at 0.9 for ProteinProphet probability (overall false positive rate below 1%) (23, 24). Only the proteins that passed the threshold and contained NXS/T motif in their peptide sequences were sorted as glycoproteins.
2.6 Data Normalization and Spectral Counting Quantification
Spectral counts were parsed from TPP (Trans Proteomics Pipeline) xml files after processing the SEQUEST data. Global normalization with protein length was used to reduce technical bias when acquiring spectral count data from different runs between and across samples. It is important to take into consideration the length of a protein when determining protein abundances using spectral counting, since small proteins tend to have fewer peptides identified per protein. If Cij is the raw spectral count for protein i in subject j, and Li is the length of protein i, then the first step in data processing is to calculate Xij using the formula
which adjusts for the total spectral count observed per sample, and for the effects of protein length. To eliminate the discontinuity in the count ratios when the spectral count for a protein is zero, the Xij data were transformed following an approach similar to that used in Old et al (25), as originally proposed by Beissbarth et al (26) for serial analysis of gene expression (SAGE). The transformation uses the logarithmic quantity
where Tj is the total of the Xij for subject j, and f is a positive constant. The value of f was set as the maximizer of the average Pearson correlation coefficient between pairs of replicate measures for the same subject, which was determined to be 0.7 (Figure S6). Finally, the data was filtered based on a minimum standard deviation threshold of 0.01, to exclude proteins with very low variability between cancer and normal groups.
2.7 Western Blotting
For Western blot analyses, urine samples from three individual patients from each independent experimental group (non-cancer and bladder cancer) were applied. 20 µg of protein was resolved on 4−20% Tris-SDS gels and transferred to PVDF membranes. Membranes were blocked with 1% BSA in PBST for 2 h at room temperature and incubated with primary antibody (mouse alpha-1-antitrypsin, mouse uromodulin from Abcam, Cambridge, MA, rabbit cell adhesion molecule 1 from Abnova, Walnut, CA) overnight at 4°C. All primary antibodies were diluted 1:1000. After three 10-min washes with PBST, membranes were incubated with anti-mouse/anti-rabbit IgG-HRP secondary antibody (Invitrogen, Carlsbad, CA) at a dilution of 1:5000 for 1 h, washed and visualized with the SuperSignal West Pico chemiluminescent substrate system (Pierce, Rockford, IL). The average time for image development was 15 seconds. Chemiluminescence was scanned on a LAS-3000 instrument using LAS-3000 Lite software, and scanned images were visualized and quantified using MultiGauge v.3.1 software (FujiFilm Life Science, Inc., Stamford, CT).
2.8 Enzyme-Linked Immunosorbent Assay (ELISA)
The level of human alpha1-antitrypsin (A1AT) was monitored in urine samples using the AssayMax Human Alpha1-Antitrypsin (A1AT) ELISA kit (Assaypro, MO). The A1AT present in standards and samples is measured by competition with a biotyinylated A1AT and detected using a streptavidin-peroxidase conjugate. 25 µl of A1AT standard or urine sample was added to each well, followed by addition of 25 µl of biotinylated A1AT. The assay was conducted according to the manufacturer’s instructions. The absorbance values were read on a microplate reader (Bio-tek, Synergy™ HT, VT) at a wavelength of 450 nm.
2.9 Statistics and Data Analysis
For the differential expression analysis of profile data we used linear mixed effects models to identify proteins that were differentially expressed between cancer and normal samples while reducing the effects of experimental batches. The mixed effects models used transformed glycoprotein abundance as the outcome variable, and included random intercepts for the batches and a fixed effect for disease status. The Z-score for the disease status variable was used to identify differentially expressed proteins, based on the False Discovery Rate (FDR) values and the Bonferroni-adjusted p-values. For ELISA data analysis, a standard curve prepared using purified A1AT was generated by regression analysis using four-parameter logistic curve-fit. Signal intensities of the A1AT disease marker were converted to concentration values by reference to the standard curve. To normalize for unknown total urine volume these values were reported as a ratio relative to urinary creatinine values (27–29). ELISA data were plotted as a scatter plot and a receiver operating characteristic (ROC) curve. The Student t-test for paired samples was used to perform pairwise comparison of AUC for A1AT signal intensities calibrated using urinary creatinine. A ROC curve is obtained by varying a decision threshold provides a direct view on how a predictive approach performs at different sensitivity and specificity levels (30, 31). In the case of a diagonal line on the ROC curve, an area under the curve (AUC) value of 0.5 is obtained, indicating that prediction between positive and negative cases is random. The other end of the scale is represented by an AUC value of 1.0, indicating perfect separation. The AUC represents an overall measure of a particular biomarker’s discriminatory potential across all thresholds. The ROC curve also allows the relationship between sensitivity and specificity to be estimated at any given point. Here, sensitivity (or true-positive rate) is the probability that a subject with bladder cancer is correctly classified as having cancer, and specificity (or false-positive rate) is the probability that a subject with bladder cancer is incorrectly classified as having cancer (specificity can be defined as one minus the false-positive rate). Differences in all analyses were regarded as significant at a p-value <0.05. Statistical analyses were performed by SPSS 13.0., and by MedCalc version 8.0 (MedCalc Software, Mariakerke, Belgium) for ROC curve analysis.
3. RESULTS
3.1 Identification of Urinary Glycoproteins by LC-MS/MS
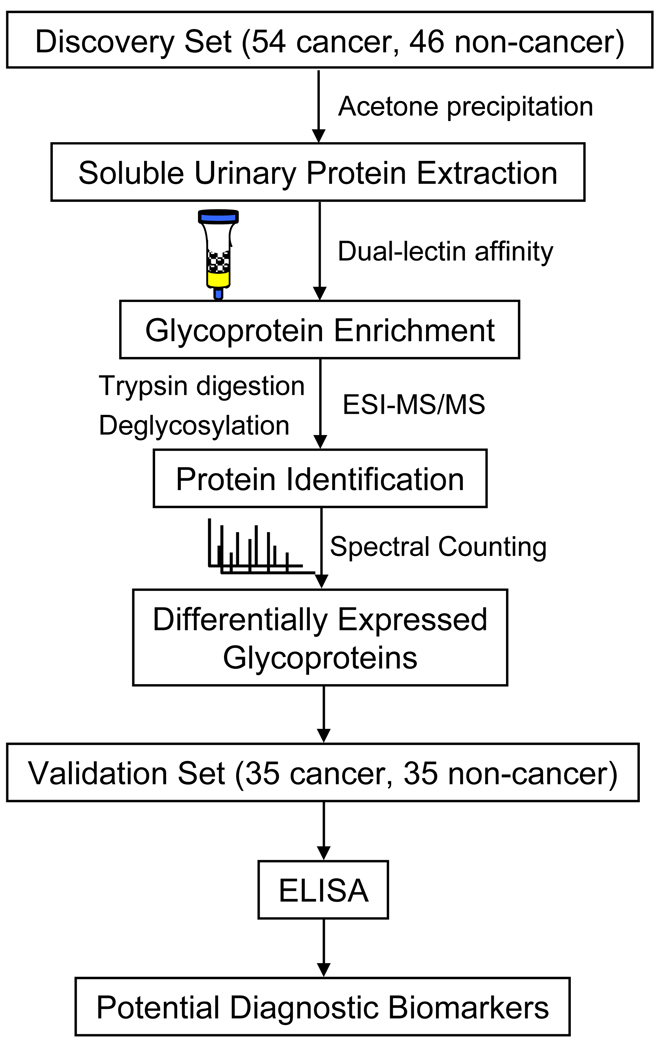
Glycoproteins from 54 bladder cancer and 46 non-cancer specimens were enriched by dual-lectins, ConA and WGA, and digested with trypsin (see workflow diagram Figure 1). After N-glycans were removed from tryptic peptide mixtures using PNGase F, they were analyzed by nanoHPLC-ESI-MS/MS using LTQ mass spectrometry and Trans-Proteomics Pipeline (TPP) software to identify glycosylated proteins. Figure S1 (supplemental data) shows a typical MS/MS spectrum of an N-glycosylated peptide from a glycoprotein, A1AT, identified in one cancer urine sample. After non-specific binding proteins and any processing artifacts were removed (such as trypsin and keratin), a total of 421 urinary proteins were identified with high confidence (ProteinProphet probability scores >= 0.9 and a false-positive rate below 1% (32)). After manual validation using the Swiss-Prot database we identified a total of 267 distinct glycoproteins. To further exclude proteins with low variability between cancer and non-cancer groups, we compiled a list of 100 unique glycoproteins with a variance standard (S.D.) > 0.01. A heat map was prepared in order to visualize the abundance of these glycoproteins across all specimens in the two groups (see supplemental Figure S2).
Figure 1.
Workflow of LC-MS/MS based identification, and immunoassay based validation.
3.2 Assessment of Reproducibility and Correlation of LC-MS/MS Data
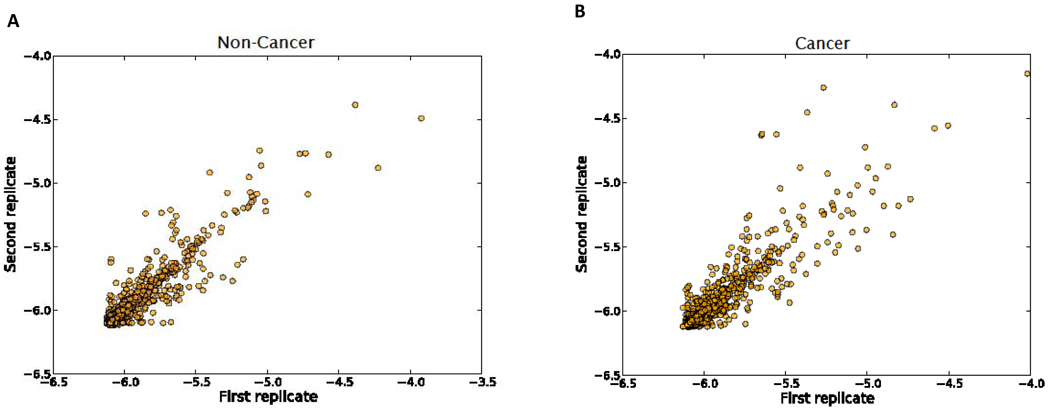
To assess the technical variation resulting from the nanoLC-MS/MS instrument, the transformed spectral counting data from technical replicates were compared. This comparison revealed that the same proteins from duplicate runs exhibited nearly identical retention time, confirming a high level of reproducibility for the LTQ instrument. An example comparison is depicted in supplemental Figure S3. Scatter plots were constructed to display the LTQ reproducibility of two consecutive analyses across all of the urinary samples from the non-cancer and the cancer groups (Figure 2). The linear regression coefficient r2 was 0.91 and 0.82 for control and cancer samples, respectively, indicating that reproducibility was high across the entire sample set. To further evaluate MS data reliability and variance among different biological samples and batches, we investigated the Pearson correlation between transformed data for all proteins, within pairs of samples. A batch analysis scatter plot (Figure S4) shows that the technical replicates were highly correlated to each other, confirming our LTQ reproducibility findings. When compared within a batch, the N/N pairs (non-cancer) and the T/T pairs (tumor-bearing) are slightly better correlated than the N/T pairs, reflecting a reasonable biological variation relationship between non-cancer and cancer specimen groups. The three pair-type comparisons are similar when compared either across different batches or within-batches. These findings show that no major LTQ analysis batch effect was evident. However, we did observe some batch variance when comparing protein extraction data (Figure S5). Therefore, our subsequent differential analyses were based on within-batch comparisons and the significance level was evaluated using a statistical model incorporating a batch factor.
Figure 2.
Scatter plots of LTQ replicates across all urine samples based on transformed spectral counts. A) Non-cancer group, B) Cancer group.
3.3 Identification of Differentially Expressed Glycoproteins
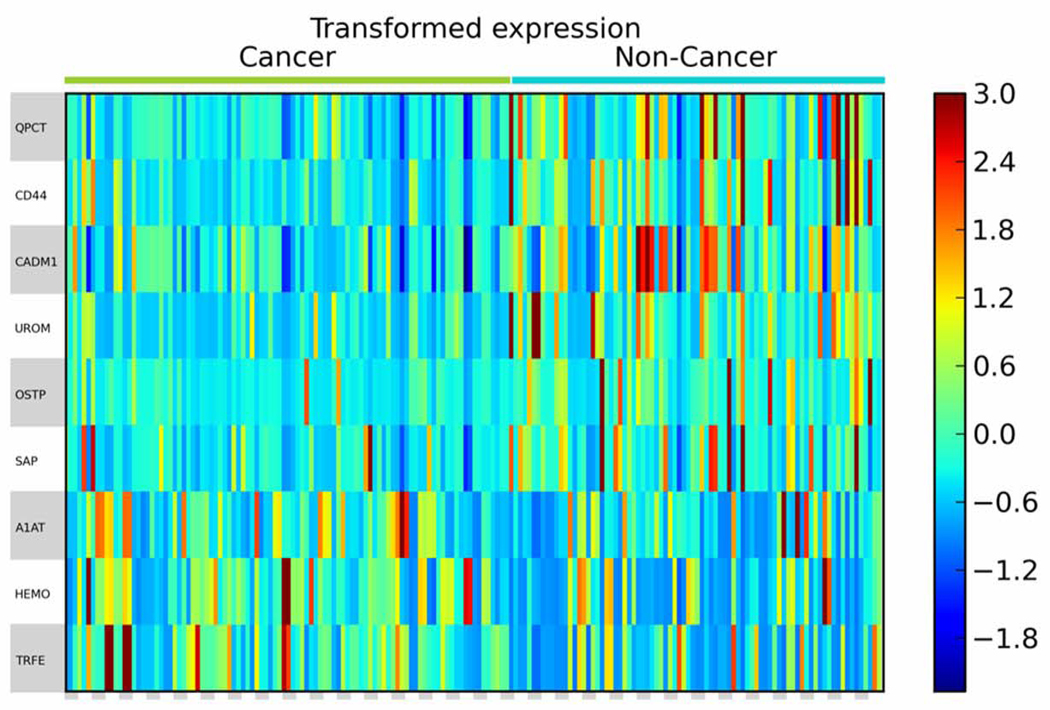
In order to find urinary glycoproteins that exhibit differential abundance between the non-cancer and cancer groups, MS data were analyzed by a label-free quantification method based on spectral counts. After protein sequence normalization and variance standard application, the quantity of each protein was transformed into a log scale. The correction factor f was determined for technical replicates to achieve the maximal correlation value (Figure S6). After transformation, statistical significance levels between the two groups were analyzed by multiple comparisons adjustment using a stringent local false discovery rate (locFDR), and considering the batch factor. To be qualified as a potential biomarker, a glycoprotein must be present in at least half of the cancer or non-cancer samples. As a result, a list of glycoproteins was found to be significantly differentially expressed with respect to the presence of bladder cancer, with a FDR value < 0.1 (Table 2). Three of the glycoproteins were expressed at significantly higher levels in urine from bladder cancer patients. A heat map was constructed to display the transformed spectral counts of these differentially expressed glycoproteins for all samples (Figure 3). Consistent with the results of spectral counting, analysis of six randomly selected urine samples (3 non-cancer, 3 bladder cancer cases) by Western-blotting showed that the levels of A1AT were elevated in the bladder cancer cases while UROM, CADM1 were found to decrease (data not shown).
Table 2.
Differentially Expressed Glycoproteins Identified in Subjects with Bladder Cancer and Non-cancera
| unique peptide No. | expression in cancer cases |
frequency* | |||||
|---|---|---|---|---|---|---|---|
| protein ID | protein name | p-value (FDR) | non-cancer | cancer | non-cancer | cancer | |
| P01009 | Alpha-1-antitrypsin | < 0.01 | 20 | 20 | up | 0.87 | 0.98 |
| P02787 | Serotransferrin | 0.04 | 20 | 25 | up | 0.62 | 0.91 |
| P02790 | Hemopexin | 0.02 | 14 | 20 | up | 0.81 | 0.96 |
| P07602 | Proactivator polypeptide | < 0.01 | 7 | 6 | down | 0.6 | 0.34 |
| P07911 | Uromodulin | 0.08 | 19 | 16 | down | 0.91 | 0.7 |
| P10451 | Osteopontin | 0.06 | 4 | 4 | down | 0.53 | 0.29 |
| P16070 | CD44 antigen | 0.06 | 4 | 3 | down | 0.96 | 0.91 |
| Q16769 | Glutaminyl-peptide cyclotransferase | < 0.01 | 6 | 3 | down | 0.51 | 0.2 |
| Q9BY67 | Cell adhesion molecule 1 | < 0.01 | 5 | 4 | down | 0.51 | 0.2 |
Spectral counts of urinary glycoproteins with significantly differential abundance (FDR-value < 0.1).
The candidate must exist in more than half of the non-cancer or cancer samples to qualify as a potential marker.
Figure 3.
Heat map of differentially expressed urinary glycoproteins identified in duplicate analyses of 103 samples. The heat map displays the transformed spectral counts data. Each column represents an independent analysis for each sample. Each row represents a specific glycoprotein identified by LC-MS/MS.
3.4 Validation of urinary A1AT abundance by ELISA
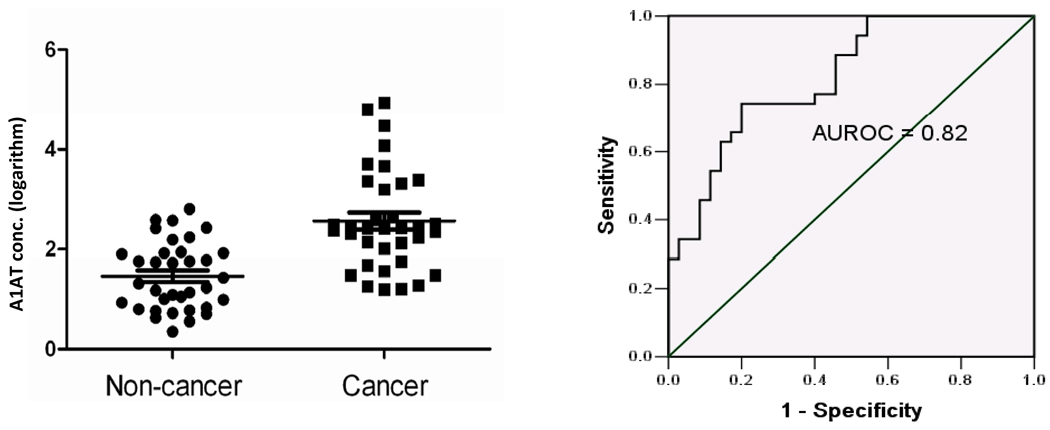
To verify the observed differential levels of the A1AT glycoprotein in bladder cancer patient samples, we measured the abundance of this target in urine samples obtained from an independent cohort (35 bladder cancer cases and 35 non-cancer controls) using a commercially available ELISA. Analyses revealed a significant association of elevated A1AT in urine samples obtained from patients with bladder cancer (Figure 4). The differential levels between groups was statistically significant (p value < 0.0001), and construction of an ROC curve illustrates the ability of A1AT to distinguish cancer and non-cancer cases (Figure 4). The area under the ROC curve (AUROC) was 0.82. From the ROC plot we can estimate the relationship between sensitivity and specificity of the assay at any given point. At a sensitivity of 74%, A1AT detection classified bladder cancer patients with a specificity of 80%. These values compare favorably with the gold standard of urine cytology, which has high specificity but a range of sensitivity from 11% to 76% (3,4). In the validation cohort, 11 of 35 (31%) of the cancer cases were positive by cytology. If we set arbitrary thresholds for the pilot A1AT ELISA data, we can estimate whether the ELISA data would have augmented the cytology data in some cases. Using a threshold of 3.2 from the data illustrated in Figure 4, the ELISA assay detected 10 cancer cases and no false-positives. Of these 10 confirmed cancer cases, 7 had a negative cytology.
Figure 4.
ELISA analysis of urinary A1AT. Left panel shows a scatter-plot of creatinine-normalized urinary A1AT concentration (logarithm M) in study subjects with (Cancer), and without (Control) bladder cancer. Differential levels were significant (p < 0.0001). Error bars denote standard deviation. Right panel shows ROC curve of A1AT data; area under the ROC (AUROC) of 0.82.
4. Discussion
Two-dimensional electrophoresis (2-DE) of proteins has been the conventional method for biomarker assessment in urological proteomics (33, 34). Irmak et al identified two proteins, orosomucoid (ORM) and zinc-alpha glycoprotein (ZAG) which were increased in urine samples of tumor-bearing patients in comparison with samples from a few healthy volunteers (17), and Pinero et al used 2D-DIGE coupled with mass spectrometry to identify regenerating protein-1 (Reg-1) and keratin 10 as being associated with bladder cancer (35). Saito et al. (36) used a 2-D PAGE approach, but focused specifically on proteins of the extracellular matrix and matrix metalloproteinases in urine. Samples enriched with gelatin-affinity beads revealed that MMP-2, MMP-9, and fibronectin (FN) fragments were present in cancer patients, but not in healthy individuals (36). To date, a limited number of proteomic studies have utilized MS technologies for the analysis of urological cancers, largely because of the lack of defined methodologies that can reduce the complexity of the sample and rapidly and accurately identify specific proteins. A number of studies have used SELDI-TOF to identify peptide signatures associated with bladder cancer, but validation of specific targets has not been forthcoming. Theodorescu et al. employed online coupling of capillary electrophoresis (CE) to an electrospray time-of-flight mass spectrometer (ESI-TOF-MS) to identify a 22-peptide signature that could discriminate between samples of bladder cancer and healthy subjects (37). The signature was able to correctly classify 31 bladder cancer patients from a cohort that included 149 samples from healthy or non-malignant urological diseases. A recent study used an iTRAQ (isobaric tag for relative and absolute quantitation) technique to discover proteins that were differentially expressed between pooled urine samples and non-tumor controls. This strategy identified 55 candidate biomarker proteins. Orthologous techniques confirmed that the level of apolipoprotein A–I (APOA1) was significantly elevated in urine samples from bladder cancer patients. Using a commercial ELISA assay, APOA1 was confirmed to have high diagnostic potential in an extended sample set (38). Collectively, these studies show the promise of MS-based urinary analysis for the discovery of biomarker panels.
The majority of reported proteomic urine profile studies have used large amounts of sample material. Hundreds of milliliters of urine are typically used for gel-based analysis, and the pooling of samples from different individuals are typically observed in the published gel analysis of normal urine proteome studies in order to increase the amount of proteins (39). Thus, a reliable and accurate profiling technique requiring only a small amount of urine is essential for investigating urinary proteomes and for marker identification in minimal samples. In order to improve efficiency and to make the glycoprotein enrichment strategy applicable to minimal sample material, we developed a nano-scale chelating Concanavalin A (Con A) monolithic capillary prepared using GMA-EDMA (glycidyl methacrylate–co-ethylene dimethacrylate) as polymeric support (40). We used this technique to identify 186 distinct N-linked glycoproteins in as little as 10ml of naturally micturated human urine. Utility for analysis of minimal biological samples was confirmed by the successful elucidation of glycoprotein profiles in mouse urine samples at the microliter scale (40). As advances in proteomics technologies continue, and findings are collated in established and curated publicly available databases, the future for the discovery and validation of urinary biomarkers of multiple diseases is encouraging.
In this study, we specifically investigated the glycoprotein component of the naturally micturated urinary proteome, and compared the profiles obtained from analysis of urine samples obtained from 54 patients with bladder cancer and form 46 non-cancer controls. We utilized a dual lectin affinity strategy to enrich N-linked glycoproteins, and incorporated a LC-MS/MS based label-free shotgun method to identify and compare glycoproteins present in the clinical sample panel. A total of 256 distinct glycoproteins were identified with high confidence, and comparative analysis confirmed that differential urinary glycoprotein profiles exist that can distinguish bladder cancer-bearing patients from individuals with no evidence of bladder cancer.
Quantitative shotgun proteomics, including both stable isotope labeled and label-free techniques has emerged as a major platform for investigating large-scale protein expression and characterization in complex biological systems (41, 42). Stable isotope labeling approaches, such as Isotope-Coded Affinity Tags (ICAT), Isobaric Tags for Relative and Absolute Quantification (iTRAQ), and other metabolic and enzymatic labeling (43, 44), can provide accurate quantitative results. However, these methods are not ideal for the initial large-scale discovery phase, due to high-cost, incomplete labeling, and artificial introduction of chemical derivatization. In recent years, an alternative shotgun proteomics strategy based on spectral counting has been developed. With this method, relative protein quantification is achieved by comparing the number of identified MS/MS spectra for a given protein species across samples (45). Previous studies have demonstrated that spectral counting has a strong linear correlation with relative protein abundance (r2= 0.9997), with a dynamic range over 2 orders of magnitude compared with sequence coverage and number of peptide (46). It was also proved to have higher reproducibility and a larger dynamic range than ion-peptide chromatogram (47). Spectral counting has the advantage of achieving cleaner, faster and simpler quantification results when it comes to large and global protein change studies (25, 48, 49).
Herein, the spectral counting method was employed for quantitative analysis in the present work, and rigorous statistical analysis and subsequent validation were designed to evaluate specific aspects of the method. Figures S3 and S4 show the high reproducibility between technical replicates observed for both non-cancer and cancer sets. In addition, to evaluate the correlation among different batches based on LTQ runs, we also examined the batch effect between protein extraction sets (Figure S5), revealing a high correlation between technical replicates. When compared within a batch, the N/N pairs were most strongly correlated, followed by the T/T pairs and the N/T pairs in order, reflecting interesting relationships between different disease stages. However, when compared across different batches, the N/N or T/T pairs were not more correlated than the N/T pairs. This suggested that some batch effect was causing variation across different batches based on sample preparation. Therefore, our differential expression analysis was grounded on mixed-effect models to reduce the effects of experimental batches. The most promising glycoprotein biomarkers had significant differential expression value when batch factor was accounted for. The subsequent validation results supported this consideration.
While a number of biomarkers identified for bladder cancer diagnosis may have utility in solid tissue or patient’s serum, urinalysis has obvious advantages for the detection of urological cancer. Most importantly, it is available for collection non-invasively. This enables patient compliance, copious sample collection and repeat sampling. The problem of protein complex formation is reduced in urine relative to serum, urinary peptides and low molecular weight proteins are generally soluble, and problems of degradation during collecting and processing experienced with other tissue and fluid analyses is avoided due to the fact that most proteolytic degradation events have already occurred before voiding (27). A limitation of urinalysis is the large dynamic range in protein and peptide concentration resulting from variations in the intake of fluid and voiding frequency. Appropriate normalization methods have to be selected to counter this problem. For MS data normalization, we applied total spectral count normalization considering protein length to adjust data deficiency and also to prevent abundant proteins from dominating the analysis.
Another challenge in urinalysis is the potential presence of blood proteins since hematuria is a common occurrence in patients with bladder cancer. With blood incursion, serum glycoproteins may be detectable during profiling in some cases. We addressed this problem in a number of ways. We excluded any visibly bloody urine samples and samples defined as having >0.010 mg/dl hemoglobin by urinalysis strip testing (normal levels, see methods). However, to confirm that candidate biomarkers were not associated with trace serum incursion we also examined the potential correlation between the glycoprotein hemoglobin and the target protein from our spectral counting data for each case. We observed no such association (see supplemental Figure S7 for scatter plot of A1AT and hemoglobin).
As in our previous study (50), we did identify a number of expected glycoproteins, including some which are under investigation as biomarkers for bladder cancer, but many glycoproteins will not have been selected by the lectin affinity approach used in this study. Specifically, we used ConA and WGA which bind alpha-linked mannose, and N-acetyl-glucosamine (GlcNAc) groups respectively. However, the strategy can be expanded to include additional lectins that are now routinely available in order to broaden the captured glycoprotein profile. The most discriminatory protein identified as being associated with bladder cancer in this study was alpha-1-antitrypsin (A1AT), also known as SERPINA1, which describes it as a member of a family of inhibitor of serine proteases. A1AT irreversibly inhibits trypsin, chymotrypsin and plasminogen activator. In humans, serpins play crucial roles in the maintenance of homeostasis, controlling multiple processes from cellular survival to blood clotting. There are a number of known serpin diseases, including emphysema and liver disease, but relatively little is known of its potential role in cancer. Genetic aberrations of A1AT have been found in some malignancies, including colorectal cancer (51). In a 2-DE and MS proteomics based analysis, Hamrita et al identified A1AT as upregulated in sera from breast cancer patients with various stages in comparison with healthy women (52). The progression from localized tumor to invasive metastasis involves matrix proteolysis. The balance between proteases and their inhibitors play a key role in this process. Thus, as well as a diagnostic biomarker, A1AT may be a useful indicator for tumor classification e.g. invasive versus superficial bladder cancer, or prognostication. A recent study in tumor tissues identified SERPINA1 expression as being correlated with malignant potential of ovarian cancer (53).
The impetus for our search for bladder cancer biomarkers comes from the idea that an accurate biomarker can reduce the number of cystoscopies performed each year, and thus, cut down the frequency of this invasive procedure. We have been able to establish a comprehensive proteomics strategy for discovery of urinary glycoproteins associated with bladder cancer using dual lectin-affinity enrichment and LC-MS/MS analysis, and performed the first comprehensive study employing the glycoprotein profiling of non-invasively obtained urine samples. Clearly, larger, confirmatory studies will be required to validate the candidate biomarkers identified in this study and to compare them with cytology and commercial assays, but the data presented here show that it is possible to detect and characterize bladder cancer based upon urinary glycoprotein profiles. In combination with other approaches, this strategy will provide targets for inclusion in multiplex biomarker panels that can be used for diagnosis, surveillance and asymptomatic screening.
Supplementary Material
Figure S1. Nano-LC-ESI-MS/MS spectrum of a single charged deglycosylated glycopeptide (at m/z 879.03) from A1AT. b- (red peaks) and y- (blue peaks) are series peak signals of fragmentation ions after CID (collision-induced dissociation). Black peaks are noise signals. The localization of the N-glycosylation site was determined by a mass increase of 1 Da on the N-X-(S/T) sequence after deamidation of asparagine residue into aspartic acid (D−N=1Da). The sequence of the peptide is LGNATAIFFLPDEGK. * indicates the N-glycosylated site.
Acknowledgements
This study was supported in by NIH/NCI Grant RO1 CA108597 (S.G.).
References
- 1.Jemal A, Siegel R, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2010. CA: A Cancer Journal for Clinicians. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 3.Trivedi D, Messing EM. Commentary: the role of cytologic analysis of voided urine in the work-up of asymptomatic microhematuria. BMC Urol. 2009;9:13. doi: 10.1186/1471-2490-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tetu B. Diagnosis of urothelial carcinoma from urine. Mod Pathol. 2009;22 Suppl 2:S53–S59. doi: 10.1038/modpathol.2008.193. [DOI] [PubMed] [Google Scholar]
- 5.Garcia Castro M, Fernandez Fernandez E, Martin Corriente MC, Garcia Hernandez S, Alvarez-Arguelles CH. [Usefulness of urine cytology for bladder carcinoma diagnosis: comparative study with biopsy] Actas Urol Esp. 2008;32:904–907. doi: 10.1016/s0210-4806(08)73958-5. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Kumar R, Gupta NP. Comparison of NMP22 BladderChek test and urine cytology for the detection of recurrent bladder cancer. Jpn J Clin Oncol. 2006;36:172–175. doi: 10.1093/jjco/hyi244. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura K, Kasraeian A, Iczkowski KA, Chang M, Pendleton J, Anai S, et al. Utility of serial urinary cytology in the initial evaluation of the patient with microscopic hematuria. BMC Urol. 2009;9:12. doi: 10.1186/1471-2490-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raitanen MP, Aine R, Rintala E, Kallio J, Rajala P, Juusela H, et al. Differences between local and review urinary cytology in diagnosis of bladder cancer. An interobserver multicenter analysis. Eur Urol. 2002;41:284–289. doi: 10.1016/s0302-2838(02)00006-4. [DOI] [PubMed] [Google Scholar]
- 9.Madeb R, Messing EM. Long-term outcome of home dipstick testing for hematuria. World J Urol. 2008;26:19–24. doi: 10.1007/s00345-007-0224-1. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez Banos JL, del Henar Rebollo Rodrigo M, Antolin Juarez FM, Garcia BM. Usefulness of the BTA STAT Test for the diagnosis of bladder cancer. Urology. 2001;57:685–689. doi: 10.1016/s0090-4295(00)01090-6. [DOI] [PubMed] [Google Scholar]
- 11.Landman J, Chang Y, Kavaler E, Droller MJ, Liu BC. Sensitivity and specificity of NMP-22, telomerase, and BTA in the detection of human bladder cancer. Urology. 1998;52:398–402. doi: 10.1016/s0090-4295(98)00219-2. [DOI] [PubMed] [Google Scholar]
- 12.Chang YH, Wu CH, Lee YL, Huang PH, Kao YL, Shiau MY. Evaluation of nuclear matrix protein-22 as a clinical diagnostic marker for bladder cancer. Urology. 2004;64:687–692. doi: 10.1016/j.urology.2004.05.038. [DOI] [PubMed] [Google Scholar]
- 13.Linder S. Cytokeratin markers come of age. Tumour Biol. 2007;28:189–195. doi: 10.1159/000107582. [DOI] [PubMed] [Google Scholar]
- 14.Gromov P, Moreira JMA, Gromova I, Celis JE. Proteomic strategies in bladder cancer: From tissue to fluid and back. Proteomics Clinical Applications. 2008;2:974–988. doi: 10.1002/prca.200780163. [DOI] [PubMed] [Google Scholar]
- 15.Topsakal M, Karadeniz T, Anac M, Donmezer S, Besisik A. Assessment of fibrin-fibrinogen degradation products (Accu-Dx) test in bladder cancer patients. Eur Urol. 2001;39:287–291. doi: 10.1159/000052455. [DOI] [PubMed] [Google Scholar]
- 16.Schmetter BS, Habicht KK, Lamm DL, Morales A, Bander NH, Grossman HB, et al. A multicenter trial evaluation of the fibrin/fibrinogen degradation products test for detection and monitoring of bladder cancer. J Urol. 1997;158:801–805. doi: 10.1097/00005392-199709000-00029. [DOI] [PubMed] [Google Scholar]
- 17.Irmak S, Tilki D, Heukeshoven J, Oliveira-Ferrer L, Friedrich M, Huland H, et al. Stage-dependent increase of orosomucoid and zinc-alpha(2)-glycoprotein in urinary bladder cancer. Proteomics. 2005;5:4296–4304. doi: 10.1002/pmic.200402005. [DOI] [PubMed] [Google Scholar]
- 18.Lokeshwar VB, Habuchi T, Grossman HB, Murphy WM, Hautmann SH, Hemstreet GP, 3rd, et al. Bladder tumor markers beyond cytology: International Consensus Panel on bladder tumor markers. Urology. 2005;66:35–63. doi: 10.1016/j.urology.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 19.Vrooman OP, Witjes JA. Urinary markers in bladder cancer. Eur Urol. 2008;53:909–916. doi: 10.1016/j.eururo.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez J, Thompson IM. Prostate-specific antigen: A review of the validation of the most commonly used cancer biomarker. Cancer. 2004;101:894–904. doi: 10.1002/cncr.20480. [DOI] [PubMed] [Google Scholar]
- 21.Carlson KJ, Skates SJ, Singer DE. Screening for ovarian cancer. Ann Intern Med. 1994;121:124–132. doi: 10.7326/0003-4819-121-2-199407150-00009. [DOI] [PubMed] [Google Scholar]
- 22.Kreunin P, Zhao J, Rosser C, Urquidi V, Lubman DM, Goodison S. Bladder cancer associated glycoprotein signatures revealed by urinary proteomic profiling. J Proteome Res. 2007;6:2631–2639. doi: 10.1021/pr0700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 25.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Beissbarth T, Hyde L, Smyth GK, Job C, Boon WM, Tan SS, et al. Statistical modeling of sequencing errors in SAGE libraries. Bioinformatics. 2004;20 Suppl 1:i31–i39. doi: 10.1093/bioinformatics/bth924. [DOI] [PubMed] [Google Scholar]
- 27.Decramer S, de Peredo AG, Breuil B, Mischak H, Monsarrat B, Bascands JL, et al. Urine in Clinical Proteomics. Molecular & Cellular Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Molecular & Cellular Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Pu FS. Urinary excretion of creatinine in normal subjects. Chinese Pharmaceutical Journal. 1992;44:235–240. [Google Scholar]
- 30.Baker SG. The central role of receiver operating characteristic (ROC) curves in evaluating tests for the early detection of cancer. J Natl Cancer Inst. 2003;95:511–515. doi: 10.1093/jnci/95.7.511. [DOI] [PubMed] [Google Scholar]
- 31.Sun Y, Goodison S. Optimizing molecular signatures for predicting prostate cancer recurrence. Prostate. 2009;69:1119–1127. doi: 10.1002/pros.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan W, Lee H, Deutsch EW, Lazaro CA, Tang W, Chen E, et al. A dataset of human liver proteins identified by protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. Mol Cell Proteomics. 2004;3:1039–1041. doi: 10.1074/mcp.D400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Tantipaiboonwong P, Sinchaikul S, Sriyam S, Phutrakul S, Chen ST. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics. 2005;5:1140–1149. doi: 10.1002/pmic.200401143. [DOI] [PubMed] [Google Scholar]
- 34.Nabi G, N'Dow J, Hasan TS, Booth IR, Cash P. Proteomic analysis of urine in patients with intestinal segments transposed into the urinary tract. Proteomics. 2005;5:1729–1733. doi: 10.1002/pmic.200401125. [DOI] [PubMed] [Google Scholar]
- 35.Orenes-Pinero E, Corton M, Gonzalez-Peramato P, Algaba F, Casal I, Serrano A, et al. Searching urinary tumor markers for bladder cancer using a two-dimensional differential gel electrophoresis (2D-DIGE) approach. J Proteome Res. 2007;6:4440–4448. doi: 10.1021/pr070368w. [DOI] [PubMed] [Google Scholar]
- 36.Saito M, Kimoto M, Araki T, Shimada Y, Fujii R, Oofusa K, et al. Proteome analysis of gelatin-bound urinary proteins from patients with bladder cancers. Eur Urol. 2005;48:865–871. doi: 10.1016/j.eururo.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 37.Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, et al. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen YT, Chen CL, Chen HW, Chung T, Wu CC, Chen CD, et al. Discovery of Novel Bladder Cancer Biomarkers by Comparative Urine Proteomics Using iTRAQ Technology. J Proteome Res. 2010;9:5803–5815. doi: 10.1021/pr100576x. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Li F, Sun W, Wu S, Wang X, Zhang L, et al. Concanavalin A-captured glycoproteins in healthy human urine. Mol Cell Proteomics. 2006;5:560–562. doi: 10.1074/mcp.D500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Feng S, Yang N, Pennathur S, Goodison S, Lubman DM. Enrichment of Glycoproteins using Nanoscale Chelating Concanavalin A Monolithic Capillary Chromatography. Analytical Chemistry. 2009;81:3776–3783. doi: 10.1021/ac900085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motoyama A, Yates JR., 3rd Multidimensional LC separations in shotgun proteomics. Anal Chem. 2008;80:7187–7193. doi: 10.1021/ac8013669. [DOI] [PubMed] [Google Scholar]
- 42.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 43.Chen EI, Yates JR., 3rd Cancer proteomics by quantitative shotgun proteomics. Mol Oncol. 2007;1:144–159. doi: 10.1016/j.molonc.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veenstra TD. Global and targeted quantitative proteomics for biomarker discovery. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:3–11. doi: 10.1016/j.jchromb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 47.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 48.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, et al. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J Proteome Res. 2009;8:3752–3759. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 49.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreunin P, Yoo C, Urquidi V, Lubman DM, Goodison S. Differential expression of ribosomal proteins in a human metastasis model identified by coupling 2-D liquid chromatography and mass spectrometry. Cancer Genomics Proteomics. 2007;4:329–339. [PMC free article] [PubMed] [Google Scholar]
- 51.Lindor NM, Yang P, Evans I, Schowalter K, De Andrade M, Li J, et al. Alpha-1-antitrypsin deficiency and smoking as risk factors for mismatch repair deficient colorectal cancer: a study from the colon cancer family registry. Mol Genet Metab. 2010;99:157–159. doi: 10.1016/j.ymgme.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamrita B, Chahed K, Trimeche M, Guillier CL, Hammann P, Chaieb A, et al. Proteomics-based identification of alpha1-antitrypsin and haptoglobin precursors as novel serum markers in infiltrating ductal breast carcinomas. Clin Chim Acta. 2009;404:111–118. doi: 10.1016/j.cca.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 53.Normandin K, Peant B, Le Page C, de Ladurantaye M, Ouellet V, Tonin PN, et al. Protease inhibitor SERPINA1 expression in epithelial ovarian cancer. Clin Exp Metastasis. 2010;27:55–69. doi: 10.1007/s10585-009-9303-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Nano-LC-ESI-MS/MS spectrum of a single charged deglycosylated glycopeptide (at m/z 879.03) from A1AT. b- (red peaks) and y- (blue peaks) are series peak signals of fragmentation ions after CID (collision-induced dissociation). Black peaks are noise signals. The localization of the N-glycosylation site was determined by a mass increase of 1 Da on the N-X-(S/T) sequence after deamidation of asparagine residue into aspartic acid (D−N=1Da). The sequence of the peptide is LGNATAIFFLPDEGK. * indicates the N-glycosylated site.