Summary
Certain mutants in Escherichia coli lacking multiple penicillin-binding proteins (PBPs) produce misshapen cells containing kinks, bends and branches. These deformed regions exhibit two structural characteristics of normal cell poles: the peptidoglycan is inert to dilution by new synthesis or turnover, and a similarly stable patch of outer membrane caps the sites. To test the premise that these aberrant sites represent biochemically functional but misplaced cell poles, we assessed the intracellular distribution of proteins that localize specifically to bacterial poles. Green fluorescent protein (GFP) hybrids containing polar localization sequences from the Shigella flexneri IcsA protein or from the Vibrio cholerae EpsM protein formed foci at the poles of wild-type E. coli and at the poles and morphological abnormalities in PBP mutants. In addition, secreted wild-type IcsA localized to the outer membrane overlying these aberrant domains. We conclude that the morphologically deformed sites in these mutants represent fully functional poles or pole fragments. The results suggest that prokaryotic morphology is driven, at least in part, by the controlled placement of polar material, and that one or more of the low-molecular-weight PBPs participate in this process. Such mutants may help to unravel how particular proteins are targeted to bacterial poles, thereby creating important biochemical and functional asymmetries.
Introduction
The bacterial cell pole is a very special, increasingly crowded place. A number of proteins, complexes and physiological processes congregate preferentially at one or both poles, with implications for protein secretion, environmental sensing, virulence and gene transfer (for reviews, see Lybarger and Maddock, 2001; Shapiro et al., 2002). For example, the Shigella flexneri IcsA protein is secreted at one pole (Goldberg et al., 1993; Charles et al., 2001; Brandon et al., 2003), creating an asymmetric distribution of IcsA that propels the organism within and between mammalian cells by nucleating the formation of an actin tail. The ActA protein of Listeria monocytogenes is similarly distributed (Kocks et al., 1993), although whether its asymmetry is created by targeted secretion at the pole remains unclear. In Vibrio cholerae, the Eps type II secretion complex is confined to the older cell pole, where it secretes cholera toxin and a haemagglutinin/protease (Scott et al., 2001); likewise, the Dot/Icm secretion system is positioned at the poles of Legionella pneumophila (Conover et al., 2003). In Agrobacterium tumefaciens, the VirD4 protein assembles a type IV apparatus at the pole to transport DNA into plant cells (Kumar and Das, 2002); and in Streptomyces, plasmids transfer to neighbouring cells by passing through specialized pores embedded in the poles (Grohmann et al., 2003).
Besides these secretory processes, bacteria display other intracellular asymmetries. One of the earliest discoveries was that methyl-accepting chemotaxis proteins (MCPs) are anchored in the cytoplasmic membrane at one pole of Caulobacter crescentus and E. coli, thereby nucleating a spatially limited assembly of chemotaxis factors (Alley et al., 1992; Maddock and Shapiro, 1993; Sourjik and Berg, 2000). Also, in C. crescentus, a set of histidine kinases and enzymes involved with cellular differentiation is drawn to one pole by the PodJ protein (Viollier et al., 2002; Hinz et al., 2003); and in Pseudomonas aeruginosa, the PilS kinase localizes to the poles (Boyd, 2000). In Bacillus subtilis, the earliest known protein to populate newly formed poles is the DivIVA protein, which attracts and holds several cell cycle proteins, including the MinCD septation inhibitor (Edwards and Errington, 1997; Marston et al., 1998; Edwards et al., 2000), the Soj (ParA) chromosomal partitioning factor (Autret and Errington, 2003) and RacA, which tethers the chromosomal origin of replication (Ben-Yehuda et al., 2003).
What is the positional information that leads to proper polar localization of these proteins?
Relatively little is known about what distinguishes the pole from the cylindrical portion of rod-shaped bacteria. One clear difference is that polar peptidoglycan is extremely long-lived, i.e. metabolically inert to normal synthesis and turnover, in contrast to the rapid replacement of peptidoglycan in the side-walls (de Pedro et al., 1997). This stability also extends to outer membrane domains associated with poles in Gram-negative bacteria, in that polar outer membrane proteins are not diluted by the addition of new material during growth (de Pedro et al., 2003). How these envelope components mature to become inert poles addresses one of the most fundamental steps in bacterial differentiation.
Recently, we discovered that several mutants lacking multiple low-molecular-weight penicillin-binding proteins (LMW PBPs) synthesize envelope fragments that are pole-like and apparently contribute to the formation of exceedingly misshapen cells (de Pedro et al., 2003). In these mutants, sites of morphological abnormalities co-localize with inert peptidoglycan and stable patches of outer membrane, characteristics normally associated only with bona fide bacterial poles (de Pedro et al., 2003). In this work, we ask whether these ectopic structures also behave as fully functional poles, by testing whether proteins that accumulate preferentially at the poles of E. coli also localize to the morphological abnormalities of PBP mutants. In fact, IcsA and EpsM localize in a specific manner to these sites, and IcsA is secreted to the outer membrane over these regions, indicating that the misplaced patches not only look like poles but mimic their functions as well. Therefore, these mutants may be valuable tools for examining the molecular processes leading to the differentiation and localization of normal poles.
Results
Escherichia coli cells lacking multiple LMW PBPs have abnormal shapes, manifested as bumps, bends, kinks and branches that display the structural characteristics of cell poles (de Pedro et al., 1997; 2003; Nelson and Young, 2000; 2001). To determine whether these deformed zones also behaved as poles, we assessed whether IcsA and EpsM, two proteins known to localize green fluorescent protein (GFP) fusions to the pole (Charles et al., 2001; Scott et al., 2001), also localized to the deformed zones of misshapen mutants. Although IcsA and EpsM are not naturally present in E. coli, when expressed in E. coli, each localizes to the pole, indicating that the positional information required for each protein’s polar localization is present in this species (Charles et al., 2001; Scott et al., 2001).
Construction of shape-deformed mutants in E. coli with a complete lipopolysaccharide (LPS)
Full-length IcsA is secreted at the pole of Shigella and E. coli and, in the presence of a complete LPS, is retained at the pole. As E. coli K-12 strains express a truncated O-antigen (Liu and Reeves, 1994), IcsA exported to the outer membrane in K-12 strains is delocalized from the poles by diffusion (Sandlin et al., 1995; Sandlin and Maurelli, 1999; Robbins et al., 2001), leading to a relatively uniform distribution of the protein on the bacterial surface. Therefore, to determine the functional characteristics of the outer membrane at morphological imperfections in shape-defective E. coli, we deleted multiple PBPs from E. coli 2443, a strain that expresses intact O8 serotype O-antigen and therefore an intact LPS (Meier and Mayer, 1985; Sandlin and Maurelli, 1999).
Mutants of strain 2443 lacking four or more PBPs grew as mixed populations of normal and aberrantly shaped cells. Surprisingly, mutants lacking seven PBPs (AG70A-6 and AG70C-12) produced a smaller fraction of abnormal cells than did mutants derived from the K-12 strain CS109. For example, ≈55–65% of cells of the CS109 derivative CS703-1, which lacks seven PBPs, exhibited one or more morphological oddities. However, when the same seven PBPs were deleted from E. coli 2443, only ≈10–20% of the cells exhibited similar abnormalities, and the deformities that arose were not quite as severe (not shown). The possibility that a complete O-antigen affects cell shape is being explored. Nonetheless, the number of aberrant 2443 cells was sufficient to test the pole-like behaviour of deformed sites.
One of the distinctive characteristics of bacterial poles is that they are stable: E. coli retains a patch of inert peptidoglycan and outer membrane over each pole, and the same phenomenon occurs at sites of morphological deformities in misshapen mutants (de Pedro et al., 2003). To confirm that this trait was retained in strains expressing intact O-antigen, we labelled the outer membrane proteins of 2443-derived mutants with Texas Red succinimidyl ester and grew the cells in the absence of dye for one or two generations. Stable membrane domains retain label, while sections diluted by diffuse insertion of new material become less fluorescent over time (de Pedro et al., 2003). To enhance the spatial discrimination of the poles and deformities, the cells were filamented by treatment with aztreonam, a specific inhibitor of the septation protein PBP 3 (FtsI). In the parental strains, CS109 (de Pedro et al., 2003) and 2443 (Fig. 1), conspicuous fluorescent patches remained over the poles. In the mutants, CS703-1 (derived from CS109) and AG70A-6 and AG70C-12 (derived from 2443), long-lived fluorescent patches were concentrated at deformities as well as at poles (Fig. 1; not shown). Thus, the poles and morphologically abnormal sites in 2443-derived strains exhibited the same traits as those observed in CS109-derived mutants.
Fig. 1.

Outer membrane stability at poles and deformities of E. coli 2443-derived strains. Cells were labelled with Texas Red succinimidyl ester and incubated in the absence of dye and in the presence of aztreonam. Stable regions of outer membrane proteins appear as brightly fluorescent patches at the poles in strain 2443 (left) and at the poles and deformities in the PBP mutant AG70C-12 (right).
IcsA pole-targeting sequences localize GFP to morphological deformities
Two regions of IcsA (IcsA1-104 and IcsA507-620) each localize a GFP fusion to the cytoplasmic face of the inner membrane at the pole of E. coli K-12 strains (Charles et al., 2001). Like full-length IcsA, IcsA1-104–GFP and IcsA507-620–GFP localize to at least one pole in 99% of cells, and to both poles in ≈50% of cells (Charles et al., 2001). A similar pattern of localization is observed for IcsA1-104–GFP and IcsA507-620–GFP fusion proteins in wild-type E. coli that expresses complete O-antigen (strain 2443) (Fig. 2A and Table 1). In strain 2443, 97 ± 3% (mean ± SD) of cells expressing IcsA1-104–GFP contained fluorescent foci at one or both poles, as did 95 ± 4% that expressed IcsA507-620–GFP (Table 1). A similar high frequency of IcsA507-620–GFP accumulated at the poles of the K-12 derivative CS109 (Table 1).
Fig. 2. Localization of IcsA–GFP fusion proteins to poles and deformities in E. coli 2443-derived strains. Direct fluorescence (left) and phase (right) micrographs of IcsA507-620–GFP (A, C and E) and IcsAΔ507-729–GFP (B, D and F).
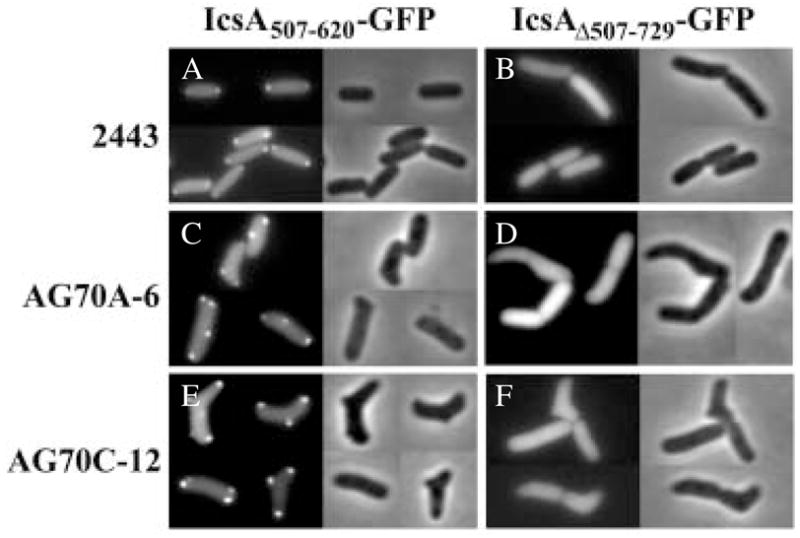
A and B. Wild-type strain 2443.
C and D. PBP mutant AG70A-6.
E and F. PBP mutant AG70C-12.
Table 1.
Localization of hybrid proteins.
| Strain | PBPs deleted | Fusion protein | Distribution of GFP signal (% of cells)
|
|||
|---|---|---|---|---|---|---|
| Foci at poles and deformities | Foci at poles only | Foci at deformities only | Diffuse | |||
| 2443a | None | IcsA1-104–GFP | NAb | 97 ± 3 | NA | 3 ± 3 |
| IcsA507-620–GFP | NA | 95 ± 4 | NA | 5 ± 4 | ||
| IcsAΔ507-729–GFP | NA | 7 ± 9 | NA | 93 ± 9 | ||
| GFP | NA | <1 | NA | >99 | ||
| AG70A-6a | 1a, 4, 5, 6, 7, | IcsA1-104–GFP | 72 ± 7 | 26 ± 8 | 1 ± 2 | 1 ± 2 |
| DacD, AmpC | IcsA507-620–GFP | 78 ± 5 | 16 ± 5 | 4 ± 2 | 2 ± 2 | |
| IcsAΔ507-729–GFP | <1 | 8 ± 4 | 1 ± 2 | 91 ± 5 | ||
| GFP | <1 | <1 | <1 | >99 | ||
| AG70C-12a | 1b, 4, 5, 6, 7, | IcsA1-104–GFP | 93 ± 3 | 7 ± 3 | <1 | <1 |
| DacD, AmpC | IcsA507-620–GFP | 83 ± 17 | 15 ± 14 | 2 ± 4 | <1 | |
| IcsAΔ507-729–GFP | 15 ± 5 | 26 ± 13 | 2 ± 2 | 57 ± 15 | ||
| GFP | <1 | <1 | <1 | >99 | ||
| CS109c | None | IcsA507-620–GFP | NA | >99 | NA | <1 |
| IcsA507-620–GFP | ||||||
| + aztreonam | NA | >99 | NA | <1 | ||
| GFP–EpsM | NA | >99 | NA | <1 | ||
| CS703-1c | 1a, 4, 5, 6, 7, | IcsA507-620–GFP | 85 | 15 | <1 | <1 |
| AmpC, AmpH | IcsA507-620–GFP | |||||
| + aztreonam | >99 | <1 | <1 | <1 | ||
| GFP–EpsM | 90 | 10 | <1 | <1 | ||
Tabulation was of 30 cells in each of three experiments for each strain expressing each fusion protein.
Not applicable: deformities were extremely rare or non-existent among cells in which no PBPs were deleted.
Tabulations for CS109 were from 150, 74 and 146 cells, respectively, and from CS703-1 were from 81, 25 and 82 cells respectively.
In addition to localizing to normal poles, these two IcsA–GFP hybrids localized to the extra poles of obviously branched cells and to other morphological deformities in aberrant cells of the shape-defective mutants AG70A-6 and AG70C-12 (Table 1). For some of the mutant cells, whether a protrusion represented a pole or a deformity was difficult to determine. Therefore, for consistency in our analysis, we defined the cell poles as the two most distal convex ends. To ensure that deformities would be highly represented in the quantification, only bacteria with grossly visible deformities were included.
Among cells with obvious morphological abnormalities, 72 ± 7% of aberrantly shaped AG70A-6 cells and 93 ± 3% of aberrantly shaped AG70C-12 cells contained IcsA1-104–GFP foci at deformities as well as at one or both poles (Table 1). Similarly, upon expression of IcsA507-620–GFP, 78 ± 5% of aberrantly shaped AG70A-6 cells (Fig. 2C) and 83 ± 17% of aberrantly shaped AG70C-12 cells (Fig. 2E) contained fluorescent foci at deformities and at one or both poles (Table 1). The sites to which the GFP foci localized were commonly branches or protrusions that were situated at a distance from the ends of the cells; these were likely to be true deformities and unlikely to be poles. Thus, IcsA1-104 and IcsA507-620 localize GFP to shape deformities as well as to cell poles.
Similar patterns and frequencies of localization of IcsA507-620–GFP to sites of deformities and poles were observed in a K-12-derived PBP mutant, CS703-1, which lacks a complete O-antigen (Fig. 3A and B and Table 1). In parallel, to enhance the spatial discrimination of poles and deformities, cells were filamented by treatment with aztreonam. In CS109 filaments, fluorescent foci were located at undivided septa and at poles (not shown), as observed previously in other strains (Janakiraman and Goldberg, 2004). As before, IcsA507-620–GFP localized to deformed sites and at the termini of extra poles in branched cells of CS703-1 (Fig. 3B). Therefore, the presence or absence of intact O-antigen did not affect localization of IcsA–GFP proteins to morphological abnormalities in PBP mutants.
Fig. 3. Localization of IcsA–GFP and GFP–EpsM in the K-12-derived PBP mutant CS703-1. Direct fluorescence (left) and phase (right) micrographs.

A. IcsA507-620–GFP.
B. IcsA507-620–GFP in cells filamented with aztreonam.
C. GFP–EpsM.
In PBP mutants expressing either of the IcsA–GFP fusion proteins, two adjacent foci were frequently observed at a single pole or deformity (Fig. 2E). This unusual localization may reflect an underlying pattern of peptidoglycan synthesis. de Pedro et al. (2003) observed split poles in similar PBP mutants, in which a sliver of metabolically active peptidoglycan bisected patches of inert peptidoglycan at poles or deformities. Thus, paired IcsA–GFP foci may mark opposite sides of this heterogeneous domain.
To be certain that localization of the IcsA–GFP fusions was specific for deformed sites and poles, we examined the distribution of GFP alone and a protein fusion that localizes inefficiently to the poles. As expected, GFP by itself was distributed diffusely in all strains (Table 1). IcsAΔ507-729–GFP, which lacks the polar targeting sequence present in IcsA507-620–GFP but contains the targeting sequence present in IcsA1-104–GFP, distributes diffusely in the cytoplasm of Shigella (Charles et al., 2001). In wild-type 2443 cells, IcsAΔ507-729–GFP was diffuse in 93 ± 9% and present as polar foci in only 7 ± 9% of the cells. (Fig. 2B and Table 1). In PBP mutants of 2443, the pattern of localization was similar: IcsAΔ507-729–GFP accumulated at poles and deformities in <1% of AG70A-6 cells and in 15 ± 5% of AG70C-12 cells (Fig. 2D and F and Table 1). And, in the K-12-derived strain CS109 and its abnormally shaped descendent CS703-1, IcsAΔ507-729–GFP was cytoplasmic and diffuse (not shown). Thus, in all strain backgrounds, IcsAΔ507-729–GFP localized significantly less efficiently to the poles than did IcsA507-620. The reason for some variability in the efficiency with which this fusion localized to the poles and deformities in different strain backgrounds is unclear. Taken together, these data indicate that localization of IcsA1-104–GFP and IcsA507-620–GFP to deformities in PBP mutants was specific rather than a generalized artifact accompanying expression of fusion proteins, and that the various abnormalities in misshapen cells with or without intact O-antigen contained the positional information that normally mediates polar localization of IcsA.
Full-length IcsA localizes to the outer membrane at morphological deformities and poles in PBP mutants with intact O-antigen
Full-length IcsA is retained at the old pole on the surface of Shigella and in E. coli that express an intact O-antigen (Goldberg et al., 1993; Sandlin et al., 1995; Sandlin and Maurelli, 1999). To test whether the deformities in PBP mutants exhibited this same function, we examined the distribution of full-length IcsA on the surface of E. coli PBP mutants that expressed intact O-antigen. For these experiments, we examined derivatives of 2443 and AG70C-12 in which ompT was disrupted, because the OmpT pro-tease cleaves mature IcsA, thereby releasing the extracellular domain from the surface (Nakata et al., 1993).
Full-length IcsA generated fluorescent signals on the surface of AG70C-12 ompT cells at sites of morphological abnormalities, as well as at poles (Fig. 4B), indicating that full-length IcsA was concentrated in the outer membrane at these sites. Enhanced signals were observed at deformities that appeared to be branches (Fig. 4B, arrows), as well as at deformities that appeared to be split poles (Fig. 4B, arrowheads). A similar distribution of signals from full-length IcsA was seen at the poles of the rod-shaped parent, 2443 ompT (Fig. 4A), a pattern identical to that described previously (Sandlin et al., 1995). In conjunction with our demonstration that stable patches of outer membrane are associated with the deformities (above), these data indicate that these sites function like poles. Moreover, if specific proteins or structures are required for the secretion of IcsA into the outer membrane at the pole of wild-type cells, then the same proteins or structures are likely to be present at deformities in the mutants.
Fig. 4. Localization of full-length IcsA to the outer membrane of E. coli 2443-derived strains. Indirect immunofluorescence (left) and phase (right) micrographs of surface-localized IcsA.
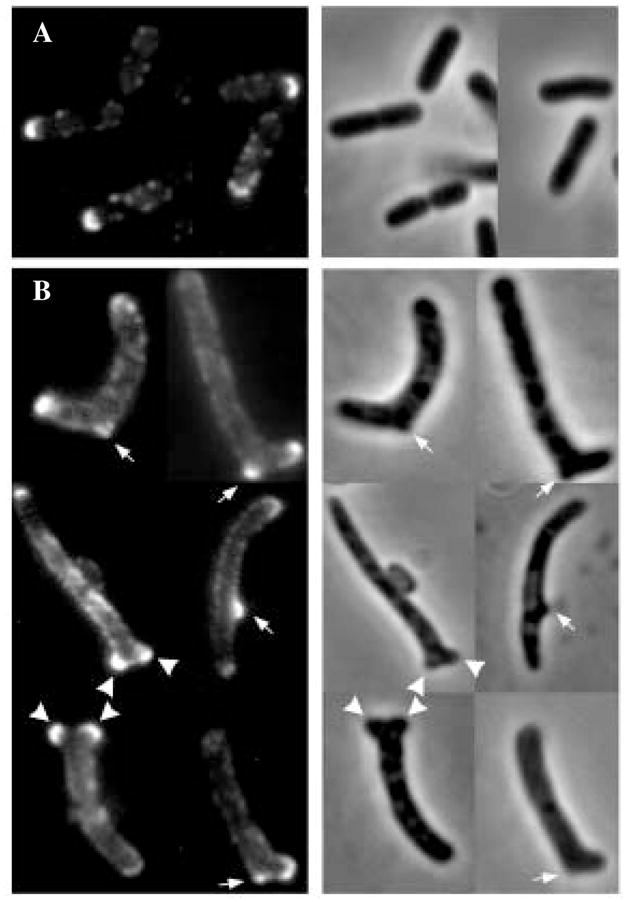
A. E. coli 2443 ompT.
B. PBP mutant AG70C-12 ompT. Arrows, IcsA localized at branches; arrowheads, IcsA localized at split poles.
IcsA507-620–GFP and full-length IcsA compete for targeting to poles and deformities
To examine whether the localization of IcsA507-620–GFP foci at poles and deformities could be an artifact of over-expression of fusion proteins, we examined the ability of full-length IcsA to interfere with this pattern of localization. Upon expression of full-length IcsA in trans in either wild-type 2443 ompT or the PBP mutant AG70C-12 ompT, the GFP signal from IcsA507-620–GFP was almost completely diffuse (Fig. 5, and data not shown). Thus, expression of full-length IcsA appeared to interfere with localization of IcsA507-620–GFP to both deformities and poles, strongly suggesting that the localization of the fusion to these sites is specific and not an artifact of overexpression or inclusion body formation.
Fig. 5. Interference of full-length IcsA with IcsA507-620–GFP localization in PBP mutant AG70C-12 ompT. Direct fluorescence (left) and phase (right) micrographs.

A. PBP mutant AG70C-12 ompT expressing IcsA507-620–GFP alone.
B. PBP mutant AG70C-12 ompT expressing both full-length IcsA and IcsA507-620–GFP.
IcsA507-620–GFP and full-length IcsA co-localize to morphological deformities and poles
The surface distribution of full-length IcsA was similar to the localization of IcsA1-104–GFP and IcsA507-620–GFP in the cytoplasm. To test whether full-length IcsA was localizing to the same sites as the cytoplasmic IcsA–GFP hybrids, we co-expressed full-length IcsA and IcsA507-620–GFP in ompT derivatives of E. coli wild-type strain 2443 and its PBP mutant derivative AG70C-12. To obtain visible GFP foci, IcsA507-620–GFP had to be expressed for 30 min, instead of 10 min, which was used in other experiments in this work, consistent with interference of full-length IcsA with the GFP fusion. Nevertheless, in some 2443 ompT cells, IcsA507-620–GFP foci and surface-labelled IcsA colocalized to the poles of the rod-shaped bacilli (Fig. 6A). Similarly, in some AG70C-12 cells, full-length IcsA was concentrated on the surface of the same morphological deformities and poles to which IcsA507-620–GFP foci were localized in the cytoplasm (Fig. 6B and C). The relatively low frequency of co-localization, only seen in two or three out of five possible poles and deformities in Fig. 6, is probably caused at least in part by interference of full-length IcsA with IcsA507-620–GFP. Taken together, these data demonstrate that the morphological deformities present in the PBP mutants contain: (i) the positional information that localizes IcsA at the cytoplasmic face of the inner membrane; (ii) the proteins or structures required for IcsA secretion into the outer membrane; and (iii) the stability required for maintenance of IcsA in the outer membrane.
Fig. 6. Co-localization of full-length IcsA and IcsA507-620–GFP to poles and sites of deformities of E. coli 2443-derived strains. Phase micro-graphs (Phase), direct fluorescence micrographs (Ics A507-620–GFP), indirect immunofluorescence micrographs (Full-length IcsA) and overlay of direct fluorescence (green) and indirect immunofluorescence (red) micrographs (Overlay).

A. Wild-type derivative 2443 ompT.
B and C. PBP mutant AG70C-12 ompT. Arrows indicate co-localization of cytoplasmic and surface IcsA.
GFP–EpsM accumulates at morphological deformities as well as poles
To test whether pole-like behaviour was a general attribute of the abnormal sites in shape mutants, we also examined the distribution of GFP–EpsM, which accumulates at the pole in the cytoplasmic membrane (Scott et al., 2001). In CS109, GFP–EpsM accumulated at the poles in essentially all cells (Table 1). Of the abnormally shaped cells produced by CS703-1, 90% exhibited GFP–EpsM foci at deformities and at one or both poles (Fig. 3C and Table 1). Thus, the distribution of GFP–EpsM was similar to that observed for IcsA507-620–GFP, indicating that the deformed sites behaved as ectopic poles with respect to more than one protein.
Discussion
The simplest manifestation of asymmetry in bacteria is the differential localization of cellular components to the poles, and discoveries in the past few years have revealed that a surprisingly large number of proteins and processes congregate at these sites (see Introduction). The breadth, ubiquity and specificity of the phenomenon argue that polar localization plays a fundamental role in several aspects of bacterial physiology.
Previously, we reported that the slender rod shape of E. coli is replaced by remarkably aberrant morphologies in mutants lacking certain PBPs (de Pedro et al., 2003). The deformities that typify these misshapen cells contain stable patches of peptidoglycan and outer membrane, structural characteristics that, to date, have been associated only with normal cell poles (de Pedro et al., 1997; 2003). We now show that these pole-like domains are biochemically functional, demonstrating that poles or pole fragments can be created at positions other than at a cell’s extreme ends. First, GFP hybrids of IcsA and EpsM localized to poles and at positions where the mutants were bent, kinked or otherwise deformed. These patterns indicate that the positional information present in the cytoplasmic membrane at the poles of wild-type bacteria was also present in the cytoplasmic membrane underlying morphological abnormalities in shape-defective mutants. Secondly, full-length IcsA was secreted to the outer membrane overlying the deformities. In wild-type bacteria, IcsA is targeted to the pole in the cytoplasm where it is secreted and inserted into the outer membrane (Brandon et al., 2003). The localization of IcsA–GFP to the inner face of the cytoplasmic membrane at morphologically abnormal sites in PBP mutants, and the export of full-length IcsA to the outer membrane at these same positions, strongly suggests that IcsA is secreted at these sites as well as at normal poles. As secretion of IcsA necessarily involves translocation across the cytoplasmic and outer membranes, as well as passage through the peptidoglycan, the results imply that, at deformities, the entire thickness of the cell envelope is functioning in concert as a pole. We conclude that these extra, non-terminal envelope regions exhibit the structural and biochemical properties of misplaced poles. The existence of mutants with defects in pole creation indicates that an active mechanism limits normal pole maturation to the extreme ends of a cell. Although the molecular details leading to the morphological deformities are unclear, our results imply that the process affects not only peptidoglycan synthesis but also the assembly or maturation of all layers of the cell envelope. The same phenomena occur during the formation of poles in wild-type and mutant cells. Because the single most distinguishing feature of normal poles is that they are descended from a previous septation event at the end of a programme of cell division, it seems likely that the propagation of aberrantly localized polar material originates during the sequence of events that normally determines the placement of the site of septation. If so, then some of the LMW PBPs must influence septal biogenesis.
How might the LMW PBPs, which are non-essential, affect septation? Among these enzymes, the loss of PBP 5 is most important for the development of visible shape abnormalities in E. coli (Nelson and Young, 2000; 2001), an effect heightened by the simultaneous absence of other PBPs (Nelson and Young, 2001; B. Meberg et al., unpublished). Deleting PBP 5 increases the number of pentapeptide side-chains in mature peptidoglycan, but the only overt phenotype associated with this change is the propensity to form grossly misshapen cells. After evaluating several mutations and conditions that result in similarly dramatic morphological changes, it was predicted that the placement or activity of FtsZ may represent the common denominator through which these disparate agents affect cell shape (Young, 2003). Such an interpretation would be consistent with the results reported here because the fundamental alteration in these deformed cells is the mis-localization of polar material, which should be generated by a cascade of events nucleated by FtsZ (Buddelmeijer and Beckwith, 2002). The simplest explanation is that the loss of certain PBPs produces an imbalance in the substrates acted on by FtsZ or FtsZ-dependent reactions, which results in the synthesis of mislocalized septal material (Young, 2003). The substrates and signals that trigger these events remain poorly defined.
In conclusion, we have shown that the maturation or positioning of cell poles can be mishandled by E. coli, thereby giving rise to misplaced polar material and nonuniform cell shapes. Understanding the creation of ectopic poles should shed light on the genesis, structure and topological regulation of normal poles. Furthermore, it may be possible to approach questions about polar targeting of specific proteins by identifying the underlying mechanisms that synthesize and position this distinctive envelope domain.
Experimental procedures
Bacteria, plasmids and general techniques
Escherichia coli CS109 (W1485 rpoS rph) is a K-12 derivative from C. Schnaitman. CS703-1 is derived from CS109 by deletion of seven PBP genes (mrcA dacA dacB dacC pbpG ampC ampH) (Meberg et al., 2001). E. coli 2443 (StrR) (thr-1 leuB6 Δ(gpt-proA)66 argE3 thi-1 rfbO8 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51), which expresses intact O-antigen, serotype 8 (Meier and Mayer, 1985), was obtained from A. T. Maurelli. Mutants derived from these two strains are listed in Table 2. Plasmids pBAD-IcsA1-104–GFP, pBAD-IcsA507-620–GFP and pBAD-IcsAΔ507-729–GFP express hybrid proteins consisting of various segments of IcsA fused in frame to GFP (Charles et al., 2001), under the control of the arabinose promoter of pBAD24 (Guzman et al., 1995). Full-length IcsA was expressed from its own promoter in a pBR322 vector backbone (Magdalena and Goldberg, 2002). To permit simultaneous expression of IcsA507-620–GFP and full-length IcsA, the coding sequence of IcsA507-620–GFP was subcloned into the chloramphenicol-resistant vector pBAD33. Plasmid pGFP-EpsM (Scott et al., 2001) was received from M. Sandkvist and M. Scott. Bacteria were grown in Luria–Bertani (LB) broth, on LB agar plates or in M9 minimal glucose medium (Miller, 1992). Where appropriate, antibiotics were added to the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 20 μg ml−1; and kanamycin 50 μg ml−1. Where indicated, aztreonam, a specific inhibitor of PBP 3, was added to cultures at a final concentration of 5 μg ml−1.
Table 2.
E. coli strains and plasmids.
| E. coli strain | Relevant genotype | PBPs deleted | Source or reference |
|---|---|---|---|
| 2443 | thr-1 leuB6 Δ(gpt-proA)66 argE3 thi-1 rfbo8 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl51 rpsL31 kdgK51 | None | A. T. Maurelli |
| AD202 | ompT::km (KanR) | Akiyama and Ito (1990) | |
| AG104-3K | E. coli 2443 ΔdacB::Kan | 4 | This work, E. coli 2443 × (P1 CS11-2K) |
| AG104-3KT | E. coli 2443 (zba-3000::Tn10) ΔdacB::Kan | 4 | This work, AG104-3K × (P1 KL743) |
| AG104-1 | E. coli 2443 (zba-3000::Tn10) ΔdacB | 4 | This work |
| AG247-1 | AG104-1 ΔpbpG | 4, 7 | This work, AG104-1 × (P1 CS9-19K) |
| AG375-1 | AG247-1 ΔdacA | 4, 5, 7 | This work, AG247-1 × (P1 CS12-7K) |
| AG456-1 | AG375-1 ΔdacC | 4, 5, 6, 7 | This work, AG375-1 × (P1 CS17-1K) |
| AG56D-1 | AG456-1 ΔdacD | 4, 5, 6, 7, DacD | This work, AG456-1 × (P1 CS18-2K) |
| AG60B-1 | AG56D-1 ΔmrcB | 1b, 4, 5, 6, 7, DacD | This work, AG56D-1 × (P1 CS16-1K) |
| AG60C-1 | AG56D-1 ΔampC | 4, 5, 6, 7, DacD, AmpC | This work, AG56D-1 × (P1 CS14-2K) |
| AG70A-6 | AG60C-1 ΔmrcA | 1a, 4, 5, 6, 7, DacD, AmpC | This work, AG60C-1 × (P1 BMCS04-1K) |
| AG70C-12 | AG60B-1 ΔampC | 1b, 4, 5, 6, 7, DacD, AmpC | This work, AG60B-1 × (P1 CS14-2K) |
| BMCS04-1K | CS109 ΔmrcA::Kan | 1a | Meberg et al. (2001) |
| CS109 | W1485 rpoS rph | None | C. Schnaitman |
| CS9-19K | CS109 ΔpbpG::Kan | 7 | Denome et al. (1999) |
| CS11-2K | CS109 ΔdacB::Kan | 4 | Denome et al. (1999) |
| CS12-7 | CS109 ΔdacA | 5 | Denome et al. (1999) |
| CS12-7K | CS109 ΔdacA::Kan | 5 | Denome et al. (1999) |
| CS14-2K | CS109 ΔampC::Kan | AmpC | Denome et al. (1999) |
| CS16-1K | CS109 ΔmrcB::Kan | 1b | Denome et al. (1999) |
| CS17-1K | CS109 ΔdacC::Kan | 6 | Denome et al. (1999) |
| CS18-2K | CS109 ΔdacD::Kan | DacD | Denome et al. (1999) |
| CS203-1B | CS109 ΔdacB ΔpbpG | 4, 7 | Denome et al. (1999) |
| CS313-1 | CS109 Δ(mrcA-yrfE-yrfF) ΔdacB ΔpbpG | 1a, 4, 7 | Denome et al. (1999) |
| CS315-1 | CS109 ΔdacB ΔdacA ΔpbpG | 4, 5, 7 | Denome et al. (1999) |
| CS477-1 | CS109 ΔdacB ΔdacA ΔpbpG ΔmepA | 4, 5, 7 | This work |
| CS604-2 | CS109 Δ[mrcA-yrfE-yrfF] ΔdacB ΔdacC ΔpbpG ΔampC ΔampH | 1a, 4, 6, 7, AmpC, AmpH | Denome et al. (1999) |
| CS617-1 | CS109 ΔmrcA ΔdacB ΔdacC ΔpbpG ΔampC ΔampH | 1a, 4, 6, 7, AmpC, AmpH | Denome et al. (1999) |
| CS703-1 | CS109 ΔmrcA ΔdacB ΔdacA ΔdacC ΔpbpG ΔampC ΔampH | 1a, 4, 5, 6, 7, AmpC, AmpH | Meberg et al. (2001) |
| CS804-1 | CS109 ΔmrcA ΔdacB ΔdacA ΔdacC ΔdacD ΔpbpG ΔampC ΔampH | 1a, 4, 5, 6, 7, DacD, AmpC, AmpH | Meberg et al. (2001) |
| KL743 | MG1655 zba-3000::Tn10 LAM− rph− | B. Backman (CGSC #6213) | |
| S17-1(λpir) | recA thi pro hsdR [res− mod+] | C. Sternberg (Kristensen et al., 1995) | |
| pJMSB8 | [RP4::2-Tc::Mu-Km::Tn7] λpir lysogen | ||
| TRI321 | 2443 ompT::km | This work, 2443 × (P1 AD202) | |
| TRI364 | AG70A-6 ompT::km | This work, AG70A-6 × (P1 AD202) | |
| TRI418 | AG70C-12 ompT::km | This work, AG70C-12 × (P1 AD202) |
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pBAD24 | PBAD expression vector (AmpR) | Guzman et al. (1995) |
| pBAD33 | PBAD expression vector (CmR) | Guzman et al. (1995) |
| pBAD24-gfp | pBAD-gfp (AmpR) | Charles et al. (2001) |
| pBAD24-icsA1-104::gfp | PBAD-icsA1-104::gfp (AmpR) | Charles et al. (2001) |
| pBAD24-icsA507-620::gfp | PBAD-icsA507-620::gfp (AmpR) | Charles et al. (2001) |
| pBAD24-icsAΔ507-729::gfp | PBAD-icsAΔ507-729::gfp (AmpR) | Charles et al. (2001) |
| pBAD33-icsA507-620::gfp | PBAD-icsA507-620::GFP (CmR) | This study |
| pGFPuv | pUC19 + gfpUV (AmpR) | BD Clontech Laboratories |
| pGFP-EpsM | pMMB66 + gfp-epsM (AmpR) | M. Sandkvist (Scott et al., 2001) |
| pJMSB8 | pUT derivative + parA (AmpR) | C. Sternberg (Kristensen et al., 1995) |
| pMBG270 | pBR322 with SphI–BamHI insert containing icsA (AmpR) | Magdalena and Goldberg (2002) |
Construction of PBP deletion mutants
PBP genes were deleted sequentially from the chromosome of E. coli 2443 by a series of P1 transductions (Miller, 1992). First, dacB, which encodes PBP 4, was deleted by transducing the ΔdacB::res-npt-res (KanR) allele from E. coli CS11-2K (Denome et al., 1999) into 2443, creating strain AG104-3K (Table 2). We then introduced a tetracycline resistance marker to the 10 min region of the chromosome by transducing the zba-3000::Tn10 insertion from KL743 to AG104-3K, creating AG104-3KT. This allowed us to select 2443-derived strains after subsequent mating steps. To prepare the strain to receive another PBP mutation, the res–npt–res cassette was excised by transient expression of the RP4 ParA resolvase (Kristensen et al., 1995; Denome et al., 1999). After conjugation, the mating mixture was plated onto LB plates with 12.5 μg ml−1 tetracycline to kill the donor and select for the 2443-derived mutant. Cells from which res–npt–res was eliminated were identified by reversion to kanamycin sensitivity. Additional PBP genes were deleted by repeating the above steps, yielding mutants lacking from one to seven PBPs, culminating in strains AG70A-6 (2443 mrcA dacA dacB dacC dacD pbpG ampC) and AG70C-12 (2443 mrcB dacA dacB dacC dacD pbpG ampC) (Table 2). PBP loss was confirmed by labelling cells with [125I]-penicillin-X and visualizing PBPs by autoradiography after SDS-PAGE (Henderson et al., 1997).
Labelling outer membrane proteins
Escherichia coli outer membrane proteins were labelled with Texas Red succinimidyl ester (100 μg ml−1 final; Molecular Probes), as described previously (de Pedro et al., 2003). Stained cells were incubated at 37°C in the absence of dye, and samples were withdrawn and fixed with 0.5% formalin for microscopy.
Expression of IcsA–GFP fusions, full-length IcsA and GFP–EpsM
Expression of IcsA–GFP proteins was performed essentially as described previously (Charles et al., 2001) except that cells were grown in LB medium, and genes were induced by 0.2% L-arabinose for either 10 min at 37°C or 60 min at 25°C. Native GFP was expressed from plasmid pGFPuv or pBAD-GFP. Because the outer membrane protease OmpT hydrolyses outer membrane IcsA, ompT was inactivated by moving the ompT::km allele from AD202 (Akiyama and Ito, 1990) into appropriate strains by P1L4 phage transduction. Surface-exposed IcsA was visualized by indirect immunofluorescence using antibody to IcsA (Goldberg et al., 1993). GFP–EpsM was expressed by growth and induction in M9 medium, as described previously (Scott et al., 2001), or by diluting cells into LB broth and adding IPTG to 5 μM.
Microscopy
Cell samples (5 μl) were placed on agarose-coated microscope slides and incubated at room temperature for 10 min to immobilize cells. Fluorescence and phase microscopy were performed using a 100× oil immersion objective on a Nikon OPTIPHOT-2 or TE300 microscope with Fryer, Nikon or Chroma Technology filters. Images were captured digitally using a black and white Photometrics Sensys charge-coupled device camera and IP LABORATORY (Scanalytics) or IMAGE-PRO PLUS software (Media Cybernetics). Colour figures were assembled by separately capturing signals with each of the appropriate filter sets and digitally pseudocolouring the images using Adobe PHOTOSHOP software.
Tabulating protein distributions
Distribution of GFP foci on fluorescence images was performed on 30 or more bacteria chosen randomly from phase images from each of three independent experiments for each strain. For PBP mutants, only cells with grossly visible deformities were included, and cell poles were defined as the two most distal convex protrusions. Individual cells not expressing or overexpressing IcsA–GFP fusion proteins (representing <5% of the population) were not tabulated: cells were not counted when the absolute level of fluorescence in the cytoplasm at mid-cell was greater than or less than two standard deviations (SDs) removed from the mean level at mid-cell for that particular strain (Charles et al., 2001).
Acknowledgments
This work was supported by grants GM61019 (to K.D.Y.) and AI35817 (to M.B.G.) from the National Institutes of Health, and a postdoctoral fellowship from the Research Council of Norway (to T.N.). We especially thank M. Sandkvist for the GFP–EpsM protein fusion construct.
References
- Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- Alley MR, Maddock JR, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Autret S, Errington J. A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis. Mol Microbiol. 2003;47:159–169. doi: 10.1046/j.1365-2958.2003.03264.x. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Boyd JM. Localization of the histidine kinase PilS to the poles of Pseudomonas aeruginosa and identification of a localization domain. Mol Microbiol. 2000;36:153–162. doi: 10.1046/j.1365-2958.2000.01836.x. [DOI] [PubMed] [Google Scholar]
- Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol Microbiol. 2003;50:45–60. doi: 10.1046/j.1365-2958.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Beckwith J. Assembly of cell division proteins at the E. coli cell center. Curr Opin Microbiol. 2002;5:553–557. doi: 10.1016/s1369-5274(02)00374-0. [DOI] [PubMed] [Google Scholar]
- Charles M, Perez M, Kobil JH, Goldberg MB. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc Natl Acad Sci USA. 2001;98:9871–9876. doi: 10.1073/pnas.171310498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 2000;19:2719–2727. doi: 10.1093/emboj/19.11.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Barzu O, Parsot C, Sansonetti PJ. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson TA, Young KD, Denome SA, Elf PK. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Janakiraman A, Goldberg MB. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc Natl Acad Sci USA. 2004;101:835–840. doi: 10.1073/pnas.0305747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Hellio R, Gounon P, Ohayon H, Cossart P. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J Cell Sci. 1993;105:699–710. doi: 10.1242/jcs.105.3.699. [DOI] [PubMed] [Google Scholar]
- Kristensen CS, Eberl L, Sanchez-Romero JM, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol. 2002;43:1523–1532. doi: 10.1046/j.1365-2958.2002.02829.x. [DOI] [PubMed] [Google Scholar]
- Liu D, Reeves PR. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Magdalena J, Goldberg MB. Quantification of Shigella IcsA required for bacterial actin polymerization. Cell Motil Cytoskel. 2002;51:187–196. doi: 10.1002/cm.10024. [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg BM, Sailer FC, Nelson DE, Young KD. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J Bacteriol. 2001;183:6148–6149. doi: 10.1128/JB.183.20.6148-6149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U, Mayer H. Genetic location of genes encoding enterobacterial common antigen. J Bacteriol. 1985;163:756–762. doi: 10.1128/jb.163.2.756-762.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics: a Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, Yoshikawa M, Sasakawa C. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol Microbiol. 1993;9:459–468. doi: 10.1111/j.1365-2958.1993.tb01707.x. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Young KD. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol. 2000;182:1714–1721. doi: 10.1128/jb.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Young KD. Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J Bacteriol. 2001;183:3055–3064. doi: 10.1128/JB.183.10.3055-3064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Young KD, Höltje JV, Schwarz H. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J Bacteriol. 2003;185:1147–1152. doi: 10.1128/JB.185.4.1147-1152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JR, Monack D, McCallum SJ, Vegas A, Pham E, Goldberg MB, Theriot JA. The making of a gradient: IcsA (VirG) polarity in Shigella flexneri. Mol Microbiol. 2001;41:861–872. doi: 10.1046/j.1365-2958.2001.02552.x. [DOI] [PubMed] [Google Scholar]
- Sandlin RC, Maurelli AT. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect Immun. 1999;67:350–356. doi: 10.1128/iai.67.1.350-356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandlin RC, Lampel KA, Keasler SP, Goldberg MB, Stolzer AL, Maurelli AT. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect Immun. 1995;63:229–237. doi: 10.1128/iai.63.1.229-237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott ME, Dossani ZY, Sandkvist M. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc Natl Acad Sci USA. 2001;98:13978–13983. doi: 10.1073/pnas.241411198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Nat Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. Bacterial shape. Mol Microbiol. 2003;49:571–580. doi: 10.1046/j.1365-2958.2003.03607.x. [DOI] [PubMed] [Google Scholar]


