Abstract
Studies on the temporal order of DNA replication are difficult due to the lack of sensitivity of methods available for replication kinetic analysis. To overcome problems associated with the current techniques, we propose a PCR-based assay to determine the replication time of any single-copy DNA sequence in complex genomes. Human cells labeled with 5-bromodeoxyuridine (BrdU) were flow sorted, according to their DNA content, at different times after synchronous release from the G1/S phase boundary. The selective removal of newly-replicated BrdU-substituted DNA was achieved by UV light irradiation followed by S1 nuclease treatment. The timing of replication of selected DNA sequences (housekeeping, tissue-specific, and non-coding loci) was determined by polymerase chain reaction (PCR) amplification using appropriate primers. DNA sequences localized in inactive replication units allowed amplification whereas those that have replicated will not be amplified by PCR. Using this sensitive and quantitative assay the replication kinetic analysis of a number of different DNA sequences can be performed from a single sorting experiment.
Full text
PDF

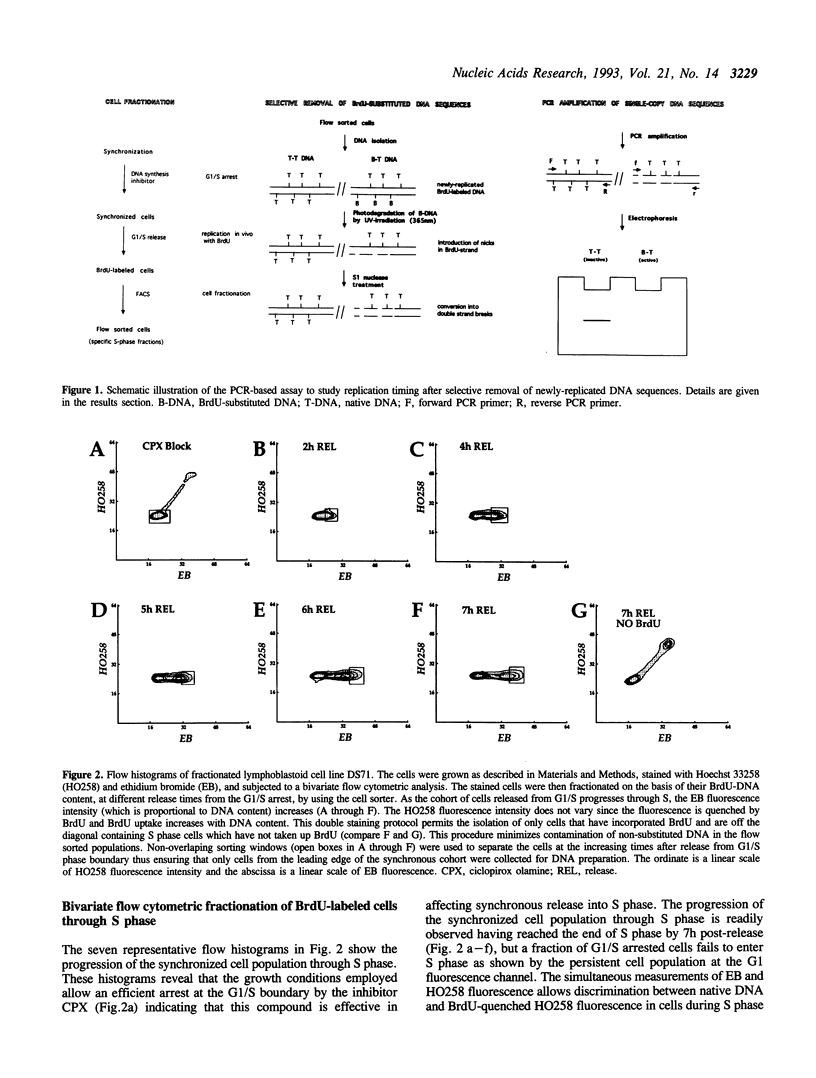



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braunstein J. D., Schulze D., DelGiudice T., Furst A., Schildkraut C. L. The temporal order of replication of murine immunoglobulin heavy chain constant region sequences corresponds to their linear order in the genome. Nucleic Acids Res. 1982 Nov 11;10(21):6887–6902. doi: 10.1093/nar/10.21.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A. D., Tantravahi U., Lalande M., Perle M. A., Latt S. A. High resolution analysis of the timing of replication of specific DNA sequences during S phase of mammalian cells. Nucleic Acids Res. 1983 Jul 25;11(14):4753–4774. doi: 10.1093/nar/11.14.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol Cell Biol. 1992 Sep;12(9):3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin R., Lemieux N., Richer C. L. Analysis of DNA replication during S-phase by means of dynamic chromosome banding at high resolution. Chromosoma. 1990 Aug;99(4):273–280. doi: 10.1007/BF01731703. [DOI] [PubMed] [Google Scholar]
- Edenberg H. J., Huberman J. A. Eukaryotic chromosome replication. Annu Rev Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- Epner E., Forrester W. C., Groudine M. Asynchronous DNA replication within the human beta-globin gene locus. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8081–8085. doi: 10.1073/pnas.85.21.8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M. A., Holmquist G. P., Gray M. C., Caston L. A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984 May 18;224(4650):686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978 Oct;15(2):317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- Hoffman B. D., Hanauske-Abel H. M., Flint A., Lalande M. A new class of reversible cell cycle inhibitors. Cytometry. 1991;12(1):26–32. doi: 10.1002/cyto.990120105. [DOI] [PubMed] [Google Scholar]
- Holmquist G. P. Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet. 1987 Feb;40(2):151–173. [PMC free article] [PubMed] [Google Scholar]
- Kubbies M., Rabinovitch P. S. Flow cytometric analysis of factors which influence the BrdUrd-Hoechst quenching effect in cultivated human fibroblasts and lymphocytes. Cytometry. 1983 Jan;3(4):276–281. doi: 10.1002/cyto.990030408. [DOI] [PubMed] [Google Scholar]
- Lalande M., Hanauske-Abel H. M. A new compound which reversibly arrests T lymphocyte cell cycle near the G1/S boundary. Exp Cell Res. 1990 May;188(1):117–121. doi: 10.1016/0014-4827(90)90285-i. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Harland R. M. Replication origins in the eucaryotic chromosome. Cell. 1981 May;24(2):283–284. doi: 10.1016/0092-8674(81)90316-0. [DOI] [PubMed] [Google Scholar]
- Latt S. A. Microfluorometric detection of deoxyribonucleic acid replication in human metaphase chromosomes. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3395–3399. doi: 10.1073/pnas.70.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray V., Martin R. F. The degree of ultraviolet light damage to DNA containing iododeoxyuridine or bromodeoxyuridine is dependent on the DNA sequence. Nucleic Acids Res. 1989 Apr 11;17(7):2675–2691. doi: 10.1093/nar/17.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawotka K. A., Huberman J. A. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol Cell Biol. 1988 Apr;8(4):1408–1413. doi: 10.1128/mcb.8.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivin C. J., Fangman W. L. Replication fork rate and origin activation during the S phase of Saccharomyces cerevisiae. J Cell Biol. 1980 Apr;85(1):108–115. doi: 10.1083/jcb.85.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Migeon B. R. Asynchronous replication of homologous loci on human active and inactive X chromosomes. Proc Natl Acad Sci U S A. 1990 May;87(10):3685–3689. doi: 10.1073/pnas.87.10.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig S., Okumura K., Ward D. C., Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992 Mar;11(3):1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spack E. G., Lewis E. D., Paradowski B., Schimke R. T., Jones P. P. Temporal order of DNA replication in the H-2 major histocompatibility complex of the mouse. Mol Cell Biol. 1992 Nov;12(11):5174–5188. doi: 10.1128/mcb.12.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetten G., Latt S. A., Davidson R. L. 33258 Hoechst enhancement of the photosensitivity of bromodeoxyuridine-substituted cells. Somatic Cell Genet. 1976 May;2(3):285–290. doi: 10.1007/BF01538967. [DOI] [PubMed] [Google Scholar]
- Stubblefield E. Analysis of the replication pattern of Chinese hamster chromosomes using 5-bromodeoxyuridine suppression of 33258 Hoechst fluorescence. Chromosoma. 1975 Dec 10;53(3):209–221. doi: 10.1007/BF00329172. [DOI] [PubMed] [Google Scholar]
- Ten Hagen K. G., Gilbert D. M., Willard H. F., Cohen S. N. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell Biol. 1990 Dec;10(12):6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P. A., Hanauske-Abel H. H., Flint A., Lalande M. Mimosine reversibly arrests cell cycle progression at the G1-S phase border. Cytometry. 1991;12(3):242–246. doi: 10.1002/cyto.990120306. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Gyapay G., Dib C., Vignal A., Morissette J., Millasseau P., Vaysseix G., Lathrop M. A second-generation linkage map of the human genome. Nature. 1992 Oct 29;359(6398):794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- Willard H. F., Latt S. A. Analysis of deoxyribonucleic acid replication in human X chromosomes by fluorescence microscopy. Am J Hum Genet. 1976 May;28(3):213–227. [PMC free article] [PubMed] [Google Scholar]
- Yunis J. J. Mid-prophase human chromosomes. The attainment of 2000 bands. Hum Genet. 1981;56(3):293–298. doi: 10.1007/BF00274682. [DOI] [PubMed] [Google Scholar]