Abstract
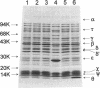
The gene encoding the theta subunit of DNA polymerase III holoenzyme, designated holE, was isolated using a strategy in which peptide sequence was used to derive a DNA hybridization probe. Sequencing of the gene, which maps to 41.43 centisomes of the chromosome, revealed a 76-codon open reading frame predicted to produce a protein of 8,846 Da. When placed in a tac promoter expression vector, the open reading frame directed expression of a protein, that comigrated with authentic theta subunit from purified holoenzyme, to 6% of total soluble protein.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebersold R. H., Leavitt J., Saavedra R. A., Hood L. E., Kent S. B. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci U S A. 1987 Oct;84(20):6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Carter J. R., Franden M. A., Aebersold R., McHenry C. S. Molecular cloning, sequencing, and overexpression of the structural gene encoding the delta subunit of Escherichia coli DNA polymerase III holoenzyme. J Bacteriol. 1992 Nov;174(21):7013–7025. doi: 10.1128/jb.174.21.7013-7025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco R., Bhatnagar S. K., Brown A., Bessman M. J. The interaction of DNA polymerase III and the product of the Escherichia coli mutator gene, mutD. J Biol Chem. 1984 May 10;259(9):5567–5573. [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The adjacent dnaZ and dnaX genes of Escherichia coli are contained within one continuous open reading frame. Nucleic Acids Res. 1986 Oct 24;14(20):8091–8101. doi: 10.1093/nar/14.20.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hawker J. R., Jr, McHenry C. S. Monoclonal antibodies specific for the tau subunit of the DNA polymerase III holoenzyme of Escherichia coli. Use to demonstrate that tau is the product of the dnaZX gene and that both it and gamma, the dnaZ gene product, are integral components of the same enzyme assembly. J Biol Chem. 1987 Sep 15;262(26):12722–12727. [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Johanson K. O., McHenry C. S. Adenosine 5'-O-(3-thiotriphosphate) can support the formation of an initiation complex between the DNA polymerase III holoenzyme and primed DNA. J Biol Chem. 1984 Apr 10;259(7):4589–4595. [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken R. S., Kornberg A. The primosomal protein n' of Escherichia coli is a DNA helicase. J Biol Chem. 1988 Apr 25;263(12):5512–5518. [PubMed] [Google Scholar]
- Lee E. H., Masai H., Allen G. C., Jr, Kornberg A. The priA gene encoding the primosomal replicative n' protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4620–4624. doi: 10.1073/pnas.87.12.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Marians K. J. Escherichia coli replication factor Y, a component of the primosome, can act as a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8345–8349. doi: 10.1073/pnas.84.23.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Horiuchi T., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. I. Amplification of the dnaE gene product and polymerase activity of the alpha subunit. J Biol Chem. 1985 Oct 25;260(24):12982–12986. [PubMed] [Google Scholar]
- Maki H., Horiuchi T., Sekiguchi M. Structure and expression of the dnaQ mutator and the RNase H genes of Escherichia coli: overlap of the promoter regions. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7137–7141. doi: 10.1073/pnas.80.23.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4389–4392. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985 Oct 25;260(24):12987–12992. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J Biol Chem. 1988 May 15;263(14):6555–6560. [PubMed] [Google Scholar]
- McHenry C. S., Crow W. DNA polymerase III of Escherichia coli. Purification and identification of subunits. J Biol Chem. 1979 Mar 10;254(5):1748–1753. [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme. Components, structure, and mechanism of a true replicative complex. J Biol Chem. 1991 Oct 15;266(29):19127–19130. [PubMed] [Google Scholar]
- McHenry C., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem. 1977 Sep 25;252(18):6478–6484. [PubMed] [Google Scholar]
- Mok M., Marians K. J. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J Biol Chem. 1987 Dec 5;262(34):16644–16654. [PubMed] [Google Scholar]
- Nurse P., DiGate R. J., Zavitz K. H., Marians K. J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M. E. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1987 Dec 5;262(34):16558–16565. [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Ostell J., Benson D. A. Alignment of Escherichia coli K12 DNA sequences to a genomic restriction map. Nucleic Acids Res. 1990 Jan 25;18(2):313–321. doi: 10.1093/nar/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R., Tam S., Burgers P. M., Lu C., Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studwell P. S., O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990 Jan 15;265(2):1171–1178. [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Tomasiewicz H. G., McHenry C. S. Sequence analysis of the Escherichia coli dnaE gene. J Bacteriol. 1987 Dec;169(12):5735–5744. doi: 10.1128/jb.169.12.5735-5744.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn B. K., Kushner S. R. Construction and analysis of deletions in the structural gene (uvrD) for DNA helicase II of Escherichia coli. J Bacteriol. 1991 Apr;173(8):2569–2575. doi: 10.1128/jb.173.8.2569-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. M., McHenry C. S. Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol. 1982 Oct;152(1):351–356. doi: 10.1128/jb.152.1.351-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. Mechanism of DNA elongation catalyzed by Escherichia coli DNA polymerase III, dnaZ protein, and DNA elongation factors I and III. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3511–3515. doi: 10.1073/pnas.73.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., Kornberg A. DNA polymerase 3 star requires ATP to start synthesis on a primed DNA. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3679–3683. doi: 10.1073/pnas.70.12.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Zechner E. L., Hughes A. J., Jr, Franden M. A., McHenry C. S., Marians K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. IV. Reconstitution of an asymmetric, dimeric DNA polymerase III holoenzyme. J Biol Chem. 1992 Feb 25;267(6):4064–4073. [PubMed] [Google Scholar]
- Wu C. A., Zechner E. L., Marians K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992 Feb 25;267(6):4030–4044. [PubMed] [Google Scholar]
- Wu C. A., Zechner E. L., Reems J. A., McHenry C. S., Marians K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. V. Primase action regulates the cycle of Okazaki fragment synthesis. J Biol Chem. 1992 Feb 25;267(6):4074–4083. [PubMed] [Google Scholar]
- Zechner E. L., Wu C. A., Marians K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase-primase interaction governs primer size. J Biol Chem. 1992 Feb 25;267(6):4054–4063. [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Identification of two Escherichia coli factor Y effector sites near the origins of replication of the plasmids (ColE1 and pBR322. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6521–6525. doi: 10.1073/pnas.77.11.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]