Abstract
Insulator elements and matrix attachment regions are essential for the organization of genetic information within the nucleus. By comparing the pattern of histone modifications at the mouse and human c-myc alleles, we identified an evolutionarily conserved boundary at which the c-myc transcription unit is separated from the flanking condensed chromatin enriched in lysine 9-methylated histone H3. This region harbors the c-myc insulator element (MINE), which contains at least two physically separable, functional activities: enhancer-blocking activity and barrier activity. The enhancer-blocking activity is mediated by CTCF. Chromatin immunoprecipitation assays demonstrate that CTCF is constitutively bound at the insulator and at the promoter region independent of the transcriptional status of c-myc. This result supports an architectural role of CTCF rather than a regulatory role in transcription. An additional higher-order nuclear organization of the c-myc locus is provided by matrix attachment regions (MARs) that define a domain larger than 160 kb. The MARs of the c-myc domain do not act to prevent the association of flanking regions with lysine 9-methylated histones, suggesting that they do not function as barrier elements.
In the eukaryotic nucleus, chromosomes occupy individual, nonoverlapping territories that are subdivided structurally and functionally into euchromatic and heterochromatic domains (8). The boundaries separating these domains have emerged as important elements in the establishment and maintenance of the chromosomal organization by preventing the encroachment of silencing heterochromatin into transcriptionally active domains. Boundary elements may also function as “gatekeepers” that either permit or prevent access of regulatory signals to the transcription machinery within a transcription domain. This organizational structure ensures that the normal transcriptional unit is shielded from the influence of neighboring enhancer regions and may also facilitate the stimulatory function of more remote locus control regions.
Structural differences distinguishing transcriptionally active and inactive chromatin have been recognized for decades; histone acetylation was described as a marker for active genes (1, 40), and differential sensitivity to DNase I was identified as a characteristic feature that distinguishes the chromatin of transcribed and silent genes (46). Correlations between N-terminal modifications of histones and the structural and functional organization of chromatin have led to the hypothesis that a “histone code” is essential for the correct tissue-specific regulation of gene expression during development (reviewed in reference 21). Consistent with this model, transcriptionally silent regions are generally organized into heterochromatic structures that typically include histone H3 methylated at lysine 9 (H3meK9) but lack acetylated H3 and H4. In contrast, histone acetylation is a property common to most transcriptionally active regions and to regulatory regions. Histone acetylation therefore may serve as a marker for regulatory and/or transcribed domains. The extent of histone acetylation can differ considerably: while at some genes the acetylated histones H3 and H4 are restricted to the promoter, histone acetylation at the murine β-globin gene extends over more than 100 kb (5, 14, 15). This extensive domain is further divided into distinct subdomains formed by local hypoacetylation at the embryonic β-globin genes (24).
The protection of transcriptionally active regions from the silencing effects of compacted chromatin is thought to require insulator elements. These elements were originally identified in Drosophila and were subsequently found in yeast and vertebrate cells (reviewed in references 18, 37, and 47). For instance, in flies both the scs/scs′ and gypsy elements are associated with DNase I-hypersensitive sites, block enhancer-promoter interaction, and provide position-independent expression of transgenes. Previous studies at the mating-type region in fission yeast have shown that specific DNA elements located at the borders of the silenced domain contribute to the establishment of boundaries separating euchromatin from heterochromatin (36). Studies of silencing mechanisms at the yeast telomeres and at the ribosomal DNA and HMR loci have led to the identification of barrier elements that protect genes from the negative influence of surrounding heterochromatin (reviewed in reference 37).
In the chicken β-globin locus, studies with the HS4 element (cHS4) have provided insight into biochemical pathways of boundary element function. The enhancer-blocking activity of cHS4 is mediated by the CTCF protein (3). This 11-zinc-finger protein was originally identified as a repressor of c-myc transcription and later was found to be necessary and sufficient for vertebrate enhancer blocking (3, 12). The barrier activity of cHS4 is associated with a peak of histone acetylation over the insulator element independent of the expression status of the β-globin gene (35). Consistent with a role for histone acetylation in the barrier function of cHS4, the ability of a yeast tRNA gene to inhibit the progression of silenced telomeric chromatin requires the recruitment of transcription activators that associate with histone-modifying complexes (10).
The structural and functional organization of a chromosomal domain is thought to involve anchoring the chromosomes to a proteinaceous scaffold (matrix) of the nucleus through matrix attachment regions (MARs). The association of MARs with some enhancers (reviewed in reference 42) suggests that they participate in the long-range control of gene expression by facilitating the activity of enhancers. However, the MARs of the chicken lysozyme gene and the human apolipoprotein B (apoB) gene are not associated with enhancer sequences (22, 39, 45). These elements have been shown to protect genes from position effects. Other experiments have shown that the establishment of position-independent expression is separate from the ability of an element to bind nuclear matrix preparations in vitro (39).
The global organization of chromosomal domains within the nucleus may depend on the boundary elements that are distributed throughout chromosomes. Elegant studies using the Drosophila gypsy element have shown that the gypsy insulator can recruit sequences normally found in different regions of the nucleus to a single nuclear subregion (17). These findings led to a model in which boundary elements associate to form insulator bodies that organize the chromosomes into functional domains. In this model, transcriptionally inert heterochromatin forms the core of these structures, whereas transcriptionally active euchromatin forms rosette-like loops located on the surface of the chromosomal territory. The insulator bodies may be attached to a structure such as the nuclear matrix or nuclear lamina that provides a scaffold for nuclear organization (reviewed references 26 and 47).
The positioning of genes within the nucleus may have a profound influence on their expression, and the identification of DNA elements that contribute to this level of organization of genes will advance our understanding of the mechanism of gene expression. We have mapped the hyperacetylated regions that lie within the human and mouse c-myc loci. These studies reveal a conserved region located 2 kb upstream of the transcribed region which exhibits insulator activity: it efficiently inhibits enhancer-promoter interaction and protects transgenes from position-effect variegation. The transcriptionally active acetylated domain of the c-myc gene is embedded within a 160-kb region that is enriched in lysine 9-methylated histone H3 and is devoid of any other expressed genes. Matrix attachment regions separate this domain from the neighboring pvt1 gene and from another gene locus of unknown identity. Thus, the 5′ and 3′ MARs of the c-myc locus define a 160-kb domain that contains both hetero- and euchromatin.
MATERIALS AND METHODS
Cell culture.
The human HL60 and Jurkat cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and the antibiotics penicillin (50 U/ml) and streptomycin. HL60 cells were induced to differentiate with 1.5% dimethyl sulfoxide (DMSO) for 2 h. The interleukin 2 (IL-2)-dependent murine CD8+ T-cell line CTLL2 was maintained in RPMI 1640 supplemented with 1 mM sodium pyruvate, 10% fetal bovine serum (FBS), 2 mM l-glutamine, antibiotics, and 50 U of rIL2 (Chiron, Emeryville, Calif.)/ml. To inhibit proliferation and to generate a resting T-cell population, CTLL2 cells were placed into medium lacking IL-2 for 8 h. For mitogenic induction, 100 U of rIL2/ml was added.
RNA analyses.
Nuclease S1 protection assays were performed as previously described (25). Briefly, total RNA was isolated from human HL-60 or murine CTLL cells. 5′ end-labeled probes were generated by extension of an oligonucleotide that had been phosphorylated with T4-kinase and [γ-32P]ATP in a 14-cycle PCR on linearized double-stranded plasmid DNA (5 μg). End-labeled single-stranded DNA was purified with a 6% polyacrylamide gel. Sequences of extension primers for human and murine c-myc probes corresponded to positions +302 to +270 and +270 to +253, respectively, relative to the P1 initiation site. Sequences of the human and mouse GAPDH oligonucleotide probes are described in reference 30. Hybridization of single-stranded probes with total RNA was performed as described previously (31). S1 protected fragments were resolved on denaturing 8% polyacrylamide gels.
DNA constructs.
The construct E-P-neo-scs′ was supplied by M. Krangel (49). In these constructs, E, P, neo, and scs′, represent the TCRδ enhancer, the Vδ promoter, the neomycin resistance gene, and the Drosophila hsp70 scs′ boundary element, respectively. Derivative constructs were generated as follows: P-neo-scs′ lacking the Eδ enhancer was generated by XbaI/ClaI digestion, treatment with T4 polymerase, and religation. All boundary and control fragments were introduced into the NotI and/or XbaI sites upstream of Eδ or the ClaI and/or SalI sites between Eδ and the Vδ promoter. The E-MINE-P-neo-scs′ construct was generated by triple ligation of a PCR-generated 1.0-kb ClaI/HindIII barrier element (BE) fragment (see below) and the 640-bp HindIII/SalI CTCF into ClaI-SalI sites between Eδ and the Vδ1 promoter. The 1.0-kb BE fragment from the c-myc locus was generated by PCR amplification of DNA from BAC clone 19.1 (forward primer, 5′ to 3′, CTATGAGATCGATGTGGACC; reverse primer, 5′ to 3′, GCTAATGAAAGTCGAACTATGG), and the 640-bp CTCF was generated by PCR amplification of cosmid DNA (forward primer, 5′-GAGTTTCATCGATGTGGGGG-3′; reverse primer, 5′-GGGGTGCGTCGACAGCATGT-3′). Control sequences derived from the chicken β-globin HS4 (1.2-kb element) and human β-globin HS5 (2.0-kb and 140-bp elements) were cloned between the enhancer and the promoter at the ClaI-SalI site by either directional restriction digest-mediated or blunt-ended cloning. The 1.2-kb chicken HS4 element was derived from an XbaI digest of the PJC5-4 construct as described by Chung et al. (7) (GenBank sequence accession number U78775). The ∼2.0-kb BamHI-EcoICRI 5′ human HS5 fragment corresponds to bp 1031 to 2872 (GenBank accession number L22754) (27). The 140-bp CTCF-containing element from human HS5 (GenBank accession number L22754; bp 611 to 755) was PCR generated with ClaI-SalI-compatible ends using primers F (ccatcgatGGGCTCCACCATCTTGGAC) and R (acgcgtcgacGGGATTATATTTCCTGA). All constructs were verified by sequencing (lowercase indicates linker sequences; underlining indicates ClaI-SalI restriction sites).
Soft agar colony-forming assay.
The transfection of Jurkat cells with each construct was performed in triplicate. Jurkat cells (107 cells) were electroporated with 1.5 pmol (∼5.0 to 10.0 μg) of linearized plasmid using a BTX (Hercules, Calif.) electroporator at 250 V, 1,700 μF, 72Ω (time constant, ∼40 ms). After the electroporation, cells were placed on ice for 20 to 30 min. The cell suspension was then transferred into 10 ml of RPMI 1640-10% fetal bovine serum and cultured at 37°C. After 48 h, 5 × 105 cells were resuspended in a 7-ml RPMI plating medium (11% FBS, 31% Jurkat-conditioned medium, 50 μl of penicillin-streptomycin [78 μg/ml], 100 μl of l-glutamine [78 U/ml], and 750 μg of active G418 [Life Technologies, Gaithersburg, Md.]/ml in RPMI). After addition of 3 ml of soft agar plating medium (1:1 volumes of 0.2% agar [Difco] and 2× RPMI 1640 medium), cells were plated and incubated for 3 to 4 weeks.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (43). Chromatin was cross-linked in the presence of 1% formaldehyde for 5 min at room temperature. After the addition of glycine to a 0.125 M concentration, the cells were washed in ice-cold phosphate-buffered saline containing protease inhibitors (Roche Molecular Biochemicals). Cells were pelleted, resuspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1), 1× protease inhibitor cocktail, and 10 mM butyrate), and sonicated 6 times for 20 s each at setting 6.5 in a Branson sonicator with a microtip. The lengths of the DNA fragments ranged from 200 to 800 bp. After centrifugation to remove cell debris, the whole-cell extract was diluted 10-fold with ChIP dilution buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 1× protease inhibitor cocktail, and 10 mM butyrate). Before immunoprecipitation, the chromatin solution was precleared with preblocked protein A agarose slurry and normal rabbit serum to reduce nonspecific background. Anti-acetylated histone H3, anti-methylated K9, or anti-CTCF antibody (Upstate Biotechnology) was added, and the mixture was incubated overnight at 4°C with agitation; a no-antibody control was also performed for each ChIP assay. Immune complexes were collected by addition of 25 μl of preblocked protein A agarose slurry that had been preincubated with bovine serum albumin and salmon sperm DNA. After incubation at 4°C for 2 h, the beads were washed several times according to the method described in reference 38. The DNA contained within the immune complexes was recovered by the addition of proteinase K and SDS (0.1%) for 1 h at 55°C followed by incubation at 65°C for at least 4 h to reverse the cross-links. The DNA was then purified by phenol-chloroform extraction and precipitated with ethanol precipitation.
Antibodies.
Anti-acetylated histone H3 (06-599; Upstate Biotechnology), anti-dimethyl-histone H3 (lysine 9) (07-212; Upstate Biotechnology), and anti-CTCF (06-917; Upstate Biotechnology) were used for ChIP assays.
PCR analysis.
DNA recovered from immunoprecipitations or from nuclear matrix preparations was analyzed by duplex PCR using c-myc-specific primers and reference primers (β-globin, apoB). Sequences of the primers are available upon request. The reactions were performed with the Failsafe Taq polymerase in a solution containing 1× buffer F (Epicenter) and 10 pmol of each c-myc primer in the presence of [32P]dCTP. Under these conditions, the amplification of fragments was linear. After electrophoresis on a native 6% acrylamide gel, the signals were quantified with the Cyclone phosphoimager. The fold enrichment in each immunoprecipitation or MAR assay was determined by calculating the ratio of the signal obtained with c-myc primers to that of the reference primers.
MAR assay.
Nuclei from Jurkat and HL60 cells were isolated as described by Izaurralde et al. (20) with some modification and were kept at −20°C in storage buffer (5 mM Tris-Cl [pH 7.5], 20 mM KCl, 125 μM spermidine, 50 μM spermine, 500 μM EDTA, 1% [vol/vol] thioglycol, 50% [vol/vol] glycerol, 0.01% digitonin, 0.2 mM phenylmethylsulfonyl fluoride, 0.7-μg/ml pepstatin) at a density of ∼108 nuclei/ml. Matrices were stable for at least 6 months stored in 50% glycerol at −20°C. To isolate matrix-bound (pellet [P]) or soluble (supernatant [S]) DNA, nuclei from Jurkat T cells were subjected to a lithium 3,5-diiodosalicylate extraction protocol described by Mirkovitch et al. (33). Nuclei of ∼107 matrices were used per assay. Nonscaffold proteins were extracted by the addition of 1.0 ml of 10 mM lithium 3,5-diiodosalicylate (D-3635; Sigma) extraction buffer, and the salt-extracted nuclei were collected and subjected to restriction enzyme digests with EcoRI, HindIII, XbaI, and ClaI (∼500 to 1,000 U/sample) and/or with the addition of EcoICR1 (Promega). After proteinase K treatment, DNA samples were then subjected to phenol-chloroform extraction and ethanol precipitation. To quantify the relative distribution of specific DNA sequences in the P and S fractions, we performed a duplex PCR assay. Two nanograms of each P or S fraction was used as a template for amplification with primer pairs corresponding to each restriction fragment (generated by HindIII, XbaI, ClaI, EcoRI, and EcoICRI) across ∼200 kb of the c-myc locus. The PCRs contained a primer set specific for the MAR at the apoB gene (2) as a positive control (forward primer, 5′-GGGTGAATGAATGCCCTATG-3′; reverse primer, 5′-TACTCCATGCGAGGTCCACT-3′). Primer sequences specific for the c-myc fragments are available upon request. The relative level of association with the pellet (%P) or with the supernatant (%S) was calculated using the following formulas: %P = IP/(IP + IS) and %S = IS/(IP + IS), where IP and IS represent the intensity of PCR signals obtained with the P or the S fraction.
RESULTS
Murine and human c-myc domain boundaries are highly conserved.
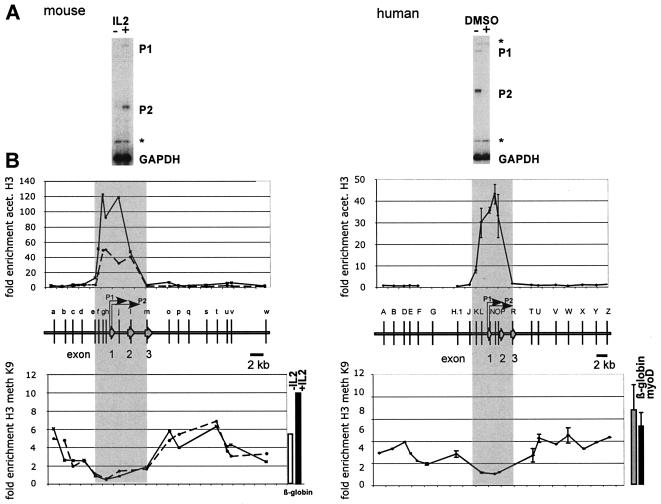
Transitions in the histone modification pattern mark the boundaries of yeast and vertebrate gene loci (29, 36). To define the euchromatic and heterochromatic domains of the mammalian c-myc gene, we generated a high-resolution map of the pattern and distribution of histone acetylation and methylation encompassing 25 kb of both the human and mouse c-myc loci using ChIP. These studies were performed in the murine T-cell line CTLL2, in which c-myc transcription is highly induced by IL-2 treatment, and in the human promyelocytic leukemia cell line HL60, in which c-myc expression is downregulated by DMSO-mediated induction of differentiation (Fig. 1A). Formaldehyde-cross-linked chromatin was prepared from each of these cell lines and immunoprecipitated with antibodies that specifically recognize acetylated lysine residues 9 (K9) and 14 (K14) or methylated K9 of histone H3. The enrichment of histone modifications was measured by duplex PCR by normalizing the c-myc-specific signal with the corresponding signal from the murine β-globin gene or from sequences 10 kb upstream of the human c-myc exon 1. The patterns of histone acetylation are very similar in both cell types. Hyperacetylated histone H3 associates with a 7.5-kb region that includes the c-myc promoter region and the transcribed sequences (Fig. 1B). A high level of hyperacetylated histone H3 is present at the transcription start site of the P1 and P2 promoters and extends into the exon 2 and intron 2 sequences. H3 acetylation decreases with increasing distance from the transcription initiation site and is barely detectable within exon 3 and 2.3 kb upstream of exon 1. Primer sets that detect sequences further upstream of −2.3.kb and downstream of exon 3 did not detect any enrichment relative to the internal standard primer sets. ChIP experiments using antibodies specific for acetylated-H4 histones revealed an essentially identical pattern of enrichment (data not shown).
FIG. 1.
The conserved pattern of histone acetylation (acet.) and methylation (meth) defines the boundaries of the mammalian c-myc locus. (A) Expression of the c-myc gene in resting (−IL-2) and mitogen-induced (+IL-2) mouse T cells (CTLL2) and in proliferating (−DMSO) and differentiating (+DMSO) human promyelocytic leukemia cells (HL60). C-myc transcripts initiated at the P1 and P2 promoters are undetectable both in resting CTLL2 cells and in differentiating HL60 cells. Total RNA was analyzed in a S1 nuclease assay with end-labeled probes specific for the mouse or the human c-myc gene. End-labeled oligonucleotides specific for the mouse and human GAPDH gene were included as controls. Asterisks denote undigested probe. (B) ChIP experiments were performed with anti-acetylated H3 and anti-methylated K9 H3 antibody. Cross-linked chromatin was isolated from resting (left panel, broken line) and IL-2-induced (left panel, solid line) mouse CTLL2 cells, and from proliferating human HL60 cells (right panels). DNA isolated from immunoprecipitated chromatin was subjected to duplex PCR to amplify DNA fragments from the mouse (left panels) or human c-myc locus (right panels). The relative positions of the amplified regions (a through w and A through Z, respectively) are indicated by vertical bars. To determine the relative levels of H3 acetylation in these regions, primers specific for the mouse β-globin gene promoter (upper left panel) or for region G (upper right panel) were included in the PCRs as internal standards. Enrichment of K9-methylated H3 was measured relative to c-myc exon 1 (human) or c-myc exon 2 (mouse) sequences. The degree of enrichment is calculated relative to the ratio of the signals obtained in the input DNA fraction using the c-myc and β-globin primer pairs. Results from the human c-myc gene are based on two independent experiments using different chromatin preparations. Data shown for the mouse c-myc gene are results from a single representative ChIP experiment. High levels of H3 acetylation are detected across promoter and transcribed regions in both species, from −2.5 kb to +4.5 kb relative to exon 1 (shaded area). The hyperacetylated regions are flanked by heterochromatin enriched in K9-methylated histones H3. The degree of hypermethylation is similar to that of the transcriptionally inactive murine β-globin gene in CTLL2 cells (see bars to the right of the lower panel) and of the human myoD and β-globin genes in HL60 cells.
ChIP assays with antibodies specific for K9-methylated histones reveal an inverse distribution along the 25-kb region of the human and murine c-myc gene loci (Fig. 1B, lower panels). The promoter region and the transcribed sequences of exon 2 are not enriched after immunoprecipitation with the antibody specific for K9-methylated histone H3. In contrast, the hypoacetylated regions upstream of −2.3 kb (relative to exon 1) and downstream of exon 3 generally are marked by K9-methylated histone H3. The level of enrichment relative to c-myc transcribed sequences ranged from three- to sixfold, a magnitude similar to that obtained for the β-globin or myoD locus. These two loci are transcriptionally silent in both human HL60 cells and mouse CTLL2 cells.
Our data indicate that the mammalian c-myc gene resides in a large hyperacetylated domain that is flanked by condensed chromatin. The physical extent of the c-myc domain in mouse and human cells is highly conserved. The hyperacetylated domain extends approximately 2 kb upstream of the c-myc exon 1 sequences and is flanked by transcriptionally inert chromatin containing K9-methylated, hypoacetylated histones H3.
Association of a CTCF binding element with the 5′ boundary.
The conserved pattern of histone acetylation and methylation at the human and mouse c-myc loci suggests that a conserved functional element may exist that acts to separate transcriptionally active from transcriptionally inactive regions. The boundary elements at the β-globin gene and at the apoB gene contain binding sites for CTCF. In vitro experiments and alignment of the c-myc sequence with a consensus CTCF binding sequence (3) identified several potential CTCF binding sites within the human c-myc gene locus (Table 1). Of these, the CTCF sites positioned near the 5′ boundary (2 kb upstream of exon 1) and at the P2 promoter (0.23 kb downstream of the beginning of exon 1) are highly conserved between the human and mouse c-myc gene (Table 1) and occur near DNase I-hypersensitive sites (44).
TABLE 1.
CTCF binding motifs within the c-myc locus
| Siteb | Sequence | Location relative to exon I (kb) | Reference | Detected by ChIP |
|---|---|---|---|---|
| I | TACTAAAAGCCAGGAGGGGAAGGGACAACACTAAGC | −5.7 | − | |
| II, 5′ boundary | TCTCTGCTGCCAGTAGAGGGCACACTTACTTTACT | −2.0 | + | |
| Murine 5′ boundary | TCTCTGtgGCCAGTAGAGGGCACACTTACTTTACT | −1.9 | + | |
| III (rev. orientation)a | TTTGGGAACCCGGGAGGGGCGCTTATGG | +0.02 | − | |
| Consensus | CCRNNAGRGG | Bell et al., 1999 | ||
| IV, site B (P1) | GAGCTGTGCTGCTCGCGGCCGCCACCGCCGGGCCCC | +0.03 | Fillipova et al., 1996 | − |
| V, site A (P2) | ATTCCAGCGAGAGGCAGAGGGAGCGAGCGGGCGGCC | +0.23 | Fillipova et al., 1996 | + |
| Murine site A (P2) | ATTCCAGCGAGAGaCAGAGGGAGtGAGCGGaCGGtt | +0.18 | Fillipova et al., 1996 | + |
| Mutant 5′ boundary | TCTCTGCTGCCAATATATGGAACACTTACTTTACT | − |
Reverse orientation.
Roman numerals I to V refer to CTCF sites within the human c-myc allele; murine sequences refer to regions homologous to the human sequence in the murine c-myc gene.
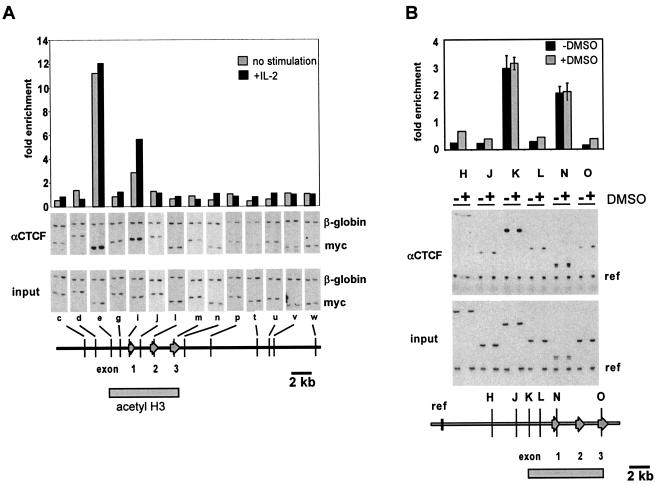
In order to determine whether the 5′ boundary of the mammalian c-myc gene associates with CTCF in vivo, we performed chromatin immunoprecipitation experiments with a CTCF-specific antibody. Formaldehyde-cross-linked chromatin from resting and mitogen-induced mouse CTLL2 cells was immunoprecipitated, and the enrichment of c-myc regions relative to the β-globin gene was measured by duplex PCR (Fig. 2). In addition, we have analyzed binding of CTCF to these sites in both proliferating and differentiating HL60 cells in which transcription of the c-myc gene is downregulated in the presence of DMSO. The quantitation of the signals obtained by primer sets specific for various regions in the human c-myc gene indicates that CTCF associates exclusively with the 5′ boundary region and the P2 promoter region (Fig. 2). A similar result was obtained in ChIP experiments with mouse CTLL2 cells in which only the regions containing the two conserved CTCF consensus sites were enriched after immunoprecipitation. The analysis of the 3′ boundary of the c-myc gene using various primer sets showed no binding of CTCF in these regions. Interestingly, the binding of CTCF to the 5′ boundary and the promoter-proximal region was not influenced by the IL-2-mediated mitogenic induction of CTLL2 cells. Under these conditions the expression of the murine c-myc gene is upregulated at least 10-fold (Fig. 1A) (34). Similarly, the inhibition of c-myc expression during the induction of differentiation of HL60 cells by DMSO also did not affect the association of CTCF with its target sites. Thus, our result that CTCF is constitutively bound to the murine and human c-myc loci is not compatible with a direct role of CTCF in transcriptional repression of the c-myc gene, as previously reported (12).
FIG. 2.
CTCF binds in vivo to selected sites of the mammalian c-myc gene loci. ChIP experiments using the anti-CTCF antibody were performed as described in Fig. 1. (A) CTCF binds to the 5′ boundary as well as to the P2 promoter region (e and i) in both resting and IL-2-induced CTLL2 cells. Signals obtained with the murine c-myc primer sets (c to w) are normalized to the signal obtained by the murine β-globin primer set. The degree of enrichment is calculated relative to the ratio obtained in the input DNA fraction as in Fig. 1. (B) CTCF binds to the homologous regions of the human c-myc gene (primer sets K and N). Binding of CTCF is constitutive and not affected by the DMSO-induced inhibition of transcriptional elongation. Note that the 3′ boundaries of the hyperacetylated c-myc regions (grey bar at bottom of figures) are not occupied by CTCF. The data shown for the human c-myc gene are averages from two independent ChIP experiments with independently prepared chromatin derived from HL60 cells, with error bars indicating the standard deviation. The data shown for the murine c-myc gene represent a single representative ChIP.
The c-myc insulator element, MINE.
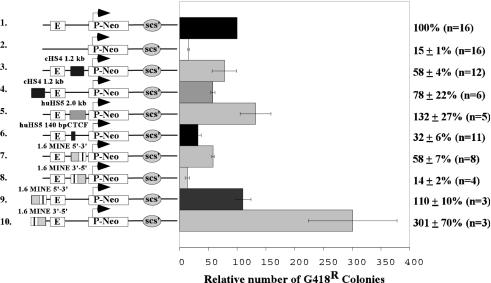
Insulator elements inhibit the spreading of heterochromatin into euchromatic regions and prevent promoter activation by an upstream enhancer element when placed between the enhancer and promoter. To determine whether this functional activity is specified by sequences at the 5′ boundary of the human c-myc gene, we employed the colony assay, an established assay of enhancer-blocking and barrier activity (7, 23, 49). In this assay, the ability of the 5′ boundary element to repress or stimulate transcription of a selectable marker construct is measured after stable integration into genomic sites. As illustrated schematically in Fig. 3, the presumed insulator sequences are placed either between the enhancer and promoter or upstream of the enhancer. The number of colonies obtained after transfection is proportional to the number of cells expressing the neomycin-resistance gene (Neor) at levels sufficient to confer resistance to G418. The Neor transcription of the reference construct is driven by the human T-cell-specific promoter Vδ and the Eδ enhancer from the TCRδβ locus (E-P-neo-scs′; Fig. 3, construct 1). In the absence of the Eδ enhancer, the reduced neomycin expression results in a sevenfold reduction in the number of G418-resistant colonies (15% versus 100%; construct 2s and 1, respectively). Consistent with previous reports (5-7, 11), the insertion of one copy of the 1.2-kb chicken β-globin HS4 insulator sequence (Fig. 3, construct 3) or the 140-bp insulator fragment derived from the HS5 of the human β-globin gene (hHS5; Fig. 3, construct 6) between the Eδ enhancer and the Vδ promoter reduces by approximately threefold the number of G418-resistant colonies after transfection into the Jurkat T-cell line. In contrast, a 2.0-kb control fragment, derived from a region flanking human HS5, does not influence the expression of the Neor reporter gene when inserted between the Eδ and the Vδ promoter (Fig. 3, construct 5). This suggests that the observed inhibition of enhancer activity by DNA fragments is sequence specific and is not affected by changes in the distance between the enhancer and the promoter. In addition, the placement of the cHS4 element upstream of the Eδ enhancer also has very little effect (Fig. 3, construct 4).
FIG. 3.
The 1.6-kb MINE impairs enhancer-promoter interaction and functions as a barrier element in colony assays. The ability of MINE to support expression or interfere with enhancer activity was determined using reporter constructs that contain a neomycin-selectable marker gene driven by the human T-cell receptor δβ promoter (P-NeoR-scs′) and the human Eδ enhancer (E). In addition, the constructs contain the Drosophila insulator element scs′ to inhibit transcriptional interference originating from the integration site at the 3′ end as described in the work of Zhong and Krangel (49). The 1.6-kb MINE fragment or the chicken β-globin insulator cHS4 was inserted in either orientation between the enhancer and promoter (constructs 3, 5, 6, 7, and 8) or upstream of the enhancer (constructs 4, 9, and 10). The constructs were transfected into the Jurkat T-cell line, and the number of G418-resistant colonies that express the reporter construct was determined by soft agar cloning (see Materials and Methods). The activity of the parent construct E-P-neo-scs′ (construct 1) was used as a reference and normalized to 100. Results are shown with standard errors, and the number of individual experiments (n) is indicated.
Similar to the results seen with cHS4 and the hHS5, a 1.6-kb fragment derived from the c-myc 5′ boundary (−1.7 kb to −3.3 kb relative to exon 1) inhibits the enhancer activity when it is placed between Eδ and Vδ (Fig. 3, construct 7), and it has no inhibitory effect when it is positioned upstream from Eδ (Fig. 3, construct 9). Because of its functional similarity to cHS4, the 1.6-kb fragment of the 5′ boundary was named MINE (the c-myc insulator element). Surprisingly, MINE blocked enhancer-promoter interaction more efficiently in the reverse orientation (Fig. 3, construct 8), in which case it reduced the number of G418-resistant colonies sevenfold, equivalent to the number of colonies obtained after deletion of the enhancer (Fig. 3, constructs 2 and 8). However, MINE had the opposite effect when inserted upstream of the enhancer (Fig. 3, construct 10); it increased the number of G418-resistant colonies by threefold, indicating that a greater percentage of cells of the transfected pool express a threshold level of Neor. These results led us to conclude that MINE provides at least two functional activities: enhancer-blocking activity and barrier activity, both integral components of the 5′ boundary of the c-myc locus.
MINE contains separable functional activities.
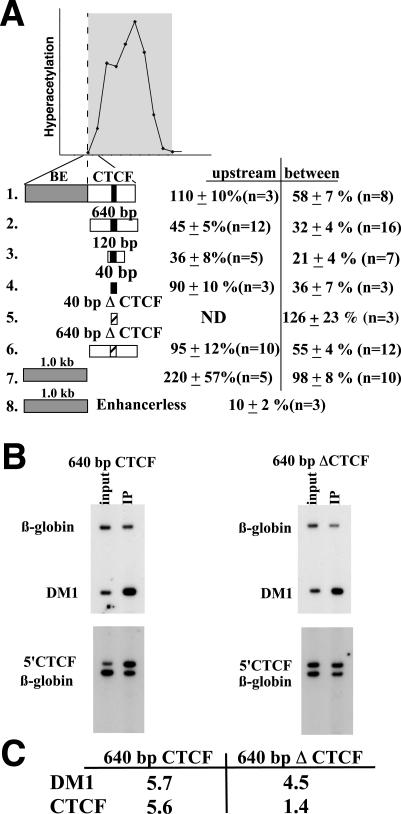
To determine whether the components of MINE that are embedded within the defined 1.6-kb fragment are experimentally separable, we have tested subfragments of MINE using the same colony assay used in the previous experiments (Fig. 4A). Distinct 40-, 120-, and 640-bp fragments of MINE encompassing the CTCF core consensus sequence were inserted between the enhancer and the promoter or upstream from the enhancer of the E-P-neo-scs′ construct. The 40-bp element reduced the level of G418-resistant colonies threefold, similar to the level obtained with the complete 1.6-kb fragment. The mutation of 4 bp within the CTCF consensus sequence (Table 1) removed this enhancer-blocking activity. Consistent with its function as an enhancer-blocking element, the 40-bp minimal CTCF binding site of MINE did not affect Neor gene expression when positioned upstream of the enhancer (Fig. 4, construct 4). In combination, these experiments indicate that enhancer-blocking activity of MINE is at least in part mediated by the 40-bp region containing the CTCF-binding sequence.
FIG. 4.
The 1.6-kb MINE mediates both barrier and enhancer-blocking activities. (A) Results of colony assays performed with subfragments of MINE. The graph above indicates the position of MINE relative to the hyperacetylated region over the c-myc gene locus (also refer to Fig. 1). Colony assays were performed as described in Fig. 3. The 640-, the 120-, and 40-bp fragments containing a functional CTCF-binding site (black bar) were inserted between the enhancer and promoter (between) or upstream of the enhancer (upstream) in the E-P-neo-scs′ construct (Fig. 3, construct 1). CTCF binding sites with point mutations (40-bp ΔCTCF and 640-bp ΔCTCF) are indicated by an open bar. (B) ChIP assay to confirm the loss of CTCF binding in the mutated 640-bp fragment. Chromatin derived from Jurkat cells transfected with constructs containing the 640-bp fragment or the 640-bp ΔCTCF fragment was used in immunoprecipitations with the CTCF antibody. Primers that are specific for the DM1 locus (13) were used as a positive control. Primers (5′CTCF) that detect the CTCF-binding region within the stably integrated transgene do not detect sequences of the endogenous c-myc gene. (C) Quantitation of ChIP experiment shown in Fig. 4B. The enrichment of CTCF sites in the transgenes E-640bp-Pneo-scs′ (640 bp CTCF) and in E-640bpΔCTCF-Pneo-scs′ (640 bp Δ CTCF) is shown compared to enrichment at the endogenous DM1 locus.
Both the 120- and the 640-bp fragments decrease transcriptional activity when positioned between the enhancer and the promoter (Fig. 4A, constructs 3 and 2; 21 and 32%, respectively). However, these elements also have an inhibitory activity when inserted upstream of the Eδ enhancer (36 and 45%, respectively; Fig. 4A). In addition, the mutation of the 640-bp fragment at four nucleotides that were found to eliminate CTCF function in the context of the 40-bp fragment only partially abrogates the enhancer-blocking function. The mutated 640-bp fragment still reduces the number of G418-resistant colonies by twofold (Fig. 4A, construct 6). To ensure that the reduced gene activity observed in the presence of the mutated 640-bp fragment is not mediated by additional cryptic CTCF sites, we have performed ChIP experiments on cell clones that were derived from pools that had been transfected with reporter constructs containing the normal and mutated 640-bp fragment (Fig. 4B). As a standard for the recovery of CTCF-bound DNA, we have determined the enrichment of myotonic dystrophy gene sequences (DM1) relative to β-globin (13). In both cell lines (E-640bp CTCF-Pneo-scs′ and E-640bpΔCTCF-Pneo-scs′), DM1 sequences cross-linked to CTCF were efficiently recovered (5.7- and 4.5-fold enrichment, respectively; Fig. 4B and C). In contrast, PCR amplification with primers specific for the 640-bp fragment did not reveal any enrichment when the CTCF consensus sequence was mutated (Fig. 4C). This result suggests that the residual inhibitory activity of the 640-bp fragment containing a mutated CTCF site may be due to CTCF-independent enhancer-blocking and/or silencing elements. However, it remains unclear whether these activities are functionally significant in the context of the full-length MINE.
Barrier activity of MINE.
The 640-bp region at the human c-myc boundary that mediates enhancer-blocking activity is highly similar to the homologous region of the mouse c-myc gene. The boundary sequences further upstream also show a high level of sequence conservation despite the absence of histone hyperacetylation or histone H3 K9 methylation. To determine the role of this region in the formation of the boundary at the c-myc locus, we inserted the 1-kb 5′ portion of MINE (BE) (Fig. 3) into the reporter construct E-P-neo-scs′ and performed colony assays. The BE fragment, in contrast to the CTCF-containing elements, did not affect the number of G418-resistant colonies when positioned between the enhancer and promoter (98% ± 8% [n = 10]) (Fig. 4, construct 7). However, when the BE sequence was positioned upstream of the enhancer, we observed a more than twofold increase in the number of colonies (220% + 57% [n = 5]) (Fig. 4, construct 7). In the absence of the Eδ enhancer, the BE was not able to enhance promoter activity (10% ± 2% [n = 3]) (Fig. 4, construct 8). The increase in the number of G418-resistant colonies suggests that the BE sequence increases the likelihood of transgene expression in various genomic sites, a hallmark feature of barrier elements. Taken together, these results demonstrate that the 1.6-kb MINE contains at least two different activities: the enhancer-blocking and barrier activities, which contribute to the boundary function.
Nuclear matrix association at the c-myc locus.
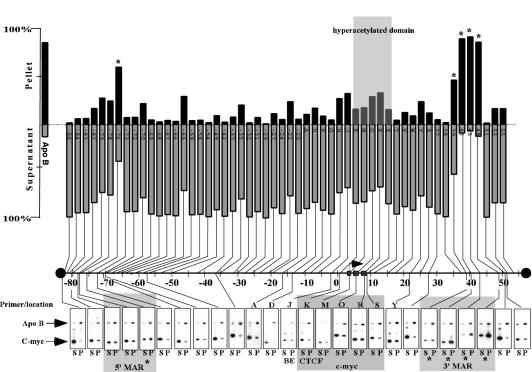
Boundary elements associate in some cases with the nuclear matrix (26, 28, 48). In addition, many MARs have been reported to enhance the activity of reporter genes (reviewed in reference 4). Thus, we speculated that the insulator activity of MINE might correlate with the formation of a structural boundary resulting from its attachment to the nuclear matrix. To test whether the MINE corresponds to a matrix attachment site, we conducted experiments in which the association of specific DNA regions with the nuclear matrix is assessed by duplex PCR (see Material and Methods). In this experiment, Jurkat cell nuclei were subjected to high-salt extraction followed by restriction enzyme digestion to release DNA that is not associated with the nuclear matrix (20, 32). The released DNA fraction in the supernatant (S) was separated from the nonsoluble DNA in the pellet (P) by centrifugation, and their distribution was quantitatively determined by duplex PCR. As a positive control we used a primer set specific for a known matrix-attached region previously characterized at the apoB gene (2). Specific primer sets were used to perform this analysis on a total of 68 genomic DNA fragments generated by a digest with the restriction enzymes HindIII/XbaI/EcoRI/ClaI (Fig. 5). In every test condition, the apoB MAR sequences were enriched in the pellet (83% ± 7%), and only small amounts were detected in the supernatant (17% ± 3%) (Fig. 5, first column). By contrast, the MINE was found predominantly in the supernatant fraction (primer set J: P, 33%; S, 67%), suggesting that its insulator function is not mediated through an attachment to the nuclear matrix. Similar to the MINE, the majority of the tested restriction fragments were enriched in the supernatant. However, five regions predominantly associated with the nuclear matrix with a strength similar to the apoB MAR: a region 76 kb upstream of the c-myc gene (P, 60%) and a cluster of three fragments within a 10-kb region downstream of c-myc were strongly attached to the nuclear matrix (at +42 kb [P = 90%], +45 kb [P = 93%], and +48 kb [P = 87%]) (Fig. 5). Interestingly, the flanking 5′ and 3′ matrix attachment regions of the c-myc gene locus separate it from neighboring gene loci; the pvt1 gene is located 60 kb downstream of the c-myc gene, and a predicted and conserved gene locus (gene locus 253372) with unknown function resides 85 kb upstream of the c-myc gene. Thus, the MARs define a >160-kb domain in which the transition of euchromatic and heterochromatic regions is maintained by insulator elements.
FIG. 5.
Matrix attachment regions define a 160-kb domain. Graphic representation of matrix-associated (Pellet) versus nonassociated (Supernatant) regions across ∼160 kb of the c-myc locus. DNA from salt-extracted and restriction enzyme-digested nuclei was purified, and the presence of DNA fragments in the pellet and supernatant fraction was determined by duplex PCR. The EcoRI/HindIII/ClaI/XbaI-generated fragments span a domain larger than 160 kb. This domain has at its 5′ end an as yet unidentified presumptive gene and at the 3′ end the pvt1 locus. Primers specific for the apoB MAR (2) serve as a positive control and consistently detect apoB sequences enriched in the matrix-associated fraction (P; 85% ± 7 [n = 68], first column in bar graph). Matrix-associated elements within the c-myc locus are indicated with an asterisk at −76 kb (P = 60%), +39 kb (P = 47%), +42 kb (P = 90%), +45 kb (P = 93%), and + 48 kb (P = 87%). The positions of the amplified regions are given relative to the HindIII site located 2,328 bp upstream of exon 1. The hyperacetylated region and the matrix-associated regions are indicated by shaded areas. Primer sets specific to the MINE, BE (J), and CTCF (K) elements are indicated below the autoradiograph.
DISCUSSION
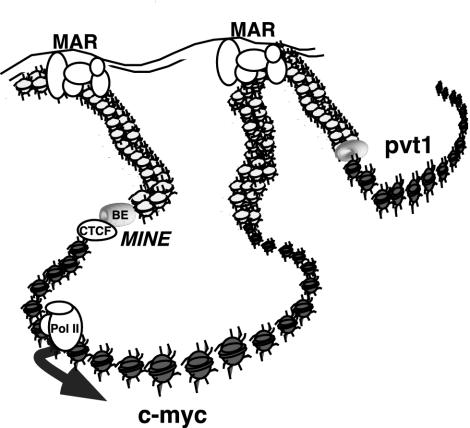
The highly acetylated state of histones at the mammalian c-myc gene defines a region of accessible chromatin independent of the transcriptional activity of the gene. The boundary that separates the hyperacetylated from the hypoacetylated domain resides within a 1.6-kb region called the myc insulator element or MINE and is located approximately 2.5 kb upstream of the c-myc transcription initiation site. This element specifies multiple activities that together form an efficient insulator. The hyperacetylated domain of the c-myc locus is flanked by regions of condensed chromatin. Finally, matrix attachment sites flank a large domain of approximately 160 kb that encompasses both the hyperacetylated c-myc transcription unit and flanking heterochromatin. These results suggest that the c-myc gene has several levels of organization, one that may render the c-myc domain topologically separate from surrounding genes as defined by MARs and a second that may define a self-contained functional transcriptional unit as defined by MINE (Fig. 6). We speculate that both levels of regulation are essential for the coordinate regulation of the c-myc gene.
FIG. 6.
Scheme of MINE and MARs contributing to the maintenance of a structurally and functionally independent c-myc domain. Matrix attachment sites located 80 kb upstream and 50 kb downstream may form a chromosomal loop that permits the association of the c-myc gene with transcriptionally active nuclear territories. The c-myc insulator element, composed of the BE and the CTCF binding element, functions as a boundary that separates active (gray) from inactive (white) chromatin.
At least two functionally separable activities are embedded within MINE. MINE and its 1.0-kb BE increased the likelihood of transgene expression at random integration sites in the genome, at least over a period of 6 weeks, after which the number of colonies was determined. The ability to protect transgenes against position effects is a characteristic of other insulator elements such as the chicken β-globin HS4. Both the barrier activity and the enhancer-blocking activity of MINE are orientation dependent. Polar behavior has been reported in other enhancer-blocking elements (reviewed in reference 47) and may result from the experimental context in which insulator elements are tested. The orientation dependence of MINE function led us to the hypothesis that both the 5′ and the promoter-proximal CTCF sites contribute to the structural and spatial organization of c-myc within the nucleus. Both CTCF sites may be important in the determination of a three-dimensional structure in which the insulator is oriented in a more effective, reversed conformation. This may be accomplished by direct interaction of two CTCF molecules with each other or recruitment of additional factors. Alternatively, CTCF may use different combinations of its 11 zinc fingers to interact simultaneously with both CTCF sites.
Several models of heterochromatin barrier activity have been proposed (reviewed in reference 9). In one model, silencer-bound complexes nucleate the spreading of histone deacetylation and the binding by heterochromatin-associated proteins to the span of the silenced region until a barrier is reached. In this passive barrier model, a multiprotein complex that is stably bound to the boundary physically interferes with the propagation of the heterochromatic structure. In a second, active barrier model, chromatin-modifying activities (e.g., acetyltransferases or nucleosome remodeling complexes) are recruited to the barrier in order to modify histones or other chromatin-associated factors. The modified nucleosomes would be less-efficient substrates for incorporation into heterochromatin. While it is unclear how chromosomal barriers exert their function, several regions within the cHS4 that interact with proteins have been identified (6). Studies at the HMR locus in Saccharomyces cerevisiae support the active model of insulator function. These experiments revealed that the barrier activity mediated by a telomeric tRNA gene may be linked to the recruitment of specific transcription activators that associate with histone-modifying complexes (10). Consistent with this notion, the cHS4 insulator region at the chicken β-globin gene locus coincides with a histone acetylation peak (35). In contrast, the barrier element of MINE at the c-myc locus is not associated with a peak of histone H3 or H4 acetylation and lies outside of the hyperacetylated domain. Thus, the mechanism of barrier function appears to be different from both that observed in the 5′ chicken β-globin HS4 region and that observed in the HMR locus in yeast.
Previous transfection experiments with heterologous reporter genes suggested that CTCF functions as a repressor of c-myc transcription (12). However, the results of our ChIP assays demonstrate constitutive CTCF binding at the 5′ boundary and at the P2 promoter region of the c-myc gene in both human HL-60 cells and mouse CTLL cells, independent of the level of c-myc transcription. Furthermore, the 40-bp CTCF-containing element does not silence a reporter gene in the absence of an enhancer (data not shown) nor when positioned upstream of an enhancer (Fig. 4). Thus, CTCF is not the sole determinant of transcriptional repression of the c-myc gene; it may rather function as an integral component of insulator function or serve as a platform onto which additional silencing factors are recruited.
Insulator elements are complex and multipartite, composed of subelements that each contribute to the function of the insulator. In addition to the barrier and enhancer-blocking activity, MINE also contains an additional activity that negatively affects the expression of the Neor reporter gene. While our data do not clearly distinguish between CTCF-independent enhancer-blocking elements or silencing activities, our results are reminiscent of those with other insulator elements, such as region II and III of the cHS4 and the Drosophila gypsy element (3, 16, 41). Indeed, deletions of any of the five subelements that together form the core of the HS4 insulator element have deleterious effects on its barrier activity. This is consistent with the proposed organization of MINE, in which the individual contributions of the subfragments are additive and in which enhancer-blocking activities or “silencing” activity may be masked by the activities contained in the other subelements.
The organization of the DNA within the nucleus may be governed by structural components that interact with chromatin (reviewed in reference 19). MARs are thought to mediate the anchoring of the chromatin fiber to the nuclear matrix by AT-rich DNA sequences. The structural and functional roles of MARs have been somewhat blurred recently by the coincidence of some insulators with MARs and the general overlap in their broad definitions as boundary elements. Although some insulators, such as the Drosophila gypsy element, are clearly associated with the nuclear matrix (17), the MINE function appears to be independent of matrix association. At the c-myc locus, we have demonstrated that the activities of the barrier-insulator element and the MARs are clearly separable entities, the former maintaining the boundary of histone acetylation of the transcriptional unit and the latter anchoring the domain to the nuclear matrix. It is important to note that both the 5′ and 3′ MARs at the c-myc locus encompass a region that contains both euchromatic and heterochromatic features.
It is attractive to speculate that the MARs that border the c-myc gene function as nuclear organizer elements similar to the model proposed for the gypsy element (17). Specific interactions between the gypsy elements that are spread along the Drosophila chromosomes are thought to organize the chromatin fiber into insulator bodies, contributing to the formation of the nuclear scaffold from which hyperacetylated domains loop out to form transcriptionally active territories. Importantly, individual sequences normally separated in the genome are relocated to other gypsy-containing chromosomal domains (17). Experiments designed to determine the nature of these matrix-associated regions that border the c-myc locus and their role as nuclear organizer are currently under way. In addition to the characterization of how MAR structure and function are related to transcriptional expression at the c-myc locus, the identification of proteins localized to these regulatory regions may provide important clues to the inherent compartmentalization of chromosomes in the eukaryotic nucleus.
Acknowledgments
This work was supported by a Research Scholar Grant from the American Cancer Society to A.K. (RSG-01-163-01-GMC). W.H.S. was supported by a grant from the NIH (CA82459).
We are especially grateful to Jim Moon and Brad Nelson (Virginia Mason Research Center) for generously providing and preparing chromatin from CTLL2 cells; Corty Thienes (Fred Hutchinson Cancer Research Center) for help with MAR assays; Mark Groudine, Susan Parkhurst, and Mike Bulger (Fred Hutchinson Cancer Research Center) for critical reading of the manuscript, and all members of the Groudine lab for discussion.
REFERENCES
- 1.Allfrey, V. G., R. M. Faulkner, and A. E. Mirsky. 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51:786-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antes, T. J., S. J. Namciu, R. E. Fournier, and B. Levy-Wilson. 2001. The 5′ boundary of the human apolipoprotein B chromatin domain in intestinal cells. Biochemistry 40:6731-6742. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Bode, J., C. Benham, A. Knopp, and C. Mielke. 2000. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr. 10:73-90. [PubMed] [Google Scholar]
- 5.Bulger, M., D. Schubeler, M. A. Bender, J. Hamilton, C. M. Farrell, R. C. Hardison, and M. Groudine. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell. Biol. 23:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, J. H., A. C. Bell, and G. Felsenfeld. 1997. Characterization of the chicken beta-globin insulator. Proc. Natl. Acad. Sci. USA 94:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, J. H., M. Whiteley, and G. Felsenfeld. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505-514. [DOI] [PubMed] [Google Scholar]
- 8.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292-301. [DOI] [PubMed] [Google Scholar]
- 9.Donze, D., and R. T. Kamakaka. 2002. Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays 24:344-349. [DOI] [PubMed] [Google Scholar]
- 10.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, C. M., A. G. West, and G. Felsenfeld. 2002. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 22:3820-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippova, G. N., S. Fagerlie, E. M. Klenova, C. Myers, Y. Dehner, G. Goodwin, P. E. Neiman, S. J. Collins, and V. V. Lobanenkov. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filippova, G. N., C. P. Thienes, B. H. Penn, D. H. Cho, Y. J. Hu, J. M. Moore, T. R. Klesert, V. V. Lobanenkov, and S. J. Tapscott. 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28:335-343. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg, E. C., and E. H. Bresnick. 2001. Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23:820-830. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gdula, D. A., T. I. Gerasimova, and V. G. Corces. 1996. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. USA 93:9378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerasimova, T. I., K. Byrd, and V. G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6:1025-1035. [DOI] [PubMed] [Google Scholar]
- 18.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 19.Ishii, K., and U. K. Laemmli. 2003. Structural and dynamic functions establish chromatin domains. Mol. Cell 11:237-248. [DOI] [PubMed] [Google Scholar]
- 20.Izaurralde, E., J. Mirkovitch, and U. K. Laemmli. 1988. Interaction of DNA with nuclear scaffolds in vitro. J. Mol. Biol. 200:111-125. [DOI] [PubMed] [Google Scholar]
- 21.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 22.Kalos, M., and R. E. Fournier. 1995. Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol. Cell. Biol. 15:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellum, R., and P. Schedl. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941-950. [DOI] [PubMed] [Google Scholar]
- 24.Kiekhaefer, C. M., J. A. Grass, K. D. Johnson, M. E. Boyer, and E. H. Bresnick. 2002. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc. Natl. Acad. Sci. USA 99:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krumm, A., L. B. Hickey, and M. Groudine. 1995. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 9:559-572. [DOI] [PubMed] [Google Scholar]
- 26.Labrador, M., and V. G. Corces. 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111:151-154. [DOI] [PubMed] [Google Scholar]
- 27.Li, Q., M. Zhang, Z. Duan, and G. Stamatoyannopoulos. 1999. Structural analysis and mapping of DNase I hypersensitivity of HS5 of the beta-globin locus control region. Genomics 61:183-193. [DOI] [PubMed] [Google Scholar]
- 28.Lipps, H. J., A. C. Jenke, K. Nehlsen, M. F. Scinteie, I. M. Stehle, and J. Bode. 2003. Chromosome-based vectors for gene therapy. Gene 304:23-33. [DOI] [PubMed] [Google Scholar]
- 29.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madisen, L., and M. Groudine. 1994. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt's lymphoma cells. Genes Dev. 8:2212-2226. [DOI] [PubMed] [Google Scholar]
- 31.Meulia, T., A. Krumm, C. Spencer, and M. Groudine. 1992. Sequences in the human c-myc P2 promoter affect the elongation and premature termination of transcripts initiated from the upstream P1 promoter. Mol. Cell. Biol. 12:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirkovitch, J., S. M. Gasser, and U. K. Laemmli. 1988. Scaffold attachment of DNA loops in metaphase chromosomes. J. Mol. Biol. 200:101-109. [DOI] [PubMed] [Google Scholar]
- 33.Mirkovitch, J., M. E. Mirault, and U. K. Laemmli. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223-232. [DOI] [PubMed] [Google Scholar]
- 34.Moon, J. J., and B. H. Nelson. 2001. Phosphatidylinositol 3-kinase potentiates, but does not trigger, T cell proliferation mediated by the IL-2 receptor. J. Immunol. 167:2714-2723. [DOI] [PubMed] [Google Scholar]
- 35.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 37.Oki, M., and R. T. Kamakaka. 2002. Blockers and barriers to transcription: competing activities? Curr. Opin. Cell Biol. 14:299-304. [DOI] [PubMed] [Google Scholar]
- 38.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 39.Phi-Van, L., and W. H. Stratling. 1996. Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry 35:10735-10742. [DOI] [PubMed] [Google Scholar]
- 40.Pogo, B. G., V. G. Allfrey, and A. E. Mirsky. 1966. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl. Acad. Sci. USA 55:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recillas-Targa, F., M. J. Pikaart, B. Burgess-Beusse, A. C. Bell, M. D. Litt, A. G. West, M. Gaszner, and G. Felsenfeld. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 99:6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheuermann, R. H., and W. T. Garrard. 1999. MARs of antigen receptor and co-receptor genes. Crit. Rev. Eukaryot. Gene Expr. 9:295-310. [DOI] [PubMed] [Google Scholar]
- 43.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 44.Siebenlist, U., L. Hennighausen, J. Battey, and P. Leder. 1984. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell 37:381-391. [DOI] [PubMed] [Google Scholar]
- 45.Wang, D. M., S. Taylor, and B. Levy-Wilson. 1996. Evaluation of the function of the human apolipoprotein B gene nuclear matrix association regions in transgenic mice. J. Lipid Res. 37:2117-2124. [PubMed] [Google Scholar]
- 46.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848-856. [DOI] [PubMed] [Google Scholar]
- 47.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 48.Zahn-Zabal, M., M. Kobr, P. A. Girod, M. Imhof, P. Chatellard, M. de Jesus, F. Wurm, and N. Mermod. 2001. Development of stable cell lines for production or regulated expression using matrix attachment regions. J. Biotechnol. 87:29-42. [DOI] [PubMed] [Google Scholar]
- 49.Zhong, X. P., and M. S. Krangel. 1997. An enhancer-blocking element between alpha and delta gene segments within the human T cell receptor alpha/delta locus. Proc. Natl. Acad. Sci. USA 94:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]