Abstract
Maspin, a 42-kDa non classical serine protease inhibitor (serpin) is expressed by epithelial cells of various tissues including the cornea. The protein localizes to the nucleus and cytosol, and is present in the extracellular space. While extracellular maspin regulates corneal stromal fibroblast adhesion and inhibits angiogenesis during wound healing in the cornea, the molecular mechanism of its extracellular functions is unclear. We hypothesized that identifying post-translational modifications of maspin, such as phosphorylation, may help decipher its mode of action. The focus of this study was on the identification of phosphorylation sites on extracellular maspin, since the extracellular form of the molecule is implicated in several functions. Multi-stage fragmentation mass spectrometry was used to identify sites of phosphorylation on extracellular corneal epithelial cell maspin. A total of eight serine and threonine phosphorylation sites (Thr50, Ser97, Thr118, Thr157, Ser240, Ser298, Thr310, Ser316) were identified on the extracellular forms of the molecule. Phosphorylation of tyrosine residues on extracellular maspin was not detected on extracellular maspin from corneal epithelial cell, in contrast to breast epithelial cells. This study provides the basis for further investigation into the functional role of phosphorylation of corneal epithelial maspin.
Keywords: maspin, cornea, phosphorylation, liquid chromatography-tandem mass spectrometry
1 Introduction
Maspin (Mammary Serine Protease Inhibitor), a 42-kDa non-classical serine protease inhibitor (serpin) and tumor suppressor was identified in mammary epithelial cells by subtractive hybridization using cDNAs from normal versus malignant mammary tissue [1]. Expression of maspin is primarily localized to epithelial cells in a wide variety of tissues including the breast, skin, prostate, small intestine and colon among others [2]. Ngamkitidechakul et al. reported that maspin was also expressed in all three major layers of the cornea, namely the epithelium, stroma and the endothelium [3].
While maspin resembles serpins in sequence and structure, and possesses a reactive site loop (RSL), it is non-inhibitory [4]. Although no target serine or cysteine proteases have been identified for maspin, it has been shown that exogenously supplied maspin regulates cell adhesion, migration, invasion and angiogenesis in a variety of cell types [3, 5-7]. Specifically, the RSL of maspin is sufficient to increase adhesion of corneal stromal fibroblasts to extracellular matrix (ECM) molecules and inhibition of invasion and migration of breast carcinoma cells [8, 9]. The anti-angiogenic effect of extracellular maspin appears to be RSL-independent. A maspin RSL deletion mutant was able to inhibit tube formation in cultured endothelial cells in response to VEGF in vitro and block neovascularization in vivo in the rat cornea pocket model [10]. These studies support the hypothesis that regions of extracellular maspin outside of the RSL may contribute to some of the observed functions of maspin.
While maspin localizes to the cytosol and nucleus intracellularly, it is also found extracellularly in conditioned medium from corneal epithelial cells. In the cornea, in vivo, extracellular maspin may function in a paracrine fashion by inhibiting the growth and migration of vascular and conjunctival endothelial cells, which do not synthesize maspin but respond to it. Previous work by Hendrix and co-workers, identified the presence of tyrosine phosphorylation on maspin obtained from immortalized and normal breast epithelial cells [11]. We focused our efforts on the identification of phosphorylation sites on extracellular maspin released by the corneal epithelium since it is hypothesized to be the ‘effector’ in several functions. To be able to perform exhaustive biochemical and functional analyses of which phosphorylation site(s) on the protein are important for its anti-angiogenic effect, the sites of phosphorylation on maspin need to be identified. Using multi-stage fragmentation mass spectrometry eight phosphorylation sites were identified on extracellular maspin. This information will enable identification of the role of specific modified forms of extracellular maspin in its observed functions.
2 Methods
2.1 Cell Culture
The immortalized human corneal epithelial cell line (HCEC; ATCC, Manassas, VA, USA) was maintained in culture at 37°C in 5% CO2 in Defined Keratinocyte Serum Free Medium (D-KSFM; Invitrogen, Carlsbad, CA, USA) with supplement, 0.1% Mito+ serum extender and 10 μg/ml ciprofloxacin. Protease and phosphatase inhibitors were not added to the culture medium to maintain the medium in as ‘natural’ a condition as possible.
2.2 Isoelectric Focusing and 2D Electrophoresis
For extracellular maspin, HCEC conditioned medium was collected, cleared of debris and concentrated using centrifugal filters (30,000 MW cut-off; Millipore, Billerica, MA, USA). The concentrated medium or cell lysates were de-salted using the ReadyPrep 2-D Cleanup Kit (BioRad, Hercules, CA, USA), and the samples were applied to IPG strips (pH 4-7; BioRad). The strips were rehydrated overnight at 50 V. Subsequently, isoelectric focusing was performed using linear voltage ramping for 4000 Vh. Following focusing the proteins were reduced and the cysteines acetylated by equilibration of the strips in Rehydration Buffers I (+DTT) and II (+iodoacetamide) (BioRad) as per the manufacturer's instructions prior to electrophoresis in the second dimension on 10% denaturing polyacrylamide gels.
In silico 2D analysis of maspin was performed using ProMoST, a protein modification analysis application developed by the Biotechnology and Bioengineering Center at the Medical College of Wisconsin (http://proteomics.mcw.edu) [12, 13]. Upon specifying the type of modification and the number of residues expected to be modified, the program generates a profile of the protein on a 2D-gel.
2.3 Western Blotting
For collection of total cell lysate, HCEC cultures were lysed in modified RIPA buffer [14] and cleared by centrifugation at 15,000×g for 10 minutes. For analysis of intracellular maspin and for detection of accumulation of extracellular maspin, total cell lysate (20 μg) and pre-cleared, concentrated medium (20×) collected over a 96 hour time course were separated on a 10% denaturing polyacrylamide gel under reducing conditions. Proteins were transferred to nitrocellulose or PVDF membranes (BioRad) and maspin was detected using a mouse monoclonal antibody (1: 10,000; BD Pharmingen, San Jose, CA, USA) followed by HRP labeled goat anti-mouse IgG (1:7500; BioRad) and a luminol based chemiluminescent system (ECL, GE Healthcare, Piscataway, NJ). Control blots were probed only with the secondary antibody to detect non-specific staining.
2.4 Immunoprecipitation (IP) of maspin
For immunoprecipitation of extracellular maspin, conditioned medium from corneal epithelial cells (HCEC) was collected and concentrated 60× using a centrifugal filter (10-kDa MW cut off; Millipore). Cell lysates and concentrated conditioned medium were immunoprecipitated using a monoclonal anti-maspin antibody (BD Pharmingen), a phosphoserine/threonine Akt substrate rabbit polyclonal antibody (Cell Signaling Technology, Danvers, MA), a phosphothreonine rabbit polyclonal antibody (Cell Signaling Technology) or 4G10 platinum phosphotyrosine murine monoclonal antibody (Invitrogen). The complexes were pulled down with Protein A/G beads (Pierce, Rockford, IL, USA). The beads were washed 3× with buffer containing: 10 mM Tris, pH 7.4, 130 mM NaCl, 0.05% Triton X-100, 0.1% BSA, 10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, 1 mM EDTA, followed by one wash each in Tris buffer with NaCl (10 mM Tris, pH 8.0, 140 mM NaCl) and without NaCl (50 mM Tris, pH 8.0) and the complexes eluted with Laemmli buffer. The samples were separated by SDS-PAGE, blotted to membranes and probed with the above antibodies.
2.5 Trypsin Digestion
The proteins in the concentrated HCEC medium were immunoprecipitated using a monoclonal antibody to maspin as described above and separated by SDS-PAGE on a 10% gel and the proteins were visualized by silver staining. Selected bands were excised and processed for trypsin digestion. Proteins were subjected to in-gel digestion using 0.02 μg/μl trypsin (Promega, Madison, WI, USA) in 100 mM ammonium bicarbonate at 37°C overnight. The resulting peptides were vacuum dried to near dryness for mass spectrometry analysis.
2.6 Instrumentation
LC - mass spectrometry experiments were performed on an LTQ mass spectrometer (Thermo Electron, Waltham, MA, USA) coupled with a Surveyor Plus HPLC system (Thermo Electron) equipped with an autosampler. The instrument was interfaced with a capillary column (100 × 0.1 mm), packed in-house with 5 μm C18 RP particles (Luna C18, Phenomenex, Torrance, CA, USA). The fused silica capillaries (Polymicro Technologies, Phoenix, AZ, USA) for the columns were pulled by a micropipette puller P-2000 (Sutter Instrument Company, CA, USA) and packed with C18 resin using a bomb-loader.
2.7 Nano-HPLC-ESI mass spectrometry
The protein digests were analyzed using an ion trap LTQ mass spectrometer interfaced with a nano-HPLC system through an electrospray ionization (ESI) source. The samples were loaded through an autosampler onto a C18 capillary column. The solvents A and B used for chromatographic separation of peptides were 5% acetonitrile in 0.1% formic acid and 95% acetonitrile in 0.1% formic acid, respectively. The peptides injected onto the microcapillary column were resolved at the rate of 200 nL/min, using the following gradient conditions: 0-30 min 0-5 % B, 30-180 min 5-35% B, 180-240 min 35-65% B, 240-250 min 65-100% B. 100% B was held for 10 min, then switched to 100% A and held for another 40 min.
The ions eluted from the column were electrosprayed at a voltage of 1.8 kV. The capillary voltage was 45 V and the temperature was kept at 200°C. No auxiliary or sheath gas was used. Helium was used in the trap, which also served as a collision gas for fragmentation of ions.
2.8 Multi-stage fragmentation (MSF) analysis
In MSF analysis, the instrument was cycled through the acquisition of a full-scan mass spectrum (m/z 300-2000) followed by MS/MS of the most abundant ion from the full-scan mass spectrum. Sequential fragmentation of the most abundant ion from the previous scan was carried through to the fourth stage fragmentation. Dynamic exclusion was enabled for 30 s. The chromatographic and mass spectral functions were controlled using the Xcalibur data system (ThermoFinnigan, Palo Alto, CA, USA).
2.9 Data Analysis
In order to search the MSn spectra using conventional search engines such as MASCOT22, the individual MSn sub-spectra were combined so as to mimic the situation in which all of the fragmentation had occurred in a single stage. To do this, the individual spectra were extracted from the raw data file as a Mascot Generic Format (.mgf) file using the extract ms program provided by Thermo Fisher and a modified version of the .mgf creation script provided by Matrix Science (Boston, MA, USA). The .mgf file was then processed using the combine_mgf program, available from the MCW Proteomics Center (http://proteomics.mcw.edu). This program collects the spectra into groups of fragmentation spectra that arise from initial MS/MS spectra. If more than one spectrum is found in a group, ie. MSn fragmentation was performed, the spectra are combined by binning the normalize intensity values from all spectra in the group into 0.01 Da mass bins. The composite spectra, and optionally the individual subspectra, are output into a new .mgf file. The resulting .mgf file produced by combine_mgf was searched using MASCOT against the Uniprot human database v49.1. The search was limited to tryptic peptides, and allowed five missed cleavages. Methionine oxidation, carbamidomethylation of cysteine, and phosphorylation on serine, threonine and tyrosine were searched for as variable modifications. Mass tolerance was set to 2 Da for parent ion mass and 0.6 Da for fragment ion mass. Peptides identified with MASCOT score of 50 or above were considered potential positive identifications. Finally, the peptides listed were manually verified for correct identification by comparing the experimental spectra with the theoretical b and y ion spectra.
2.10 Phosphatase Treatment
Concentrated conditioned HCEC medium was dephosphorylated by treatment with either 50 units of calf intestinal alkaline phosphatase (CIP) (10,000 U/ml; NEB, Ipswich, MA, USA) in 1× reaction buffer (Buffer 3; NEB, Ipswich, MA, USA) or with 100 units of lambda phosphatase (Buffer for PMP + 1mM MnCl2; NEB) at 37°C for 1 hour. The samples were either separated on a 10% Tris-Urea (6M) gel or by two dimensional electrophoresis as described above. Following electrophoresis, the proteins were transferred to PVDF membranes (BioRad) and immunoblotted with mouse monoclonal anti- maspin (1:10,000; BD Pharmingen) and a goat anti-mouse IgG secondary antibody (1:7500; BioRad). Control blots were probed with secondary antibody only. The intensity of the maspin bands was quantified by densitometry and the data was plotted using Microsoft Excel.
3 Results
3.1 Two-dimensional electrophoresis reveals multiple species of extracellular maspin from cultures of corneal epithelial cells
Conditioned medium collected at various times from immortalized human corneal epithelial cells (HCEC) was concentrated and analyzed by western blotting. A band was detected at the expected size of approximately 42 kDa that increased in density over time in culture. Maspin was also detected in cell lysates at the same molecular mass (Figure 1A). Proteins were de-salted and precipitated from concentrated conditioned medium from HCEC cultures for analysis by iso-electric focusing between pH 4 and 7 followed by SDS-PAGE electrophoresis in the second dimension and western blotting for maspin. We detected a train of spots at approximately 42kDa, the expected molecular mass for maspin (Figure 1B). The calculated pI for unmodified maspin is 5.77. The spots detected for maspin indicate a population of maspin molecules at the predicted pI with spots tending towards lower pH values, which suggest that these forms of maspin are modified by the addition of a negatively charged molecule. Similar results were obtained for maspin in conditioned medium from primary corneal epithelial cells (Data not shown). Spots were also detected tending to higher pH values indicating that a small amount of maspin might have undergone modifications that add positive charges, but these modifications were not pursued in this study. Figure 1C (open oval) shows the spot predicted to be at the PI of maspin using the phosphorylation prediction program ProMoST. Eight more negatively charged spots approximate the positions predicted for maspin with up to eight serines and threonines modified by phosphorylation.
Figure 1.
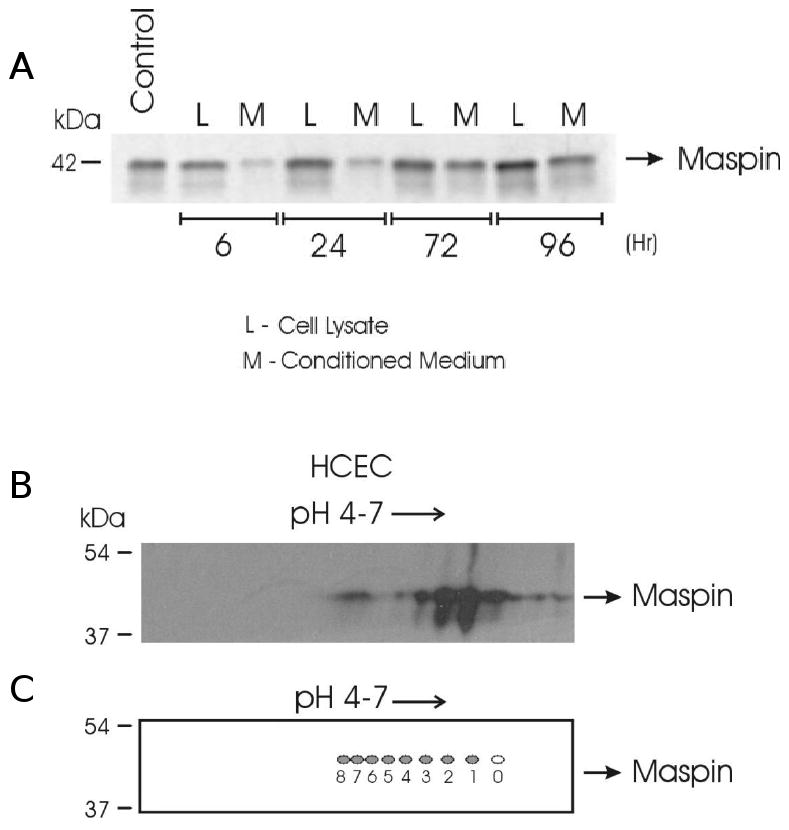
One- and two-dimensional electrophoresis of intra- and extracellular maspin. (A) Maspin-positive (Control) and experimental lysates (L) and conditioned medium (M) from cultures of human corneal epithelial cells (HCEC) were separated by SDS-PAGE under reducing conditions, and (B) conditioned medium from HCEC was focused on a pH 4-7 IPG strip. Maspin was detected by western blotting using a monoclonal maspin antibody and an HRP tagged anti-mouse IgG secondary antibody. (C) Two-dimensional representation of maspin after in silico analysis of its amino acid sequence using ProMoST, a protein modification analysis application.
3.2 Identification of phosphorylation sites on extracellular maspin by multi-stage fragmentation tandem mass spectrometry
Maspin was immunoprecipitated from epithelial cell conditioned medium and separated by SDS-PAGE (Figure 2). The band at ∼42 kDa (maspin) was cut and subjected to in-gel tryptic digestion. The resulting peptides were analyzed by multi-stage fragmentation analysis, which uses a four-stage fragmentation protocol set to fragment the most abundant ion from the previous scan. Based on the scan counts, we found about 1/3 of the maspin peptides to be phosphorylated compared to their non-phosphorylated forms. The data (Figure 3A) indicated that extracellular maspin is potentially phosphorylated on eight residues, four serines and four threonines. Although the identified phosphorylated serine and threonine residues appear to be spread out in the linear amino acid sequence of maspin (T50, S97, T118, T157, S240, S298, S310, S316), all but of one of these residues are located in one region of the molecule indicated on the solved molecular structure of maspin (Figure 3B). It is not expected that all eight sites are phosphorylated at all times on all maspin molecules. It is unclear at this point as to how many populations of maspin exist with different combinations of sites phosphorylated. The sequential mass spectra for the peptide GDpTANEIGQVLHFENVK are shown in Figure 3C. Data combined from three separate analyses of extracellular maspin from HCEC cells provided 91% coverage of tryptic peptides in the maspin molecule. Mass spectra for all phosphorylated peptides are given in the supplemental data. Peptides with all five tyrosine residues were identified but no tyrosine residues were found to be phosphorylated.
Figure 2.
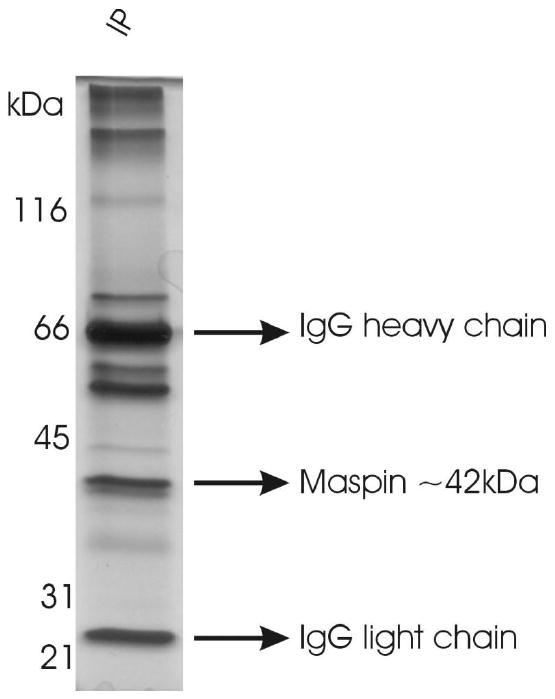
IP of extracellular maspin. Conditioned medium from HCEC cultures was concentrated, maspin was immunoprecipitated using a monoclonal maspin antibody, complexes separated by SDS-PAGE under reducing conditions and visualized by silver staining.
Figure 3.
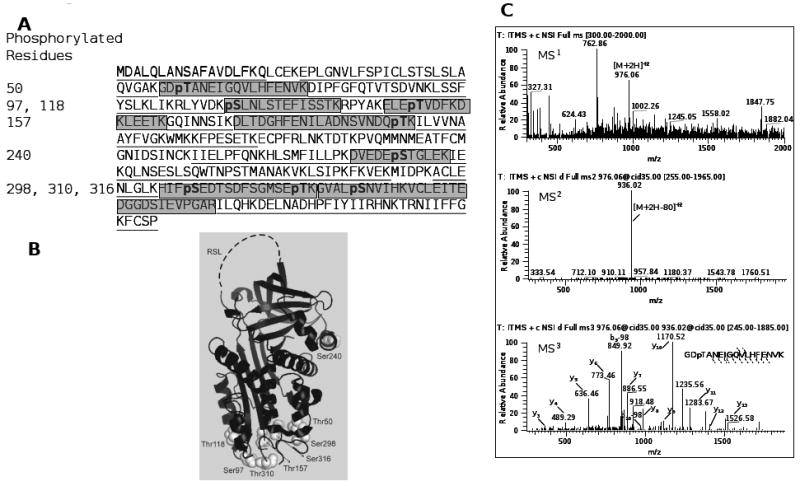
Maspin is phosphorylated on multiple serine and threonine sites. (A) Location of phosphorylation sites on the linear amino acid sequence of maspin. Underlined and shaded sequences indicate coverage of the protein in three separate MS/MS/MS analyses. Shaded areas indicate peptides detected with phosphorylated residues (pS/pT). (B) Projection of the eight phosphorylation sites on the crystal structure of maspin (PDB: 1xu8). (C) Representative MS3 spectra of the maspin phosphopeptide GDpTANEIGQVLHFENVK, residues 48-65. Spectra for additional phosphorylated peptides are given in Supplementary Materials.
3.3 Dephosphorylation of extracellular maspin
Because the phosphorylated forms of maspin are not significantly larger than the unphosphorylated forms, it is difficult to determine dephosphorylation of maspin based on a difference in molecular mass on a denaturing gel. To study the dephosphorylation of maspin, concentrated HCEC conditioned medium was dephosphorylated by treatment with lambda phosphatase and separated by two dimensional electrophoresis and analyzed by western blotting. Lambda phosphatase treatment decreased the amount of acidic maspin forms with an increase in the pI 5.7 form (Figure 4A). Additional samples of HCEC lysate samples were dephosphorylated with calf intestinal alkaline phosphatase (CIP), separated on a Tris-urea gel and analyzed by western blotting. It was expected that more negatively charged (phosphorylated) molecules of maspin would migrate faster towards the anode. Two bands with different rates of migration were detected for maspin (Figure 4B; labeled 1 and 2 for control, 3 and 4 for CIP treated). Bands 1 and 3 represent the more negatively charged species of maspin, while bands 2 and 4 represent the less negatively charged forms of maspin. CIP treatment of the conditioned medium and densitometry analysis indicated a loss of band intensity in the more negative forms (band 3 vs band 1) and an increase in the less negative forms (band 4 vs band 2) (Figure 4B). These differences were quantified and found to be statistically significant as shown in Figure 4D and E. The residual band detected for maspin could either be due to partial dephosphorylation or owing to modifications of maspin other than phosphorylation that are not removed by CIP. A reason for incomplete dephosphorylation under non-denaturing conditions, as indicated by various views of the space-fill model of the maspin molecule (Figures 4F-I), is that many of these residues are not surface accessible.
Figure 4.
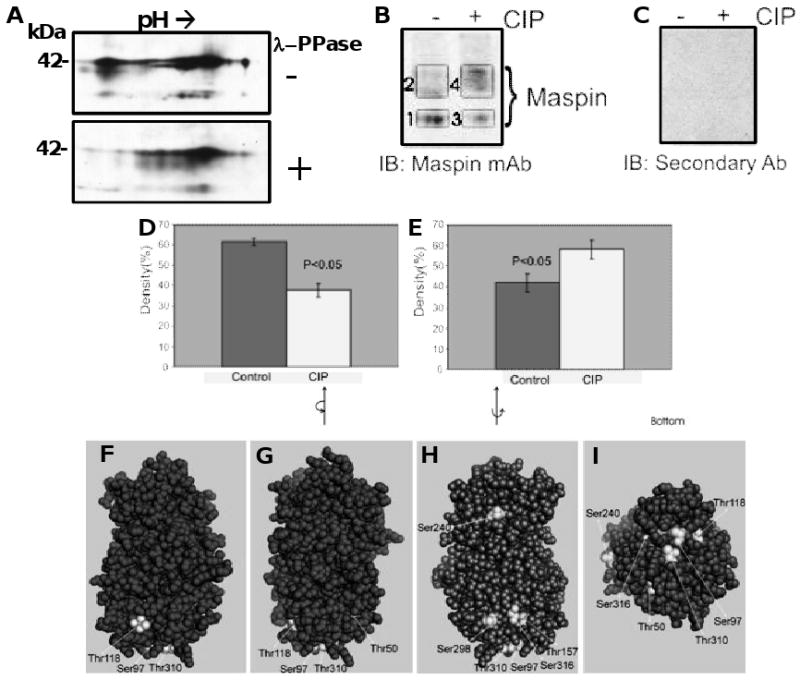
Partial dephosphorylation of maspin and surface accessibility of phosphorylation sites. Proteins in concentrated conditioned medium from HCEC were dephosphorylated using λ-phosphatase and the proteins separated by 2D electrophoresis, western blotted and probed for maspin (A). Proteins in concentrated conditioned medium from HCEC were dephosphorylated using calf intestinal phosphatase (CIP), the proteins separated on a 10% Tris-urea gel under non-reducing condition and maspin detected by western blotting using a monoclonal maspin antibody and an HRP tagged anti-mouse IgG secondary antibody (B). Non-specific staining detected using secondary antibody only (C). Quantification of the intensity of maspin bands with and without CIP treatment from A plus three additional blots (D and E). D: Control = Band 1, CIP = Band 3, E: Control = Band 2, CIP = Band 4. Spacefill model of maspin at different orientations (4F-I) highlighting the lack of surface accessibility of certain phosphorylation sites.
3.4 Intracellular maspin, unlike extracellular maspin, contains phosphotyrosine residues
HCEC lysates were separated by iso-electric focusing between pH4 and 7 followed by SDS-PAGE electrophoresis in the second dimension and western blotting for maspin. A train of at least eight spots were observed at about 42 kDa with a similar pattern as those observed for extracellular maspin (Figure 1B, C vs Figure 5A). The lysates were immunoprecipitated with antibodies to phosphotyrosine, phosphoserine/threonine and phosphotyrosine and the precipitates used for western blotting using a mouse monoclonal to maspin. Maspin was present in the lysate immunoprecipitates generated by the three antibodies (Figure 5B). In contrast, maspin was not detected in the immunoprecipitate of the concentrated conditioned medium with the phosphotyrosine antibody (Figure 5C). These results suggest that HCEC intracellular maspin exits as a mixture of post-translational modified products with at least some representing phosphorylated products. These maspin forms present in the lysate are phosphorylated on tyrosine residues but extracellular maspin does not contain phosphate groups on tyrosine residues.
Figure 5.
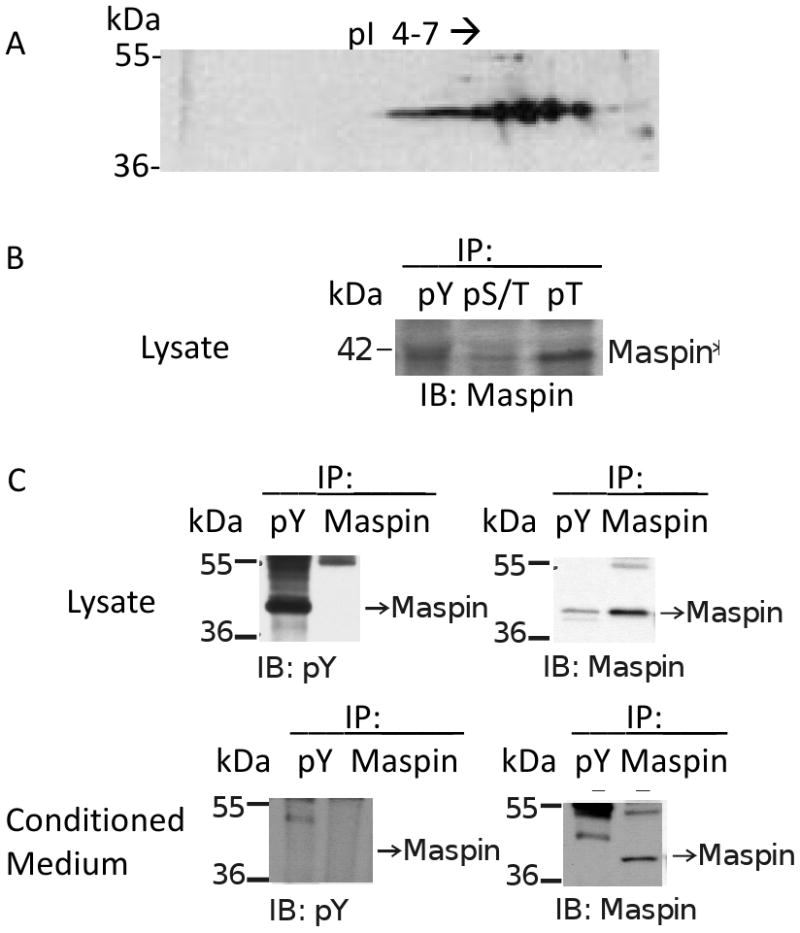
Cellular HCEC produced maspin is phosphorylated. (A) Proteins in lysates from HCEC were separated by 2D electrophoresis, western blotted and probed for maspin. (B) HCEC lysates were immunoprecipitated using mouse monoclonal antibodies to phospho-tyrosine (pY) and rabbit polyclonal antibodies to phospho-serine/threonine (pS/T) and phospho-threonine (pT). The precipitates were separated by SDS-PAGE, western blotted and probed with a mouse monoclonal antibody to maspin. (C) Concentrated HCEC lysates and conditioned medium were immunoprecipitated using mouse monoclonal antibodies to phospho-tyrosine or maspin. The precipitates were separated by SDS-PAGE, western blotted and probed for phospho-tyrosine and maspin.
4 Discussion
The current study has identified eight potential sites of phosphorylation on extracellular corneal epithelial cell maspin. Maspin, found extracellularly in conditioned medium from corneal epithelial cells, is phosphorylated on a combination of serine and threonine residues. These sites were identified by multi-stage fragmentation mass spectrometry from three biological replicate experiments. We propose that maspin exists in a mixture of differentially phosphorylated populations and that not all eight sites on maspin are phosphorylated at all times. The placement and location of these sites are note-worthy. When the sites of phosphorylation are placed on the maspin molecule (Figure 3B) seven out of the eight potential sites are located at the “bottom” of the molecule in the region opposite from the RSL. Space fill representations of the molecule (Figure 4E-H) indicate that a majority of these sites are buried in the molecule raising the question of how these sites may come in contact with a kinase. There is the possibility that some of these sites may be phosphorylated during translation and folding of the protein. We also speculate that the initial phosphorylation of one or more sites may lead to a conformational change in the protein causing a particular combination of sites to be phosphorylated.
Our data suggest that extracellular maspin from corneal epithelial cells is phosphorylated on serine and threonine residues while maspin from breast epithelial cells is reported to be phosphorylated on tyrosine residues [11]. In our study, all five tyrosine residues were identified on extracellular maspin, but no tyrosine residues were found to be phosphorylated. Neither was maspin immunoprecipitated by phosphotyrosine antibodies. This same antibody immunoprecipitated intracellular maspin. However, we do not rule out the possibility that under our experimental conditions a tyrosine phosphorylated form of the protein is not detected in the population of extracellular maspin molecules. The use of TiO2/ ZrO2 [15, 16] enrichment did not improve the detection of tyrosine phosphorylated residues. We detected multiple forms of intracellular corneal epithelial maspin with different iso-electric points and tyrosine phosphorylation, as well as, serine/ threonine phosphorylation. Mass spectrometry studies to identify sites of phosphorylation are currently underway. It is possible that maspin is regulated by diverse post-translational modifications in different cell types.
Mass spectrometry based approaches have identified modifications on maspin in several cell types. A recent study used the biotin switch method and mass spectrometry using the LTQ-Orbitrap to identify an S-nitrosylated form of maspin among 116 targets in prostate epithelial cells [17]. Maspin was also identified using nano-LC-nano-ESI-MS/MS in a study looking at pro-apoptotic proteins produced by normal breast epithelial cells [18]. MALDI-TOF analysis was employed to identify maspin as a marker for adenoid cystic carcinomas of the salivary glands [19], and in a human hepatoma revertant cell line [20]. An increase in the acidic form of maspin was detected in MCF10A using DIGE/MS cells upon treatment with TGF-β [21, 22], but the nature of the modification that caused this shift was not identified.
To our knowledge, the current study is the first to describe the post-translational modification of extracellular maspin released by corneal epithelial cells. This study employed MSF analysis for the identification on phosphorylation sites on extracellular maspin. In MSF analysis, the most abundant ion from an MS scan is fragmented using a multiple stage fragmentation protocol. The data-dependent neutral loss (DDNL) scanning method, routinely used on quadrupole and ion-trap instruments, for the identification of phosphopeptides scans for a loss of m/z corresponding to the phosphate moiety [23, 24]. In MSF analysis, the most abundant ion from the MS scan is excited for future fragmentation, irrespective of the presence or absence of a neutral loss molecule. In our laboratory, we found that the MSF method significantly improves detection of phosphopeptides on an ion-trap mass spectrometer, and detects phosphopeptides missed by standard DDNL scanning. Detection of both monophosphopeptides and multiply phosphorylated peptides is improved using the MSF method. Comparison of the MSF method with other traditional phosphopeptide identification methods is beyond the scope of this manuscript.
We speculate that extracellular maspin maybe phosphorylated by more than one kinase, both in the cell and in the extracellular milieu. Glycogen synthase is an example of a molecule phosphorylated on nine different serine residues by at least six different kinases [25]. Analysis of the maspin sequence, for identification of consensus sequences (www.hprd.org) containing the serine and threonine residues identified in this study, indicates that maspin could be phosphorylated by casein kinase II (Thr50, Thr310), PKA (Thr50, Thr118, Ser298) and PKC (Thr50) among others. Two of these kinases, casein kinase II and PKA, were shown to produce the phosphorylated anti-angiogenic forms of the related serpin, pigment epithelial derived factor (PEDF) [26]. These kinases may also produce a form of maspin that is important for its anti-angiogenic activity. Interestingly, casein kinase II and PKA are an exo- and ecto-kinase, respectively [26, 27], and have been shown to phosphorylate PEDF extracellularly. This raises the possibility that a population of maspin may also be phosphorylated in the conditioned medium. If mass spectrometry based experiments identify the same sites of serine and threonine phosphorylation on intracellular corneal epithelial maspin as on the extracellular form, in addition to tyrosine phosphorylation, maspin phosphorylation may occur intracellularly before release from the cell.
Although maspin lacks a bona fide secretion signal, the N-terminus of maspin has a hydrophobic region which is assumed to function as an uncleaved, facultative secretion signal allowing entry of some of the molecule in to the ER to be processed for secretion [2]. Information is sparse on how maspin enters the ER and what modifications it might undergo during processing. A recent report suggests that maspin is an obligate intracellular protein in a normal mammary epithelial and a prostate epithelial cell line, and the detection of maspin in conditioned medium can only be attributed to release of the protein from dead or dying cells [28]. However, several unconventional pathways exist for delivering proteins extracellularly, including translocation across the plasma membrane, secretion via exosomes and secretory lysosomes [29-31]. This is consistent with previous reports that suggest that secreted proteins such as FGF-2 [32, 33] and Tubby [34] that either lack a secretion signal or possess a cryptic signal sequence may employ one or more unconventional means for secretion. Studies have shown that maspin is present in the extracellular space and can be delivered extracellularly by exosomes [35, 36]. One or more of these unconventional pathways may be involved in the export of maspin from corneal epithelial cells.
The anti-angiogenic potential of extracellular maspin has been investigated in a number of systems including the skin [37], cornea [10] and mammary tumors [38]. A number of anti-angiogenic factors are produced by the cornea to prevent neovascularization and maintain transparency. Extracellular maspin functions as an anti-angiogenic molecule in the cornea as evidenced by experiments using the rat cornea pocket model [10]. Extracellular maspin may also prevent the invasion of vessel-forming endothelial cells from the conjunctival tissue adjacent to the cornea. The molecular mechanism for the anti-angiogenic effect of maspin and what role phosphorylation might play in enhancing this effect remains at large. Based on the importance of phosphorylation on the the anti-angiogenic activity of PEDF, the identification of post-translational modifications of extracellular maspin is the logical first step towards understanding maspin's anti-angiogenic role in the cornea.
Phosphorylation of proteins is a fundamental mechanism leading to a variety of effects on protein function. To our knowledge, this study is the first report on the identification of the phosphorylation sites on maspin. This information will help elucidate the molecular mechanism of phosphorylated maspin released from corneal epithelial cells in several important processes such as regulation of wound healing and prevention of angiogenesis in the cornea. The function of phosphorylated intracellular maspin in the maintenance of homeostasis in the corneal epithelium will be elucidated in future studies.
Supplementary Material
Acknowledgments
This study was funded by grants from the Eye Institute of the National Institutes of Health (RO1- EY12731, RO1-14168 and P30-EY01931). The LTQ-XL mass spectrometer was funded by grants from the National Institutes of Health (N01-HV-28182) and Advanced Healthier Wisconsin (AHW-5520058).
Abbreviations
- MSF
Multi-stage fragmentation
- LTQ
linear trap quadrupole
- HCEC
human corneal epithelial cells
- ECM
extracellular matrix
Footnotes
The authors declare no conflict of interest.
Supplemental Data: MS3 spectra and fragmentation patterns of maspin phosphopeptides A) pSLNLSTEFISSTK, residues 97-109; B) ELEpTVDFKDKLEETK, residues 115-129; C) DLTDGHHFENILADNSVNDQpTK, residues 138-158; D) DVEDEpSTGLEK, residues 235-245; E) HIFpSEDTSDFSGMSETK, residues 295-311; F) HIFSEDTSDFSGMSEpTK, residues 295-311 and G) GVALpSNVIHK, residues 312-340. Peptides with multiple serine and threonine residues are adequately fragmented with 98 Da losses to enable the identification of the specific phosphorylated residue. The * in peptide HIFpSEDTSDFSGMSETK (E) refers to an oxidized methionine.
References
- 1.Zou Z, Anisowicz A, Hendrix MJ, Thor A, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 2.Pemberton PA, Tipton AR, Pavloff N, Smith J, et al. Maspin is an intracellular serpin that partitions into secretory vesicles and is present at the cell surface. J Histochem Cytochem. 1997;45:1697–1706. doi: 10.1177/002215549704501213. [DOI] [PubMed] [Google Scholar]
- 3.Ngamkitidechakul C, Burke JM, O'Brien WJ, Twining SS. Maspin: synthesis by human cornea and regulation of in vitro stromal cell adhesion to extracellular matrix. Invest Ophthalmol Vis Sci. 2001;42:3135–3141. [PubMed] [Google Scholar]
- 4.Pemberton PA, Wong DT, Gibson HL, Kiefer MC, et al. The tumor suppressor maspin does not undergo the stressed to relaxed transition or inhibit trypsin-like serine proteases. Evidence that maspin is not a protease inhibitory serpin. J Biol Chem. 1995;270:15832–15837. doi: 10.1074/jbc.270.26.15832. [DOI] [PubMed] [Google Scholar]
- 5.Yin S, Lockett J, Meng Y, Biliran H, Jr, et al. Maspin retards cell detachment via a novel interaction with the urokinase-type plasminogen activator/urokinase-type plasminogen activator receptor system. Cancer Res. 2006;66:4173–4181. doi: 10.1158/0008-5472.CAN-05-3514. [DOI] [PubMed] [Google Scholar]
- 6.Cher ML, Biliran HR, Jr, Bhagat S, Meng Y, et al. Maspin expression inhibits osteolysis, tumor growth, and angiogenesis in a model of prostate cancer bone metastasis. Proc Natl Acad Sci U S A. 2003;100:7847–7852. doi: 10.1073/pnas.1331360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham S, Zhang W, Greenberg N, Zhang M. Maspin functions as tumor suppressor by increasing cell adhesion to extracellular matrix in prostate tumor cells. J Urol. 2003;169:1157–1161. doi: 10.1097/01.ju.0000040245.70349.37. [DOI] [PubMed] [Google Scholar]
- 8.Ngamkitidechakul C, Warejcka DJ, Burke JM, O'Brien WJ, Twining SS. Sufficiency of the reactive site loop of maspin for induction of cell-matrix adhesion and inhibition of cell invasion. Conversion of ovalbumin to a maspin-like molecule. J Biol Chem. 2003;278:31796–31806. doi: 10.1074/jbc.M302408200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Shi Y, Magit D, Furth PA, Sager R. Reduced mammary tumor progression in WAP-TAg/WAP-maspin bitransgenic mice. Oncogene. 2000;19:6053–6058. doi: 10.1038/sj.onc.1204006. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 11.Odero-Marah VA, Khalkhali-Ellis Z, Schneider GB, Seftor EA, et al. Tyrosine phosphorylation of maspin in normal mammary epithelia and breast cancer cells. Biochem Biophys Res Commun. 2002;295:800–805. doi: 10.1016/s0006-291x(02)00764-7. [DOI] [PubMed] [Google Scholar]
- 12.Halligan BD, Ruotti V, Jin W, Laffoon S, et al. ProMoST (Protein Modification Screening Tool): a web-based tool for mapping protein modifications on two-dimensional gels. Nucleic Acids Res. 2004;32:W638–44. doi: 10.1093/nar/gkh356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halligan BD. ProMoST: a tool for calculating the pI and molecular mass of phosphorylated and modified proteins on two-dimensional gels. Methods Mol Biol. 2009;527:283–98. doi: 10.1007/978-1-60327-834-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cella N, Contreras A, Latha K, Rosen JM, Zhang M. Maspin is physically associated with [beta]1 integrin regulating cell adhesion in mammary epithelial cells. FASEB J. 2006;20:1510–1512. doi: 10.1096/fj.05-5500fje. [DOI] [PubMed] [Google Scholar]
- 15.Cantin GT, Shock TR, Park SK, Madhani HD, Yates JR., 3rd Optimizing TiO2-based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Anal Chem. 2007;79:4666–4673. doi: 10.1021/ac0618730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kweon HK, Hakansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 17.Lam YW, Yuan Y, Isaac J, Babu CV, et al. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PLoS One. 2010;5:e9075. doi: 10.1371/journal.pone.0009075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toillon RA, Lagadec C, Page A, Chopin V, et al. Proteomics demonstration that normal breast epithelial cells can induce apoptosis of breast cancer cells through insulin-like growth factor-binding protein-3 and maspin. Mol Cell Proteomics. 2007;6:1239–1247. doi: 10.1074/mcp.M600477-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima D, Uzawa K, Kasamatsu A, Koike H, et al. Protein expression profiling identifies maspin and stathmin as potential biomarkers of adenoid cystic carcinoma of the salivary glands. Int J Cancer. 2006;118:704–713. doi: 10.1002/ijc.21318. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Q, An J, Liu DG, Sun L, et al. Proteomic analysis of differential protein expression in a human hepatoma revertant cell line by using an improved two-dimensional electrophoresis procedure combined with matrix assisted laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis. 2004;25:1160–1168. doi: 10.1002/elps.200305769. [DOI] [PubMed] [Google Scholar]
- 21.Friedman DB, Wang SE, Whitwell CW, Caprioli RM, Arteaga CL. Multi-variable Difference Gel Electrophoresis and Mass Spectrometry: A Case Study on TGF-beta and ErbB2 signaling. Mol Cell Proteomics. 2007;6:150–169. doi: 10.1074/mcp.D600001-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Wang SE, Narasanna A, Whitell CW, Wu FY, et al. Convergence of p53 and transforming growth factor beta (TGFbeta) signaling on activating expression of the tumor suppressor gene maspin in mammary epithelial cells. J Biol Chem. 2007;282:5661–5669. doi: 10.1074/jbc.M608499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 24.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 2009;44:861–878. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 25.Nakielny S, Campbell DG, Cohen P. The molecular mechanism by which adrenalin inhibits glycogen synthesis. Eur J Biochem. 1991;199:713–722. doi: 10.1111/j.1432-1033.1991.tb16175.x. [DOI] [PubMed] [Google Scholar]
- 26.Maik-Rachline G, Seger R. Variable phosphorylation states of pigment-epithelium-derived factor differentially regulate its function. Blood. 2006;107:2745–2752. doi: 10.1182/blood-2005-06-2547. [DOI] [PubMed] [Google Scholar]
- 27.Maik-Rachline G, Shaltiel S, Seger R. Extracellular phosphorylation converts pigment epithelium-derived factor from a neurotrophic to an antiangiogenic factor. Blood. 2005;105:670–678. doi: 10.1182/blood-2004-04-1569. [DOI] [PubMed] [Google Scholar]
- 28.Teoh SS, Whisstock JC, Bird PI. Maspin (SERPINB5) is an obligate intracellular serpin. J Biol Chem. 2010;285:10862–10869. doi: 10.1074/jbc.M109.073171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 30.Nickel W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic. 2005;6:607–614. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 31.Nickel W. The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur J Biochem. 2003;270:2109–2119. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 32.Torrado LC, Temmerman K, Muller HM, Mayer MP, et al. An intrinsic quality-control mechanism ensures unconventional secretion of fibroblast growth factor 2 in a folded conformation. J Cell Sci. 2009;122:3322–3329. doi: 10.1242/jcs.049791. [DOI] [PubMed] [Google Scholar]
- 33.Temmerman K, Ebert AD, Muller HM, Sinning I, et al. A direct role for phosphatidylinositol-4,5-bisphosphate in unconventional secretion of fibroblast growth factor 2. Traffic. 2008;9:1204–1217. doi: 10.1111/j.1600-0854.2008.00749.x. [DOI] [PubMed] [Google Scholar]
- 34.Caberoy NB, Li W. Unconventional secretion of tubby and tubby-like protein 1. FEBS Lett. 2009;583:3057–3062. doi: 10.1016/j.febslet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 36.Chavez-Munoz C, Kilani RT, Ghahary A. Profile of exosomes related proteins released by differentiated and undifferentiated human keratinocytes. J Cell Physiol. 2009;221:221–231. doi: 10.1002/jcp.21847. [DOI] [PubMed] [Google Scholar]
- 37.Nickoloff BJ, Lingen MW, Chang BD, Shen M, et al. Tumor suppressor maspin is up-regulated during keratinocyte senescence, exerting a paracrine antiangiogenic activity. Cancer Res. 2004;64:2956–2961. doi: 10.1158/0008-5472.can-03-2388. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Shi HY, Zhang M. Targeted expression of maspin in tumor vasculatures induces endothelial cell apoptosis. Oncogene. 2005;24:2008–2019. doi: 10.1038/sj.onc.1208449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


