Abstract
Respiratory nitric oxide reductase (NOR) was purified from membrane extract of Pseudomonas (Ps.) nautica cells to homogeneity as judged by polyacrylamide gel electrophoresis. The purified protein is a heterodimer with subunits of molecular masses of 54 and 18 kDa. The gene encoding both subunits was cloned and sequenced. The amino acid sequence shows strong homology with enzymes of the cNOR class. Iron/heme determinations show that one heme c is present in the small subunit (NORC) and that approximately two heme b and one non-heme iron are associated with the large subunit (NORB), in agreement with the available data for enzymes of the cNOR class. Mössbauer characterization of the as-purified, ascorbate-reduced and dithionite-reduced enzyme confirms the presence of three heme groups (the catalytic heme b3, and the electron transfer heme b and heme c) and one redox-active non-heme Fe (FeB). Consistent with results obtained for other cNORs, heme c and heme b in Ps. nautica cNOR were found to be low-spin while FeB was found to be high-spin. Unexpectedly, as opposed to the presumed high-spin state for heme b3, the Mössbauer data demonstrate unambiguously that heme b3 is, in fact, low-spin in both ferric and ferrous states, suggesting that heme b3 is six-coordinated regardless of its oxidation state. EPR spectroscopic measurements of the as-purified enzyme show resonances at the g ~ 6 and g ~ 2–3 regions very similar to those reported previously for other cNORs. The signals at g = 3.60, 2.99, 2.26 and 1.43 are attributed to the two charge-transfer low-spin ferric heme c and heme b. Previously, resonances at the g ~ 6 region were assigned to a small quantity of uncoupled high-spin FeIII heme b3. This assignment is now questionable because heme b3 is low-spin. On the basis of our spectroscopic data, we argue that the g = 6.34 signal is likely arising from a spin-spin coupled binuclear center comprising the low-spin FeIII heme b3 and the high-spin FeBIII. Activity assays performed under various reducing conditions indicate that heme b3 has to be reduced for the enzyme to be active. But, from an energetic point of view, the formation of a ferrous heme-NO as an initial reaction intermediate for NO reduction is disfavored because heme [FeNO]7 is a stable product. We suspect that the presence of a sixth ligand in the FeII-heme b3 may weaken its affinity for NO and thus promotes, in the first catalytic step, binding of NO at the FeBII site. The function of heme b3 would then be, to orient the FeB-bound NO molecules for the formation of the N-N bound and to provide reducing equivalents for NO reduction.
Keywords: Nitric oxide, denitrification, heme protein, non-heme iron, binuclear Fe center, EPR, Mössbauer spectroscopy
Denitrifying bacteria can be found in a wide variety of environments ranging from streams, soils, to human intestines. Ps. nautica 617 is a halofile bacterium found in marine sediments contaminated with effluents from oil refineries (1, 2). It is able to reduce nitrate to dinitrogen by utilizing the catalytic functions of four specific enzymes (3): The Mo-pterin containing nitrate reductase, the cytochrome cd1 nitrite reductase, the cytochrome bc complex nitric oxide reductase and the Cu-containing nitrous oxide reductase. The nitrate reductase, cytochrome cd1 and nitrous oxide reductase from Ps. nautica have been subjected to extensive biochemical and structural studies (4–7), while detailed characterization of nitric oxide reductase (NOR) from Ps. nautica has not been reported.
There are three classes of NOR identified in bacteria, all membrane bound. The first class, called cNOR, uses a soluble cytochrome c or a cupredoxin as its electron donor, whereas the two other classes, qNOR and qCuNOR, are quinol oxidases. The cNOR’s are composed of two subunits: a small subunit (NORC, ~17 kDa) that contains a c-type heme, and a large subunit (NORB, ~56 kDa) that contains two b-type hemes and a non-heme iron. Enzymes belonging to this class have been isolated from four denitrifying bacteria: Ps. stutzeri (8, 9), Paracoccus (Pa.) denitrificans (10, 11), Halomonas (H.) halodenitrificans (12, 13) and more recently Ps. aeruginosa (14). The enzymes of the qNOR class consist of only one subunit of 84.5 kDa. Similar to the NORB subunit of cNOR, each molecule of qNOR contains two b-type hemes and one non-heme iron. With the exception of the qNOR isolated from the denitrifying bacteria Ralstonia eutropha (15) all other purified qNOR’s reported so far were identified in pathogenic microorganisms lacking the other denitrifying enzymes (16, 17). The qCuNOR has been isolated from the Gram positive denitrifying Bacillus azotoformans (18). It is a heterodimer, composed of a small subunit of 16 kDa containing a CuA center, and a large subunit of 57 kDa containing two b-type hemes and a non-heme iron center, similar to those in qNOR and in NORB of cNOR.
The NORs of all three classes catalyze the two electron reduction of nitric oxide to nitrous oxide, a reaction that involves the formation of a N-N bond:
Biochemical and spectroscopic investigations of cNORs, together with site-directed mutagenesis studies, have provided evidence for a binuclear catalytic site, comprising a heme b (heme b3) and a non-heme iron center (FeB) (19–21). In the resting state of the enzyme (oxidized form), both Fe sites are suggested to be high-spin ferric and antiferromagnetically coupled, most probably, through a μ-oxo bridge (22), resulting in a diamagnetic ground state and consequently EPR silent. The mechanism of NO reduction remains a subject of intense interest and has been reviewed recently (23–26). Currently two alternative mechanisms are being considered, namely, the cis and trans mechanism. In the cis mechanism, one of the two iron sites of the binuclear center binds both NO molecules and the other Fe site plays the role of electron transfer and/or assist in abstracting the O atom from one of the NO molecules. In the trans mechanism, one NO molecule is bound to each one of the iron sites of the binuclear center. Ultra-rapid freeze quench EPR studies under single turnover conditions showed simultaneous appearance of EPR signals that are characteristics of ferrous heme–NO and non-heme ferrous–NO complexes at 0.5 ms after introduction of NO (14), providing evidence for the proposed trans mechanism. A recent spectroscopic study of a functional model complex also shows a reaction intermediate that exhibits both a S = 3/2 [FeBNO]7 and a S = 1/2 heme Fe(II)-NO EPR signal, supporting the trans mechanism (27). On the other hand, ferrous heme–NO complexes are highly stable, making it improbable for ferrous heme–NO to be a reaction intermediate. Also, a redox titration study of the Pa. denitrificans cNOR shows that the mid-point redox potential of heme b3 is more than 200 mV lower than that of FeB (28), indicating the presence of a thermodynamic barrier to prevent the reduction of heme b3 and thus avoid the formation of a highly stable, probably inhibitory, ferrous heme b3-NO complex. The presence of such a thermodynamic barrier has led to the proposal that the three electron-reduced cNOR with an oxidized heme b3 is the active form of the enzyme (28). In support of this proposal, studies of cyanide binding to various redox states of the enzyme indicated that affinity for CN− for the three electron-reduced state is 1000-fold higher than that for the fully reduced or oxidized state (29). But, interestingly, more recent studies with H. halodenitrificans cNOR (12) showed that NO can bind to both heme b3 and FeB in their oxidized as well as reduced states, and that the key determining factor in preventing NO binding to the binuclear center is the presence of the μ-oxo bridge. Either a weakening or a rupturing of the oxo-bridge would permit the binding of NO irrespective of the redox state of the heme b3-FeB center. Here we report a detailed biochemical and spectroscopic characterization of cNOR isolated from the denitrifying bacterium Ps. nautica. In contrast to previously assumed high-spin state for heme b3 in cNORs isolated from other organisms, our results indicate that, for Ps. nautica cNOR, heme b3 is low-spin in both oxidized and reduced states and that heme b3 has to be reduced for the enzyme to be active.
EXPERIMENTAL PROCEDURES
Cell growth and NOR purification
Pseudomonas nautica 617 cells were grown as described in Prudêncio et al. (6). For 57Fe growth, FeSO4 was replaced by the 57Fe isotope salt. Cells were harvested by centrifugation at the end of the stationary phase. The yield in wet weight was 1 g of cells per liter of culture medium. Cells were ressuspended in 100 mM Tris-HCl pH = 7 buffer and disrupted on a French Press. The crude extract was centrifuged at 8000×g for 20 minutes to separate the intact cells, and only then ultracentrifuged twice at 125,000×g for 90 min to recover the membrane fraction, which was frozen at −80°C. Membranes were washed as described in Girsh et al.(19), except for the sonication step that was done in 500 mL portions, in 4 cycles of 10 minutes in ice. All ultracentrifugation steps were performed at 180,000×g for 90 minutes. Purification of nitric oxide reductase was carried out at 4°C, following a modified procedure from the one described in Girsh et al.(19). Proteolysis inhibitors (1 mM PMSF and 10 mM benzamidine) were added to the supernatant of the extraction with 0.6% of DDM. The percentage of detergent used was judged to be effective as no significant NOR activity remained in the membrane fraction. This supernatant was applied to a DEAE-Biogel A column (Bio-Rad, 5.5×25.5 cm, 140 mL of resin per gram of protein) equilibrated with 50 mM Tris-HCl pH = 8, 0.02% DDM, 0.01% (v/v) 2-phenylethanol. The column was washed with 460 mL of the same buffer. A linear gradient between 0 and 500 mM NaCl was used, with a total volume of 2 L. The presence of NOR was assessed by activity assays and UV/visible spectroscopy. The fractions with specific activity higher than 40%, eluted between 250 and 400 mM NaCl, were pooled. After a concentration step in a Diaflo apparatus (Amicon) with a YM30 membrane, the buffer was exchanged to 10 mM potassium phosphate buffer (KPB) pH 7, 400 mM NaCl and the fraction was frozen in 340 mg aliquots of protein. Each aliquot was then further purified in a Macro-prep Ceramic Hydroxylapatite CHT-I 20μm column (Bio-Rad, 2.6×27cm) coupled to a ÄKTA basic HPLC system (Amersham Biosciences). A linear gradient was applied using NaCl (0.4 – 0 M) and potassium phosphate buffer (KPB, 0.010 – 1.50 M), pH 7, with a flow of 2 mL/min for 3 h. A second chromatographic step was performed using a Bio-Scale CHT20-I Pre-packed column with the same gradient. NOR was eluted between 300 and 350 mM KPB. For the Ceramic Hydroxylapatite columns, the concentration of detergent had to be increased both in the fraction injected (to 0.1%) and in the buffers (to 0.05%) in order to prevent further spreading of the protein in the column. In these last steps of purification, NOR containing fractions that have absorbance ratio between 410 nm and 280 nm higher than 1 were pooled. Purity of the isolated enzyme was verified by tricine SDS-PAGE electrophoresis. The protein was then concentrated and the buffer exchanged to 100 mM KPB pH 7, 0.02% dodecyl maltoside, 0.01% 2-phenylethanol.
Tricine sodium dodecyl sulfate electrophoresis
SDS-PAGE was performed using a method developed by Shagger and von Jagow (30), which uses tricine instead of glicine buffer and has the advantage of conveying the same degree of separation as in the Laemmli method with lower acrylamide concentrations. To avoid smearing of the NOR bands, and aggregation, samples were incubated at 40°C for 30 min before loading.
Spectroscopic methods
UV/visible spectra were recorded in a Shimadzu UV-265 split-beam spectrophotometer connected to an IBM PC compatible computer. Low temperature EPR measurements were performed in an X-band Brucker EMX spectrometer equipped with an Oxford Instruments liquid helium flow cryostat. Weak-field Mössbauer spectra were recorded on a spectrometer equipped with a Janis 8DT variable-temperature cryostat. The Mössbauer spectrometer operates at a constant acceleration mode in transmission geometry. The zero velocity refers to the centroid of a room temperature spectrum of a metallic iron foil. Analysis of the Mössbauer spectra were performed with the program WMOSS (SEE co.).
Enzymatic assay
NO reductase activity was measured with an ISO-NO Mark II electrode with a 2 mm sensor in an appropriate reaction vessel (World Precision Instruments). NO aqueous solution, 100 μM at 20 °C (19), was prepared from a 5% NO/95% He mixture (Air liquide). The NO/He mixture was previously washed by bubbling in a 5 M KOH solution and through non-buffered water, at pH 3, to prevent NO2− formation. The reaction vessel, with the electrode fitted on top, was degassed prior to addition of solution. Degassed solution of 20 mM KPB, pH 6, 0.02% (w/v) DDM containing the electron donor was introduced. NO aqueous solution was then added and the assay was initiated by enzyme addition (70 nM final protein concentration). ISO-NO Mark II electrode was coupled to a Quad-16/EFA-400, and data acquisition/treatment was performed by Data-Trax™ Data Acquisition software (World Precision Instruments). Different cytochromes from Ps. nautica, purified as described by Saraiva, L. M. et al (31, 32), were used as electron donors, each at a concentration of 3 μM in the assay solution.
Protein and metals determination
Protein determination was performed using the BCA method (Sigma) or by the Lowry method (33) using bovine heart cytochrome c as standard. Total iron content was determined by Inductive Coupled Plasma Emission analysis. Heme b and c content was determined by the piridine-hemocromogen method. The results were analyzed according to Berry and Trumpower (34). Non-heme iron content was determined by the TPTZ method (35). Amino acid analysis was performed as described by Moore et al. (36) after a 24 h or 48 h hydrolysis with 6N HCl at 110 °C with a Pharmacia Alpha Plus Aminoacid analyzer.
NOR gene coding sequence determination
Ps. nautica 617 cells were grown for 16 h, at 30 °C, 220 rpm, in the absence of nitrate. Genomic DNA was isolated using a standard CTAB extraction protocol (37). To obtain the sequence of the NOR-encoding gene, a first PCR was performed, using degenerate primers, designed by comparison of known NORB protein sequences. PCR was performed in a thermal cycler (Biometra). A product of ~ 980 bp was obtained, purified by gel extraction using the QIAquick Gel Extraction Kit (QIAGEN) and ligated into the pGEM-TEasy commercial vector (Promega) according to the manufacturer’s instructions. The ligation product was used to transform Epicurian coli XL1-Blue competent cells (Stratagene). Positive clones (white colonies) were grown overnight in Luria Broth medium with 100 μg/mL ampicilin at 37°C. Plasmid DNA was isolated and sequenced in a Amersham Pharmacia Biosciences ALF Express II automated sequencer. The remaining sequence of NORB subunit and the sequence of the NORC subunit were obtained applying the Universal Genome Walker Kit (Clontech) according to the manufaturer’s instructions. All primers were synthesized by Sigma-Genosys.
Redox titration
Redox titrations were carried out in an anaerobic chamber. The redox potential was measured directly in the cell using a combined silver/silver chloride and platinum electrode (Crison) calibrated with quinhydrone at controlled pH. Several different dye mediators at 0.10 μM (anthraquinone-2-sulphonate, 2-hydroxy-1,4-naftoquinone, phenazine, duroquinone, 5′-hydroxy-1,4-naftoquinone, 1,4-naftoquinone, phenazine metasulphate, 1,2-naftoquinone, tetramethyl-p-phenylene-diamine, 1,2-naftoquinone-4-sulphonic acid, potassium ferricyanide, phenazine ethasulphate and 2,5-dimethyl-benzoquinone) were added to the protein sample, selected in order to minimize their interference at wavelengths of interest. The reaction mixture was buffered with the mix described above. Reduction was accomplished by adding aliquots of concentrated solutions (15 to 25 mM) of sodium ascorbate or dithionite. Visible spectra (395–900 nm) were monitored with a J&M Tidas diode array spectrophotometer.
RESULTS
General Biochemical and Physical Properties
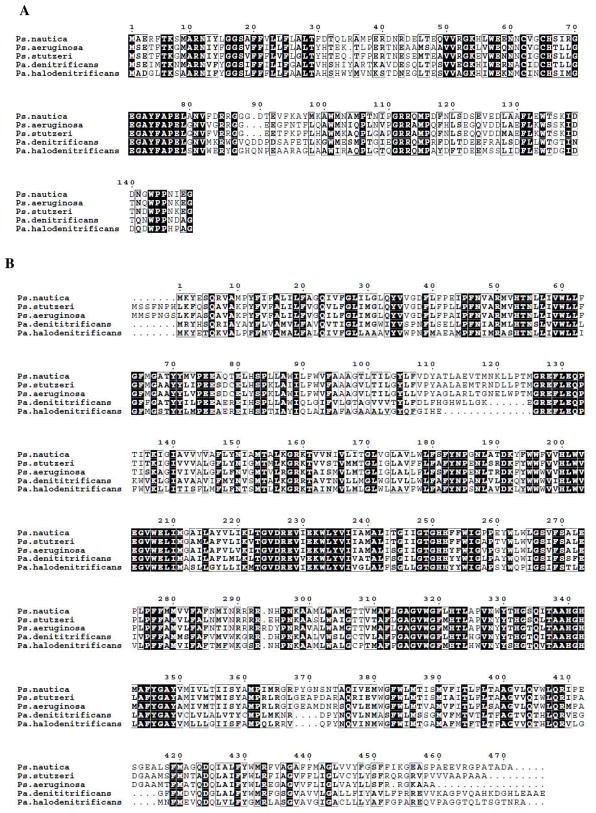
The DNA sequence of the gene that codifies Ps. nautica cNOR was determined (see Figure S1, supplementary material). Figure 1 compares the Ps. nautica cNOR primary sequence, deduced from the gene sequence, with those of other bacterial cNORs. The histidine residues that have been identified as ligands of the heme iron and non-heme iron atoms (H53 and H342 for heme b; H340 for heme b3; H200, 251 and 252 for FeB), and the catalytically essential glutamate residues (E128, E131, E204, E208 and E273) (38–41) are all conserved in Ps. nautica cNOR. In addition, two other histidine (H322 and H332) and five other glutamate residues (E75, E76, E80, E228, and E231) are also conserved. From the deduced amino acid composition and Fe cofactor content (one heme c in NORC; two heme b moieties and a non-heme Fe in NORB), molecular masses of 17,590 Da and 54,380 Da were calculated, respectively, for Ps. nautica NORC and NORB subunits. On SDS-PAGE gel, purified Ps. nautica cNOR shows two bands with apparent molecular masses of 17 kDa (NORC subunit) and 36 kDa (NORB subunit). The apparent molecular mass of NORC subunit determined from SDS-PAGE is similar to that calculated from its gene-coding primary sequence. The apparent molecular mass of NORB determined from SDS-PAGE is substantially different from that deduced from the amino-acid composition, however. This discrepancy had been reported for other cNORs and was attributed to the hydrophobic character of NORB (9, 13, 19). Heme staining methodology revealed that only the low molecular weight band (i.e., NORC subunit) had a covalently ligated heme. A total iron content of 3.5 ± 0.5 Fe/cNOR molecule was obtained by plasma emission analysis. Calcium content was determined to be 0.7 ± 0.2 Ca/cNOR. No other transition metal was detected. Values of 1.0 ± 0.1 Fe and 1.7 ± 0.2 Fe per NORB subunit were obtained for non-heme iron and heme b, respectively, while a value of 1.0 ± 0.1 Fe/NORC subunit was obtained for heme c, consistent with a cofactor stoichiometry of 1:2:1 for heme c: heme b: non-heme iron.
Figure 1.
Amino-acid sequence comparision of the small (A) and large (B) subunits of Ps. nautica cNOR with other cNORs isolated from other bacteria.
UV/visible Absorption Spectra
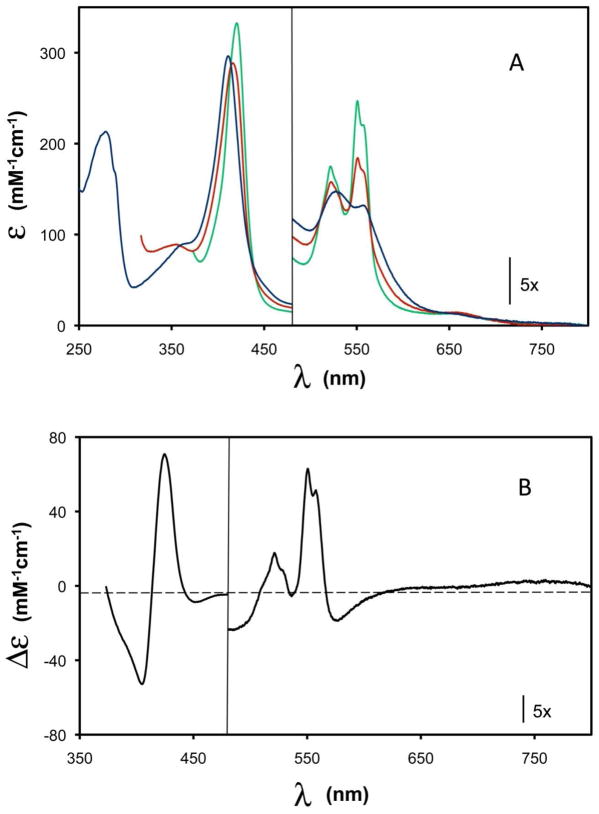
The as-purified Ps. nautica cNOR, at pH 7, displays a UV/visible spectrum (Figure 2A, blue line) typical of heme-containing proteins and very similar to those of other purified cNORs. The spectrum shows a protein peak at 280 nm, a Soret band at 411 nm and a broad band at around 550 nm with two small absorption peaks at 530 nm and 560 nm in the β and α regions, respectively. The appearance of the small absorption peaks indicate the presence of a small amount of reduced heme, in accord with the Mössbauer data presented below. The absorbance ratio of the Soret band at 411 nm to the protein peak at 280 nm was determined to be 1.4. Protein determinations by both Lowry and BCA methods revealed interference from detergent present in the buffer. Thus, protein quantifications derived from amino acid composition were used to estimate an extinction coefficient of 295 mM−1cm−1 at 411 nm for the oxidized cNOR. This value was then used to estimate protein concentration in all assays. It is important to note that while cNORs isolated from Pa. denitrificans and H. halodenitrificans exhibit an absorption band around 595 nm (19, 42), such a band is absent from the Ps. nautica cNOR. On the basis of a resonance Raman investigation, which suggests the presence of a μ-oxo group bridging heme b3 and FeB in the oxidized cNOR (22), the 595 nm band has been attributed to a ligand-to-metal charge transfer band associated with a high-spin ferric heme b3 without the proximal His ligand (43). The absence of a 595 nm band in the as-purified Ps. nautica cNOR therefore suggests a different spin state and/or a different coordination environment for heme b3 (see below). A low-intensity broad band at around 662 nm is observed, but shows no redox dependence, indicating minor presence of inactive protein.
Figure 2.
(A) UV/visible spectra of Ps. nautica cNOR in 100 mM pH 7 KPB, 0.02% dodecyl maltoside, 0.01% PE (as-isolated: blue line; ascorbate-reduced: red line; dithionite-reduced: green line). (B) Difference spectrum between the ascorbate-reduced spectrum and the dithionite-reduced spectrum (green minus red).
Reducing the enzyme with ascorbate red-shifts the maximum of the Soret band to 418 nm, and sharpens the optical bands in the α/β absorption region with maxima at 524 nm and 552 nm and a shoulder at 558 nm (Figure 2A, red line), consistent with reduction of low-spin heme groups. The absorption band at 552 nm has been assigned to heme c and the shoulder at 558 nm to heme b (11, 42, 44). Thus the optical data indicate both heme b and heme c are reduced by ascorbate. Upon reduction with sodium dithionite, the Soret maximum is further shifted to 422 nm and the α and β bands become more intense, indicating reduction of more heme groups (Figure 2A, green line). Figure 2B displays a difference spectrum (black line) between that of the dithionite- and ascorbate-reduced enzyme. On the basis of the following Mössbauer data, which indicate that only heme b3 is oxidized in the ascorbate-reduced enzyme and that it can be reduced by dithionite, this difference spectrum represents absorption changes arising from the reduction of heme b3 and indicates clearly that the reduced heme b3 is low-spin. Re-oxidation of the ascorbate- or dithionite-reduced enzyme produces a spectrum identical to that of the as-isolated enzyme, indicating reversible oxidation-reduction processes. In the presence of PMS, ascorbate can reduce the enzyme to a redox state equivalent to that of the dithionite-reduced cNOR as judged by optical spectroscopy (Figure S2).
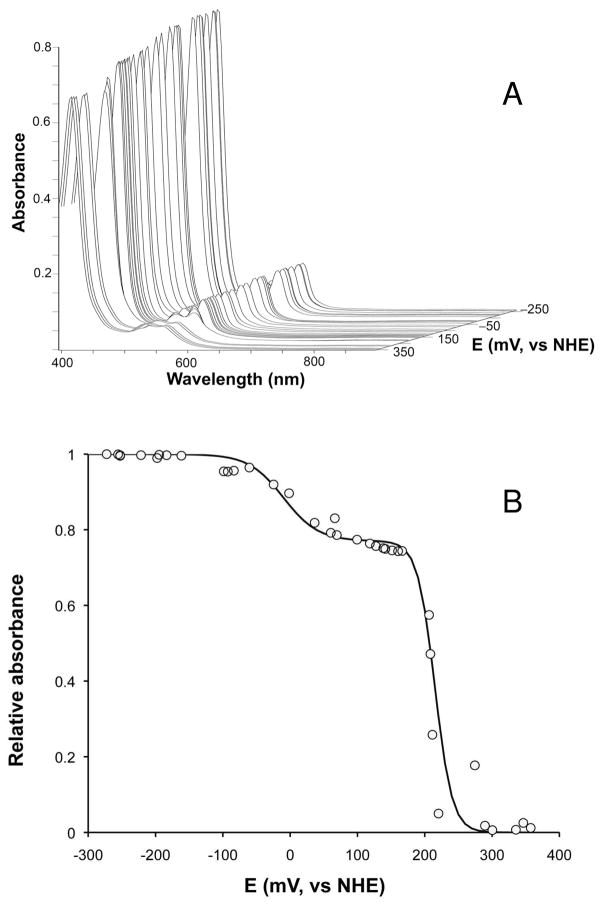
Optical Oxidation/Reduction Titrations
The UV/visible spectrum of the as-purified Ps. nautica cNOR showed no pH dependence in the range of pH 5 to 9. To determine the redox properties of the metallocofactors, redox potentiometric titrations monitored by optical absorptions were performed at two pH values 6 and 7. The resulting spectra as a function of the redox potential at pH 7 (Figure 3A) were globally fitted (SPECFIT, Spectrum Software Associates) and spectral components deconvoluted. Good fits of the data were obtained by using two independent Nernst equations of different mid-point redox potentials that differ by approximately 200 mV. At pH 7, the midpoint redox potentials were determined to be EI = +215 ± 10 mV and EII = −38 ± 10 mV and, at pH 6, EI = +232 ± 10 mV and EII = −16 ± 10 mV. To illustrate the goodness of our fits, in Figure 3B, we plotted the absorbance at 551 nm as a function of redox potential and compared the experimental data (circles) with the theoretical values (solid line) calculated by using the parameters obtained from the fits. On the basis of the Mössbauer and EPR data presented below, EI is assigned to both heme c and heme b, and EII is assigned to heme b3. As no absorption features are attributed to FeB, the redox potential of FeB is not determined from this measurement. Optical redox titration has also been performed on Pa. denitrificans cNOR (28). Although the mid-point potentials determined for the heme groups in Pa. denitrificans enzyme are about 100 mV higher than those reported here, the mid-point redox potentials for heme c (310 mV) and heme b (345 mV) in the Pa. denitrificans enzyme are also found to be comparable and are more than 200 mV above that of heme b3 (60 mV). In other words, the large redox-potential difference between the reactive heme b3 and the electron-transfer heme groups, a functionally important factor, is conserved among cNOR’s.
Figure 3.
(A) UV/visible spectra of Ps. nautica cNOR as a function of redox potential at pH 7. (B) Absorbance at 551 nm as a function of redox potential at pH 7 (100 mM pH 7 KPB, 0.02% dodecyl maltoside, 0.01% PE). The solid line is the result calculated by using the parameters obtained from globally fitting the spectra shown in (A).
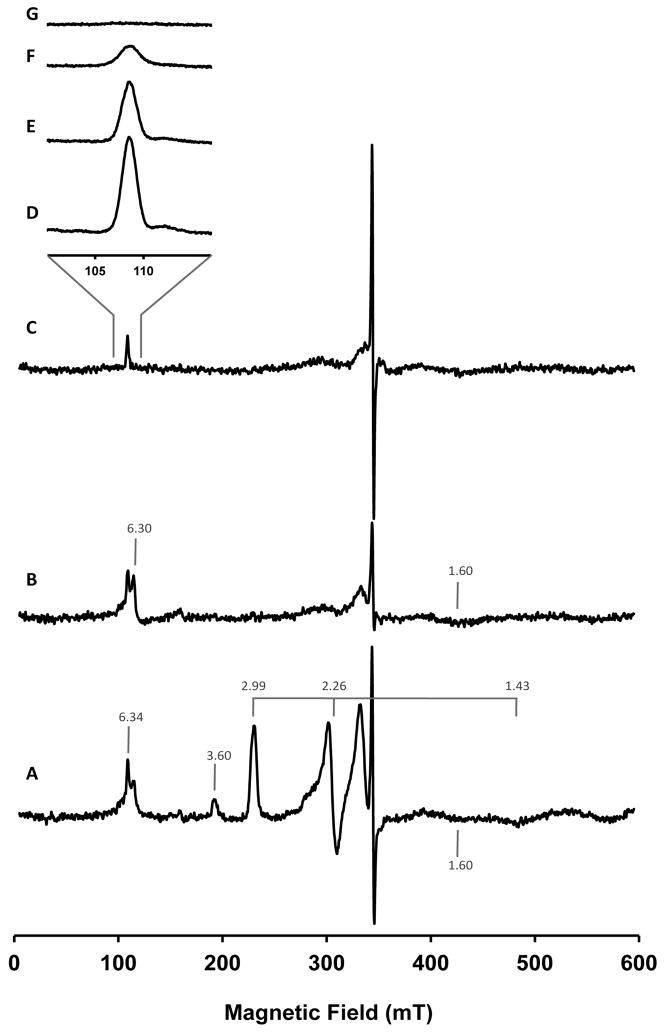
EPR Results
The as-purified Ps. nautica cNOR exhibits an EPR spectrum (Figure 4A) that is similar to those reported for other cNOR’s (19, 21, 42). Two sets of low-spin ferric heme signals are clearly visible. The prominent set of signals, with g values of 2.99, 2.26 and 1.43, have been attributed to the histidine-histidine ligated heme b of the NORB subunit (21). The highly anisotropic set of signals, from which only the gmax value of 3.60 can be observed, have been attributed to the heme c of the NORC subunit (45). Spin quantification of these two low-spin ferric heme signals were done using the Aasa and Vanngard method (46) modified according to De Vries and Albracht (47), together with spectral simulation. Approximately 0.9 spin/molecule and 0.7 spin/molecule were determined for the heme b and heme c signals, respectively (In our spin quantification estimations, gmid = 1.43 and gmin = 1.00 were assumed for heme c). In addition, a broad signal at g = 1.60, two derivative type signals at g = 2.01 and 2.05, and two overlapping signals at around g = 6.30 region are detected. The signals at g = 2.05 and 1.60, although not yet assigned, are present in other purified enzymes such as Pa. denitrificans and Ps. aeruginosa enzymes (14, 26). The current assumption is that in the oxidized enzyme, the catalytic center is composed of a high-spin ferric heme b3 antiferromagnetically coupled to a high-spin ferric FeB, resulting in a diamagnetic S = 0 ground state that yields no EPR resonances. The signals at the g = 6.30 region have been assigned to a small quantity of decoupled ferric high-spin heme b3 and the signal at g = 2.01 to FeB or an organic radical (19, 42). The Mössbauer data, presented below, reveal that in the as-purified Ps. nautica NOR, heme b3 is in fact low-spin. Consequently, a re-evaluation of the above assignment for signals at the g = 6.3 region is required, because a decoupled ferric low-spin heme b3 would not produce signals at the g = 6 region.
Figure 4.
EPR spectra at 9.653 GHz of the as-isolated (A), ascorbate-reduced (B) and dithionite-reduced (C) Ps. nautica cNOR (265 μM in 100 mM KPB, pH 7, 0.02% DDM, 0.01% PE). Experimental conditions of spectra A to C: temperature = 12 K, microwave power = 0.2 mW, modulation frequency = 100 kHz, modulation amplitude = 0.5 mT, receiver gain = 1×105, conversion time = 163.84 ms and time constant = 81.92 ms; and of spectra D to E: microwave power = 6.3 mW, modulation frequency = 100 kHz, modulation amplitude = 0.4 mT, receiver gain = 2×105, conversion time = 81.92 ms and time constant = 40.96 ms. Spectrum D was collected at 5.5 K, E at 7.0 K, F at 12 K and spectrum at 22.5 K.
Reduction of Ps. nautica cNOR with sodium ascorbate (1 mM) leads to the disappearance of the ferric low-spin signals (g = 3.60, 2.99, 2.26 and 1.45), while the signals around g = 6.30, 2.05, 2.01 and 1.60 are retained (Figure 4B). The disappearance of the ferric low-spin signals is consistent with the optical data (presented above) and the Mössbauer results (presented below), which show that heme c and heme b can be readily reduced by ascorbate to diamagnetic low-spin FeII states.
Further reduction with sodium dithionite eliminates the derivative-type EPR signal at the g = 6 region but the sharp absorption-type signal at g = 6.34 is still clearly detectable (Figure 4C). Temperature dependence of this signal is characteristic of an electronic ground state system (Figure 4D to G). The Mössbauer data (presented below) show that in the ascorbate- and dithionite-reduced enzyme, significant portions of FeB and heme b3 remain in their oxidized states. It is therefore possible that the g = 6.34 signal could be associated with the oxidized heme b3-FeB center.
Mössbauer Characterization
Mössbauer spectroscopy was used to further characterize the electronic states of the Fe centers and to provide direct and quantitative measurements on the compositions of the oxidation states of the Fe centers in various redox forms of the enzyme. Figure 5 shows the Mössbauer spectra of the as-purified (A), ascorbate-reduced (B) and dithionite-reduced (C) samples recorded at 180 K in a 50 mT magnetic field applied parallel to the γ-radiation. At this temperature, the electronic relaxation of Fe centers in proteins are generally fast in comparison with the 57Fe nuclear precession, causing cancellation of the internal magnetic field at the 57Fe nucleus and resulting in quadrupole doublets for the Fe centers. Thus, these spectra are composed of only quadrupole doublets arising from different Fe species, facilitating the analysis of the spectra. On the other hand, there are eight possible redox states for the four Fe sites in cNOR. Consequently, analysis of these spectra remains challenging, and a global analysis including all three spectra is applied.
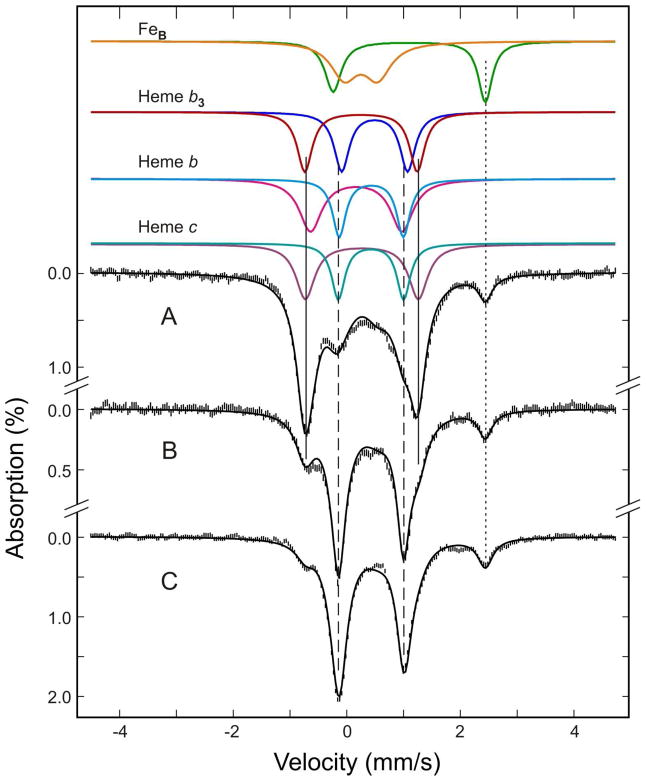
Figure 5.
Mössbauer spectra of the as-purified (A) ascorbate-reduced (B) and dithionite-reduced (C) Ps. nautica cNOR (805 μM in 100 mM KPB, pH 7, 0.02% DDM, 0.01% PE). The spectra (vertical bars) were collected at 180 K in a weak magnetic field of 50 mT. The color lines displayed above the data are the de-convoluded spectra for the for Fe cofactors in the enzyme: FeBIII, orange; FeBII, green; FeIII-heme b3, red; FeII-heme b3, blue; FeIII-heme b, magenta; FeII-heme b, cyan; FeIII-heme c, purple; FeII-heme c, blue-green. The back solid lines overlaid with the experimental spectra are composite spectra simulated with the parameters and percent absorptions listed in Tables 1. The dotted vertical line indicates the position of the high-energy line of the FeBII doublet. The two solid vertical lines and the two dashed vertical lines, respectively, mark the general line positions of the doublets arising from the low-spin FeII-heme and FeIII-heme cofactors.
First, the absorption peak at ~ 2.5 mm/s (indicated by a dotted line) can be assigned to the high-energy line of the quadrupole doublet arising from the high-spin ferrous FeB. Since this peak is well resolved from the rest of the absorptions, it allows for a reliable quantification on the amount of reduced FeB in the sample. It is therefore interesting to note that all three forms of the enzyme contain detectable amounts of reduced FeB, including the as-purified sample. Second, the most prominent feature observed in the spectrum of the as-purified sample (Figure 5A) is a broad quadrupole doublet (line positions indicated by two vertical lines) with apparent parameters (ΔEQ ~ 2 mm/s and δ ~ 0.2 mm/s) that are indicative of S = 1/2 low-spin ferric heme species. A rough estimate of its absorption area indicates that this doublet accounts for approximately 70% of the total Fe in the sample. Since heme b and heme c can contribute up to a maximum of only 50%, the remaining contribution has to arise from heme b3. In other words, the oxidized heme b3 in Ps. nautica cNOR is a low-spin ferric heme, in contrast to the high-spin ferric states reported for heme b3 of cNOR’s from other organisms (42, 43). Upon addition of sodium ascorbate, the intensity of this low-spin ferric-heme doublet decreases to about 25% of the total Fe absorption, and a new doublet (indicated by two vertical dashed lines), accounting for approximately 50% of the Fe absorption, is detected (Figure 5B). The apparent parameters for this doublet (ΔEQ ~ 1.1 mm/s and δ ~ 0.4 mm/s) are characteristics for low-spin ferrous hemes. Taking into consideration the mid-point redox potentials determined for the three heme groups (see above), only the electron-transfer heme c and heme b are reducible by ascorbate. Thus, this new doublet is assigned to the reduced heme b and heme c. With this assignment, it becomes obvious that in the spectrum of the as-purified enzyme (Figure 5A) there are absorptions arising from these reduced hemes. In other words, heme b and heme c are partially reduced in the as-purified enzyme, in agreement with the optical data described above. Observation of partial reduction of heme b and heme c in the as-purified enzyme is consistent with the high mid-point redox potential (215 – 230 mV) determined for these heme groups. Addition of sodium dithionite further decreases the intensity of the low-spin ferric heme doublet and increases the intensity of the low-spin ferrous heme doublet (Figure 5C), indicating that the active site heme b3 is partially reduced by dithionite and that the reduced heme b3 is also low-spin.
With the above understanding, it is possible to analyze these three spectra simultaneously using one set of parameters. To reduce the number of variable parameters, the following rational assumptions were made: (1) the total absorption intensity (oxidized plus reduced) for each one of the four Fe sites is 1/4 of the total Fe absorption, (2) the line-widths of a quadrupole doublet arising from a heme group are the same, and (3) for each sample, the [oxidized]/[reduced] ratio of heme b and heme c is the same (This assumption is based on the observed similar mid-point redox potentials for heme b and heme c; see above.). During the analysis, we also realized that the electronic relaxation rate of ferric FeB is not much faster than the 57Fe precession rate, resulting in a broad and featureless spectrum. A broad quadrupole doublet was therefore used to approximate the spectrum of ferric FeB. The results of the least-squares fit are presented in Figure 5 and Table 1. The parameters obtained are consistent with high-spin states for the oxidized and reduced FeB, as expected for most non-heme Fe cofactors in proteins, and low-spin states for all three heme groups regardless of their redox states, suggesting 6-coordinate environments for both ferric and ferrous hemes. The Mössbauer percent absorptions determined for the quadrupole doublets (Table 1) provide a quantitative assessment of the redox status of all four Fe cofactors in the three different oxidation states of the enzyme: heme b and heme c are partially reduced (3/25 = 12%) in the as-purified enzymes and can be completely reduced by ascorbate. Heme b3 is fully oxidized in the as-purified enzyme and cannot be reduced by ascorbate alone. This observation is consistent with the 200 mV redox-potential difference detected between heme b3 and the two electron transfer heme b and heme c. In the as-purified enzyme, FeB is significantly reduced (~30% reduction), and yet in the dithionite-reduced enzyme, it is only partially reduced. The level of reduction (60%), interestingly, is identical to that of heme b3, suggesting the presence of interactions between FeB and heme b3 which could complicate their redox behaviors. The Mössbauer results, taken together with results obtained from enzymatic activity studies (presented below), provide information for determining the “active” redox-state of the enzyme, information that is essential for understanding enzyme mechanism.
Table 1.
Mössbauer parameters and percent absorptions of Fe cofactors in the as-purified, ascorbate-reduced, and dithionite-reduced cNOR from Ps. nautica
| Cofactor | δ (mm/s) | ΔEQ (mm/s) | As-purified | Absorption in percent | |
|---|---|---|---|---|---|
| Ascorbate-reduced | Dithionite-reduced | ||||
| FeBII | 1.10 | 2.68 | 8 (1) | 11 (1) | 15 (2) |
| FeBIII | 0.25 | 0.59 | 17 (2) | 14 (2) | 10 (1) |
| FeII-heme b3 | 0.49 | 1.17 | 0 | 0 | 15 (2) |
| FeIII-heme b3 | 0.25 | 1.99 | 25 (3) | 25 (3) | 10 (1) |
| FeII-heme b | 0.43 | 1.13 | 3 (1) | 25 (3) | 25 (3) |
| FeIII-heme b | 0.23 | 2.04 | 22 (2) | 0 | 0 |
| FeII-heme c | 0.43 | 1.15 | 3 (1) | 25 (3) | 25 (3) |
| FeIII-heme c | 0.17 | 1.61 | 22 (2) | 0 | 0 |
The values in parentheses give the uncertainties in the last significant digits estimated for the percent absorptions. Due to the complexity of the spectra, the Mössbauer parameters (δ and ΔEQ) were fixed during the least-squares fittings. The line-widths of the quadrupole doublets used in our fits are 0.28 mm/s for FeII-heme b and FeII-heme c, 0.4 mm/s (left line) and 0.3 mm/s (right line) for FeIII-heme b, 0.3 mm/s (left) and 0.5 mm/s (right) for FeIII-heme c, 0.28 mm/s (left) and 0.35 mm/s (right) for FeII-heme b3, 0.36 mm/s (left) and 0.43 mm/s (right) for FeIII-heme b3, 0.35 mm/s (left) and 0.3 mm/s (right) for FeBII.
Enzymatic Activities
As a reference, specific activity of the as-purified enzyme (Table 2) was first determined by using the established assay developed by Girsh et al.(19), in which the assay solution contains 10 mM sodium ascorbate, 100 μM PMS and 20 μM horse heart cytochrome c in 20 mM KPB at pH 6. The specific activity values obtained are comparable to reported values for Pa. denitrificans and Ps. aeruginosa enzymes, at this pH (28, 51).
Table 2.
Specific activity of Ps. nautica cNOR as a function of electron donor and assay conditions.a
| Electron donor | Specific activity (μmol−1 min−1 mg−1) |
|---|---|
| none | |
| Ascorbate + PMS + oxidized horse heart cyt. c | 3.8 |
| Ascorbate + PMS | 4.3 |
| PMS + reduced horse heart cyt. c | 0.7 |
| Ascorbate | 0.7 |
| PMS | < 0.1 |
| Reduced horse heart cyt. c | < 0.1 |
| Reduced Ps. nautica cyt. c552 | 3.5 |
| Reduced Ps. nautica cyt. c552 | 0.6b |
| Reduced Ps. nautica cyt. c549 | < 0.1b |
| Reduced Ps. nautica cyt. c551 | < 0.1b |
| Reduced Ps. nauticacyt. c553 | < 0.1b |
| Reduced Ps. nautica cyt. c553(548) | < 0.1b |
| Reduced Ps. nautica cyt. c4 | < 0.1b |
Errors between duplicates are approximately ±20%. Concentrations used in the assay solutions are: [cNOR] = 70 nM; [ascorbate] = 10 mM; [PMS] = 100 μM; [cyt.] = 20 μM.
[cyt.] = 3 μM in the assay solution (see Text).
Examinations on the effects of various electron donors revealed that horse heart cytochrome c had no effect on the enzymatic activity and was ineffective as an electron donor. Presence of ascorbate and PMS in the assay solution was sufficient to produce full activity of the enzyme. Interestingly, Ps. nautica cytochrome c552 was found to be an effective electron donor and could produce fully active enzymes (Table 2). In an effort to obtain further evidence to support c552 as the physiological electron donor for Ps. nautica cNOR, a set of enzymatic assays was performed with six different low-spin cytochromes obtained from Ps. nautica (cytochromes c551, c552, c459, c553, c553(548) and membrane bound cytochrome c4) (31, 32, 48). In this series of experiments, due to limits in the amounts of some of the purified cytochromes, the experiments were carried out with 3 μM of cytochromes. Only the periplasmatic cythocrome c552 was found to show a significant value of specific activity. All other cytochromes were ineffective in providing electrons to Ps. nautica cNOR (last six rows, Table 2).
It is interesting to note that the mid-point redox potential of Ps. nautica c552 is about +250 mV at pH 7.6 (32), which is more than 200 mV higher than the reduction potential for heme b3. From a purely thermodynamical consideration, this suggests that c552 may not be a good electron donor for cNOR. But, as presented above, we have shown that c552 is the only small cytochrome that is capable to catalyze the reduction of NO. In fact, the same concern can also be raised for heme c and heme b, because their midpoint redox potentials are also much higher than that of heme b3. Taking these observations into consideration, we suggest that the initial reductive activation of heme b3 and the transfer of electrons from cytochrome c552, heme b and heme c to the diiron active site for NO reduction are separate events that could have distinct electron transfer pathways.
DISCUSSION
Spin State of Heme b3
We have purified Ps. nautica cNOR to homogeneity and characterized the purified enzyme in detail by using biochemical and spectroscopic methods. The purified enzyme exhibits an optical spectrum with an absorbance ratio of A411/A280 = 1.4, one of the highest purity preparations ever reported for cNOR. Heme and Fe content determinations show a cofactor stoichiometry of 1:2:1 for heme c: heme b: non-heme iron per molecule of cNOR, indicating full occupancy of metal centers in the purified enzyme. Regarding metal content, Ps. nautica cNOR was found to contain Ca. While the value obtained is substoichiometric, the presence of calcium was also observed in the X-ray crystallographic structure of the Ps. aeruginosa enzyme (51), indicating that Ca presence might be a common feature between anaerobic and microaerobic respiratory enzymes. As shown in Figure 1, arginine residues 50 and 433 (Ps. nautica numbering) are conserved in the five Pseudomonas and Paracoccus sequences. These residues are also present in Ca binding in cbb3 cytochrome oxidase from Ps. stutzeri (59).
Consistent with results reported for cNORs isolated from other organisms, the spectroscopic data indicate that the spin states of the electron transfer heme c and heme b are low-spin, and, for the non-heme iron FeB, it is high-spin. Contrary to the conventionally assumed high-spin state for heme b3, the Mössbauer data revealed that the spin state of the catalytic heme b3 is in fact low-spin. The UV/visible difference spectrum between the spectra of ascorbate-reduced and dithionite-reduced enzyme (Figure 2B), which displays the difference arising from changes in the oxidation state of heme b3, supports the low-spin designation and indicates further that at room temperature the reduced heme b3 remains low-spin. In support of our finding, a recent X-ray crystallographic structure of the Ps. aeruginosa enzyme, published online by Hino and coworkers (51) while this manuscript was in the reviewing process, shows that heme b3 is indeed hexacoordinated. It is axialy coordinated to a His residue (H347, Ps. stutzeri numbering) and a μ-oxo (or a possible μ-hydroxo) bridge to FeB. On the basis of temperature-dependent MCD and low-temperature EPR data, Cheesman et al. (21) concluded that the spin state of heme b3 in oxidized Ps. stutzeri cNOR underwent a spin transition from high-spin at room temperature to low-spin at 4.2 K. Our spectroscopic analysis clearly shows that heme b3 is low-spin in the ferric state between 4.2 K and 180 K, and in the ferrous active form from 4.2 K up to room temperature.
Re-Consideration of the g = 6.34 Assignment
In the oxidized enzyme, the ferric heme b3 is bridging to the ferric FeB via an oxo group (22, 51). Prior to this investigation, heme b3 was assumed to be high-spin ferric (42, 45) antiferromagnetically coupled to the ferric high-spin (S = 5/2) FeB, forming a diamagnetic (S = 0) ground state. Thus, no EPR signal was expected to arise from the oxidized catalytic diiron center. The signals at the g = 6 region were then assigned to a small quantity of uncoupled heme b3. Also, previously, a g = 4.3 signal was detected, and had been assigned to a minor quantity of the uncoupled ferric FeB. For the as-purified Ps. nautica cNOR, however, this g = 4.3 signal is almost non-observable (Figure 4A). We therefore proposed that the previously detected g = 4.3 signal may not arise from an uncoupled FeB, but rather, represent minor adventitiously bound ferric impurity, which is not present in our as-purified Ps. nautica cNOR.
Here, we present spectroscopic data establishing unambiguously that heme b3 is low-spin at low-temperature (4.2 to 180 K). Spin-spin coupling of a low-spin ferric (S = 1/2) heme b3 with a high-spin ferric (S = 5/2) FeB would yield an integer-spin ground state with S = 2 or S = 3 that may not be EPR silent (49). On the basis of the following observations, we propose that the g = 6.34 signal may arise from the oxidized binuclear heme b3-FeB site. (1) The absorption-type signal detected at g = 6.34 for cNORs is distinct from the derivative-type g = 6 signal expected for a high-spin ferric heme (Figure 4D–G). (2) In the as-purified enzyme, there are four Fe cofactors that could be detected by EPR. The electron-transfer ferric low-spin heme c and heme b are completely accounted for by the two sets of low-spin ferric EPR signals. Thus, the g = 6.34 signal has to be arising from either heme b3 or FeB or both. Because an isolated low-spin ferric heme b3 cannot yield signals at the g = 6 region, and because no additional signals are detected that may be assigned to an isolated low-spin ferric heme b3 or to an isolated ferric high-spin FeB, a probable explanation would be that heme b3 and FeB are spin coupled to form an integer-spin system that yields the g = 6.34 signal. And, (3) the g = 6.34 signal is detected in all three oxidation states of the enzyme (as purified, ascorbate-reduced and ditionite-reduced). According to the Mössbauer data (Table 1), the only common oxidation state for all the Fe cofactors that is present in all three oxidation states of the enzyme and may be detectable by EPR is the FeIII-heme b3-FeBIII site. Taken together, these observations argue favorably for the g = 6.34 signal to arise from a spin-coupled oxidized heme b3-FeB center. A definitive assignment for the g = 6.34 signal and a complete understanding of the nature of the proposed heme b3-FeB spin-spin coupling, however, will require further detailed spectroscopic investigations, such as, parallel mode EPR studies coupled with low-temperature-high-field Mössbauer measurements. Finally, it is important to point out that the g = 6.34 signal have been observed for all as-purified cNORs that have been characterized by EPR. Consequently, if the g = 6.34 signal is indeed arising from a spin coupled binuclear center comprising a low-spin ferric heme b3 and a high-spin ferric FeB, then heme b3 is most likely low-spin in all cNORs in the experimental conditions studied.
Catalytic Site Coordination
In general, high-spin heme complexes are five coordinated, while low-spin heme complexes are six coordinated, except for the low-spin [FeNO]7 heme complexes, which can be either five- or six-coordinated. The fact that heme b3 is low-spin in both its oxidized and reduced states implies that it is six-coordinated, rather than five-coordinated as previously assumed. Based on aminoacid sequence analysis and spectroscopic data comparison, we believe that H340 (Ps. nautica numbering) and a μ-oxo bridge to FeB (for ferric heme b3 states) or a hydroxide anion (in the fully reduced enzyme) are responsible for the observed low-spin state of heme b3. The presence of a sixth ligand for heme b3 is consistent with fast kinetic measurements for CO binding (60), where it was found that the initial binding of CO to FeII-heme b3 is approximately three orders of magnitude slower than photolysis induced CO recombination, presumably reflecting the need to displace a blocking axial ligand. By comparison with the known ligands of FeB in Ps. aeruginosa NOR, we attribute residues H200, H251, H252 and E204 as ligands to the non-heme iron.
Redox State of the Active Enzyme
An important information that is absolutely required for understanding enzyme mechanism is the redox state of the active enzyme. For cNOR, both the three-electron reduced state (heme b, heme c and FeB are reduced while heme b3 is oxidized) (28) and the fully reduced state (14, 19, 50) have been proposed to be the active redox state. By exploiting the large mid-point redox potential differences between heme b3 and the electron transfer heme c and heme b, we attempted to address this important question by preparing a stable three-electron reduced enzyme for activity assay. We reduced the as-purified enzyme by using a mild reductant, ascorbate, and characterized the oxidation states of the cofactors by using Mössbauer spectroscopy. The results show that heme b and heme c are completely reduced while heme b3 remains oxidized (Table 1). Approximately half of FeB is reduced. Activity assay indicates that the ascorbate-reduced cNOR is inactive (Table 2). The PMS and ascorbate (that have been used by several authors to reduce the enzyme in the activity assay) has been proved by UV/visible spectroscopy to yield a redox state similar to the one achieved by dithionite reduction (Figure S2). We therefore conclude that heme b3 has to be reduced for the enzyme to be active and that the fully reduced state is likely the active state of the enzyme.
Mechanistic Consideration
Currently, there are two proposed mechanisms for NO reduction by cNOR: the cis- and trans-mechanism (see Introduction). A major distinction between these two mechanisms is at the initial step of coordinating two NO molecules at the binuclear center. In the cis-mechanism, both NO molecules are coordinated either to the reduced FeB or to heme b3 forming a FeII-dinitrosyl complex (28). In the trans-mechanism, one NO molecule is bound to each reduced Fe site of the binuclear center forming a complex (14). The cis-mechanism with two NO molecules bound to a reduced heme b3 has been questioned based on the fact that this type of complexes are too stable for turnover.
To prevent the binding of NO to a FeII-heme b3 forming a stable [FeNO]7-heme complex in the initial NO binding step, it has been proposed that the three-electron reduced state with an oxidized heme b3 is the active state of the enzyme (28). Our conclusions that the fully reduced state is the active form of the enzyme, and that heme b3 is six-coordinated suggest an alternative possibility that favors NO binding to FeB in the initial catalytic step: The presence of a sixth ligand weakens the affinity of NO binding to heme b3 and promote the formation of a FeII-dinitrosyl complex at FeB. In such a scenario, the initial function of heme b3 could be (1) to assist in adjusting the orientation of the FeB-bound NO molecules and subsequent formation of the N–N bond, and (2) to provide the essential electron for the reduction of NO. Without further kinetic data, we cannot exclude that at a later mechanistic step heme b3 will somehow bind to NO (or to a reaction intermediate). Recently, Lachmann and coworkers (52) demonstrated that CO binds to reduced heme b3 (probably displacing the sixth ligand initially present) and that flash-induced dissociation causes heme b3 oxidation and catalysis. It is also known that for the CO-bound enzyme, NO can still bind to the FeB site (14). Both observations support the possibility of an initial binding of NO to FeB. It is important to comment that we believe that the cis-mechanism (with ligation of NO to FeB) is energetically favorable. The NO dissociation rate constants for reduced b-type ferrous porphyrin complexes are in the order of 10−3 to 10−5 s−1, demonstrating the stability of this type of nitrosyl complexes (53–56). To our knowledge very few hemic proteins form unstable/transient nitrosyl complexes: NO synthase, cytochrome cd1-type nitrite reductase, and cytochrome P450 all exhibit fast NO dissociation rate constants (2 to 100 s−1). One should mention that even with NO synthase, an enzyme that catalyzes the formation of NO, the release of NO is only productive from a FeIII-heme-NO catalytic intermediate, and very slow from the reduced FeII-heme-NO form (57). In the case of cytochrome cd1 nitrite reductase from denitrifying bacteria the fast NO dissociation from the ferrous heme form is thought to be associated with the involvement of heme d1. Fungal NOR, a member of the cytochrome P450 family, contains a heme b with axial cysteinate coordination. The NO substrate binds to the ferric state of heme b leading to the formation of a 6-coordinated ferric-heme-nitrosyl first intermediate. The FeIII-NO complex is unstable and the dissociation of NO is due to the presence of the cysteinyl ligand and to the ferric state of the heme, such as in NO synthase.
It is worth noting that Ps. nautica NOR is also able to reduce molecular oxygen (58). Although it is not established how widespread such bifunctionality is among cNOR, the fact that it was not reported for all characterized enzymes might be an indication that an alternative mechanism cannot be excluded.
Supplementary Material
Acknowledgments
We would like to thank Márcia Guilherme for her help in the purification of Ps. nautica NOR.
Abbreviations
- NOR
nitric oxide reductase
- EPR
electron paramagnetic resonance
- PMSF
phenylmethylsulfonyl fluoride
- DDM
sodium n-dodecyl α-D-maltoside
- DEAE
diethylaminoethyl cellulose
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- BCA
bicinchoninic acid
- TPTZ
2,4,6-tripyridyl-s-triazine
- CTAB
cetyl trimethylammonium bromide
- PMS
phenazine methosulfate
- PE
2-phenylethanol
Footnotes
This work was supported in part by funds from PDTC/QUI/64638/2006 grant (I.M.), SFRH/BPD/14863/2003 (C.G.T.), SFRH/BD/17840/2004 (C.E.M.), SFRH/BD/39009/2007 (A.G.D.) and National Institutes of Health grant GM 47295 (B.H.H.)
SUPPORTING INFORMATION AVAILABLE
Figure S1 shows the DNA and deduced aminoacid sequence of Ps. nautica NOR and Figure S2 compares the UV/visible spectrum of dithionite-reduced cNOR with that of the enzyme reduced by ascorbate plus PMS.
This information is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Bonin P, Bertrand JC, Giordano G, Gilewicz M. Specific sodium dependence of a nitrate reductase in a marine bacterium. FEMS Microbiology Letters. 1987;48:5–9. [Google Scholar]
- 2.Bonin P, Gilewicz M, Denis M, Bertrand JC. Salt requirements in the denitrifying bacterium Pseudomonas nautica 617. Res Microbiol. 1989;140:159–169. doi: 10.1016/0923-2508(89)90050-8. [DOI] [PubMed] [Google Scholar]
- 3.Tavares P, Pereira AS, Moura JJ, Moura I. Metalloenzymes of the denitrification pathway. J Inorg Biochem. 2006;100:2087–2100. doi: 10.1016/j.jinorgbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Brown K, Djinovic-Carugo K, Haltia T, Cabrito I, Saraste M, Moura JJ, Moura I, Tegoni M, Cambillau C. Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. J Biol Chem. 2000;275:41133–41136. doi: 10.1074/jbc.M008617200. [DOI] [PubMed] [Google Scholar]
- 5.Lopes H, Besson S, Moura I, Moura JJ. Kinetics of inter- and intramolecular electron transfer of Pseudomonas nautica cytochrome cd1 nitrite reductase: regulation of the NO-bound end product. J Biol Inorg Chem. 2001;6:55–62. doi: 10.1007/s007750000159. [DOI] [PubMed] [Google Scholar]
- 6.Brown K, Tegoni M, Prudencio M, Pereira AS, Besson S, Moura JJ, Moura I, Cambillau C. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat Struct Biol. 2000;7:191–195. doi: 10.1038/73288. [DOI] [PubMed] [Google Scholar]
- 7.Correia C, Besson S, Brondino CD, Gonzalez PJ, Fauque G, Lampreia J, Moura I, Moura JJ. Biochemical and spectroscopic characterization of the membrane-bound nitrate reductase from Marinobacter hydrocarbonoclasticus 617. J Biol Inorg Chem. 2008;13:1321–1333. doi: 10.1007/s00775-008-0416-1. [DOI] [PubMed] [Google Scholar]
- 8.Heiss B, Frunzke K, Zumft WG. Formation of the N-N bond from nitric oxide by a membrane-bound cytochrome bc complex of nitrate-respiring (denitrifying) Pseudomonas stutzeri. J Bacteriol. 1989;171:3288–3297. doi: 10.1128/jb.171.6.3288-3297.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastrau DH, Heiss B, Kroneck PM, Zumft WG. Nitric oxide reductase from Pseudomonas stutzeri, a novel cytochrome bc complex. Phospholipid requirement, electron paramagnetic resonance and redox properties. Eur J Biochem. 1994;222:293–303. doi: 10.1111/j.1432-1033.1994.tb18868.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoglen J, Hollocher TC. Purification and some characteristics of nitric oxide reductase-containing vesicles from Paracoccus denitrificans. J Biol Chem. 1989;264:7556–7563. [PubMed] [Google Scholar]
- 11.Carr GJ, Ferguson SJ. The nitric oxide reductase of Paracoccus denitrificans. Biochem J. 1990;269:423–429. doi: 10.1042/bj2690423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakurai T, Nakashima S, Kataoka K, Seo D, Sakurai N. Diverse NO reduction by Halomonas halodenitrificans nitric oxide reductase. Biochem Biophys Res Commun. 2005;333:483–487. doi: 10.1016/j.bbrc.2005.05.149. [DOI] [PubMed] [Google Scholar]
- 13.Sakurai T, Sakurai N, Matsumoto H, Hirota S, Yamauchi O. Roles of four iron centers in Paracoccus halodenitrificans nitric oxide reductase. Biochem Biophys Res Commun. 1998;251:248–251. doi: 10.1006/bbrc.1998.9451. [DOI] [PubMed] [Google Scholar]
- 14.Kumita H, Matsuura K, Hino T, Takahashi S, Hori H, Fukumori Y, Morishima I, Shiro Y. NO reduction by nitric-oxide reductase from denitrifying bacterium Pseudomonas aeruginosa: characterization of reaction intermediates that appear in the single turnover cycle. J Biol Chem. 2004;279:55247–55254. doi: 10.1074/jbc.M409996200. [DOI] [PubMed] [Google Scholar]
- 15.Cramm R, Pohlmann A, Friedrich B. Purification and characterization of the single-component nitric oxide reductase from Ralstonia eutropha H16. FEBS Lett. 1999;460:6–10. doi: 10.1016/s0014-5793(99)01315-0. [DOI] [PubMed] [Google Scholar]
- 16.Hendriks J, Oubrie A, Castresana J, Urbani A, Gemeinhardt S, Saraste M. Nitric oxide reductases in bacteria. Biochim Biophys Acta. 2000;1459:266–273. doi: 10.1016/s0005-2728(00)00161-4. [DOI] [PubMed] [Google Scholar]
- 17.Householder TC, Fozo EM, Cardinale JA, Clark VL. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect Immun. 2000;68:5241–5246. doi: 10.1128/iai.68.9.5241-5246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suharti, Heering HA, de Vries S. NO reductase from Bacillus azotoformans is a bifunctional enzyme accepting electrons from menaquinol and a specific endogenous membrane-bound cytochrome c551. Biochemistry. 2004;43:13487–13495. doi: 10.1021/bi0488101. [DOI] [PubMed] [Google Scholar]
- 19.Girsch P, de Vries S. Purification and initial kinetic and spectroscopic characterization of NO reductase from Paracoccus denitrificans. Biochim Biophys Acta. 1997;1318:202–216. doi: 10.1016/s0005-2728(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 20.Hendriks J, Warne A, Gohlke U, Haltia T, Ludovici C, Lubben M, Saraste M. The active site of the bacterial nitric oxide reductase is a dinuclear iron center. Biochemistry. 1998;37:13102–13109. doi: 10.1021/bi980943x. [DOI] [PubMed] [Google Scholar]
- 21.Cheesman MR, Zumft WG, Thomson AJ. The MCD and EPR of the heme centers of nitric oxide reductase from Pseudomonas stutzeri: Evidence that the enzyme is structurally related to the heme-copper oxidases. Biochemistry. 1998;37:3994–4000. doi: 10.1021/bi972437y. [DOI] [PubMed] [Google Scholar]
- 22.Moenne-Loccoz P, Richter OMH, Huang HW, Wasser IM, Ghiladi RA, Karlin KD, de Vries S. Nitric oxide reductase from Paracoccus denitrificans contains an oxo-bridged heme/non-heme diiron center. J Am Chem Soc. 2000;122(38):9344–9345. [Google Scholar]
- 23.Moenne-Loccoz P. Spectroscopic characterization of heme iron-nitrosyl species and their role in NO reductase mechanisms in diiron proteins. Nat Prod Rep. 2007;24(3):610–620. doi: 10.1039/b604194a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zumft WG. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J Inor Biochem. 2005;99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 25.Watmough NJ, Field SJ, Hughes RJ, Richardson DJ. The bacterial respiratory nitric oxide reductase. Biochem Soc Trans. 2009;37:392–399. doi: 10.1042/BST0370392. [DOI] [PubMed] [Google Scholar]
- 26.Field SJ, Thorndycroft FH, Matorin AD, Richardson DJ, Watmough NJ. The respiratory nitric oxide reductase (NorBC) from Paracoccus denitrificans. Meth Enzym. 2008;437:79–101. doi: 10.1016/S0076-6879(07)37005-5. [DOI] [PubMed] [Google Scholar]
- 27.Collman JP, Dey A, Yang Y, Decreau RA, Ohta T, Solomon EI. Intermediates Involved in the Two Electron Reduction of NO to N(2)O by a Functional Synthetic Model of Heme Containing Bacterial NO Reductase. J Am Chem Soc. 2008;130:16498–16499. doi: 10.1021/ja807700n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gronberg KL, Roldan MD, Prior L, Butland G, Cheesman MR, Richardson DJ, Spiro S, Thomson AJ, Watmough NJ. A low-redox potential heme in the dinuclear center of bacterial nitric oxide reductase: implications for the evolution of energy-conserving heme-copper oxidases. Biochemistry. 1999;38:13780–13786. doi: 10.1021/bi9916426. [DOI] [PubMed] [Google Scholar]
- 29.Gronberg KL, Watmough NJ, Thomson AJ, Richardson DJ, Field SJ. Redox-dependent open and closed forms of the active site of the bacterial respiratory nitric-oxide reductase revealed by cyanide binding studies. J Biol Chem. 2004;279:17120–17125. doi: 10.1074/jbc.M400824200. [DOI] [PubMed] [Google Scholar]
- 30.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 31.Saraiva LM, Besson S, Moura I, Fauque G. Purification and preliminary characterization of three c-type cytochromes from Pseudomonas nautica strain 617. Biochem Biophys Res Commun. 1995;212:1088–1097. doi: 10.1006/bbrc.1995.2081. [DOI] [PubMed] [Google Scholar]
- 32.Saraiva LM, Fauque G, Besson S, Moura I. Physico-chemical and spectroscopic properties of the monohemic cytochrome C552 from Pseudomonas nautica 617. Eur J Biochem. 1994;224:1011–1017. doi: 10.1111/j.1432-1033.1994.01011.x. [DOI] [PubMed] [Google Scholar]
- 33.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein Measurement with the Folin Phenol Reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 34.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 35.Fischer DS, Price DC. A simple serum iron method using the new sensitive chromogen tripyridyl-S-triazine. Clin Chem. 1964;10:21–31. [PubMed] [Google Scholar]
- 36.Moore S, Stein WH. Chromatographic determination of amino acids by the use of automatic recording equipment. Methods in Enzymology. 1963;6:819–831. [Google Scholar]
- 37.Ausubel FM, Brent R, Kingston RE, Seidman JG, Smith JA, Struhl K, editors. Published by Greene Pub Associates and Wiley-Interscience. J. Wiley; New York: 1989. Current protocols in molecular biology. [Google Scholar]
- 38.Butland G, Spiro S, Watmough NJ, Richardson DJ. Two conserved glutamates in the bacterial nitric oxide reductase are essential for activity but not assembly of the enzyme. J Bacteriol. 2001;183:189–199. doi: 10.1128/JB.183.1.189-199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flock U, Thorndycroft FH, Matorin AD, Richardson DJ, Watmough NJ, Adelroth P. Defining the proton entry point in the bacterial respiratory nitric-oxide reductase. J Biol Chem. 2008;283:3839–3845. doi: 10.1074/jbc.M704615200. [DOI] [PubMed] [Google Scholar]
- 40.Thorndycroft FH, Butland G, Richardson DJ, Watmough NJ. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem J. 2007;401:111–119. doi: 10.1042/BJ20060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flock U, Lachmann P, Reimann J, Watmough NJ, Ädelroth P. Exploring the terminal region of the proton pathway in the bacterial nitric oxide reductiase. J Inorg Biocherm. 2009;103:845–850. doi: 10.1016/j.jinorgbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Sakurai N, Sakurai T. Isolation and characterization of nitric oxide reductase from Paracoccus halodenitrificans. Biochemistry. 1997;36:13809–13815. doi: 10.1021/bi971070u. [DOI] [PubMed] [Google Scholar]
- 43.Field SJ, Prior L, Roldan MD, Cheesman MR, Thomson AJ, Spiro S, Butt JN, Watmought NJ, Richardson DJ. Spectral properties of bacterial nitric-oxide reductase - Resolution of pH-dependent forms of the active site heme b(3) J Biol Chem. 2002;277(23):20146–20150. doi: 10.1074/jbc.M112202200. [DOI] [PubMed] [Google Scholar]
- 44.Fujiwara T, Fukumori Y. Cytochrome cb-type nitric oxide reductase with cytochrome c oxidase activity from Paracoccus denitrificans ATCC 35512. J Bacteriol. 1996;178:1866–1871. doi: 10.1128/jb.178.7.1866-1871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oubrie A, Gemeinhardt S, Field S, Marritt S, Thomson AJ, Saraste M, Richardson DJ. Properties of a soluble domain of subunit C of a bacterial nitric oxide reductase. Biochemistry. 2002;41:10858–10865. doi: 10.1021/bi026140y. [DOI] [PubMed] [Google Scholar]
- 46.Aasa R, Vanngard T. EPR signal intensity and powder shapes: a reexamination. J Mag Res. 1975;19:308–315. [Google Scholar]
- 47.De Vries S, Albracht SPJ. Intensity of highly anisotropic low-spin heme EPR signals. Biochim Biophys Acta. 1979;546:334–340. doi: 10.1016/0005-2728(79)90050-1. [DOI] [PubMed] [Google Scholar]
- 48.Saraiva LM, Besson S, Fauque G, Moura I. Characterization of the dihemic cytochrome c549 from the marine denitrifying bacterium Pseudomonas nautica 617. Biochem Biophys Res Commun. 1994;199:1289–1296. doi: 10.1006/bbrc.1994.1371. [DOI] [PubMed] [Google Scholar]
- 49.Hendrich MP, Debrunner PG. Integer-spin electron paramagnetic resonance of iron proteins. Biophys J. 1989;56:489–506. doi: 10.1016/S0006-3495(89)82696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moenne-Loccoz P, de Vries S. Structural characterization of the catalytic high-spin heme b of nitric oxide reductase: A resonance Raman study. J Am Chem Soc. 1998;120:5147–5152. [Google Scholar]
- 51.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 52.Lachmann P, Huang Y, Reimann J, Flock U, Adelroth P. Substrate Control of Internal Electron Transfer in Bacterial Nitric-oxide Reductase. J Biol Chem. 2010;285:25531–25537. doi: 10.1074/jbc.M110.123984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinaldo S, Arcovito A, Giardina G, Castiglione N, Brunori M, Cruruzzollà F. New insights into the activity of Pseudomonas aeruginosa cd1 nitrite reductase. Biochem Soc Trans. 2008;36:1155–1159. doi: 10.1042/BST0361155. [DOI] [PubMed] [Google Scholar]
- 54.Ford PC, Lorkovic IM. Mechanistic aspects of the reaction of nitric oxide with transition-metal complexes. Chem Rev. 2002;102:993–1017. doi: 10.1021/cr0000271. [DOI] [PubMed] [Google Scholar]
- 55.Cooper CE. Nitric oxide and iron proteins. Biochim Biophys Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 56.Goodrich LE, Paulat F, Praneeth VKK, Lehnert N. Electronic structure of heme-nytrosyls and its significance for nitric oxide reactivity, sensing, transport, and toxicity in biological systems. Inorg Chem. 2010;49:6293–6316. doi: 10.1021/ic902304a. and references therein. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z-Q, Wei C-C, Stuehr DJ. How does a valine residue that modulates heme-NO binding kinetics in inducible NO synthase regulate enzyme catalysis? J Inorg Biochem. 2010;104:349–356. doi: 10.1016/j.jinorgbio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Cordas CM, Pereira AS, Martins CE, Timoteo CG, Moura I, Moura JJG, Tavares P. Nitric oxide reductase: Direct electrochemistry and electrocatalytic activity. Chembiochem. 2006;7:1878–1881. doi: 10.1002/cbic.200600253. [DOI] [PubMed] [Google Scholar]
- 59.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The Structure of cbb(3) Cytochrome Oxidase Provides Insights into Proton Pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 60.Hendriks JH, Prior L, Baker AR, Thomson AJ, Saraste M, Watmough NJ. Reaction of carbon monoxide with the reduced active site of bacterial nitric oxide reductase. Biochemistry. 2001;40:13361–13369. doi: 10.1021/bi011428t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.