Abstract
Sexual reproduction in heterothallic ascomycete fungi is controlled by a single mating-type locus called MAT1 with two alternate alleles or idiomorphs, MAT1-1 and MAT1-2. These alleles lack sequence similarity and encode different transcriptional regulators. A large number of phytopathogenic fungi including Alternaria spp. are considered asexual, yet still carry expressed MAT1 genes. The molecular evolution of Alternaria MAT1 was explored using nucleotide diversity, nonsynonymous vs. synonymous substitution (dn/ds) ratios and codon usage statistics. Likelihood ratio tests of site-branch models failed to detect positive selection on MAT1-1-1 or MAT1-2-1. Codon-site models demonstrated that both MAT1-1-1 and MAT1-2-1 are under purifying selection and significant differences in codon usage were observed between MAT1-1-1 and MAT1-2-1. Mean GC content at the third position (GC3) and effective codon usage (ENC) were significantly different between MAT1-1-1 and MAT1-2-1 with values of 0.57 and 48 for MAT1-1-1 and 0.62 and 46 for MAT1-2-1, respectively. In contrast, codon usage of Pleospora spp. (anamorph Stemphylium), a closely related Dothideomycete genus, was not significantly different between MAT1-1-1 and MAT1-2-1. The purifying selection and biased codon usage detected at the MAT1 locus in Alternaria spp. suggest a recent sexual past, cryptic sexual present and/or that MAT1 plays important cellular role(s) in addition to mating.
Introduction
Sexual reproduction in heterothallic ascomycete fungi is initiated when strains of opposite mating type interact, and this interaction is controlled by a single regulatory locus called MAT1 [1]. The MAT1 locus has two alternate alleles, MAT1-1 and MAT1-2, which lack sequence similarity and have been termed idiomorphs [2]. All ascomycete MAT1 idiomorphs encode proteins with confirmed or putative DNA binding motifs (e.g., MAT1-1-1 = alpha box; MAT1-2-1 = high mobility group [HMG] box), suggesting that MAT1 genes encode transcriptional regulators which control the expression of additional genes required for sexual reproduction and possibly other cellular processes [3]. Little is known about the targets of these regulators in filamentous fungi [1], [4], however, these mating regulators have been well- studied in yeast [5]. MAT1 loci are also found in putatively asexual ascomycete species [6]–[10]. It has also been demonstrated that genes at the MAT1 locus are expressed in the asexual species, Alternaria alternata, and this locus was able to functionally complement a MAT1-null mutant in the closely related sexual species Cochliobolus heterostrophus [7].
A large number of phytopathogenic fungi are considered to be asexual (mitosporic) because they have no known teleomorph, yet still carry functional MAT1 genes [6]–[7]. The role of MAT1 genes in asexual fungi is unknown although most or all may be recombining through cryptic meiosis or a parasexual cycle [8]–[9], [11]. Direct observation of microbial reproduction in nature is difficult and most studies that have inferred recombination in putatively asexual fungi have relied on indirect inference using gametic disequilibrium or parsimony tree length comparisons among molecular markers [8]–[9], [11]–[12].
Alternaria is considered to be a largely asexual genus; however, A. infectoria has been connected to a Lewia teleomorph [13] and occupies a basal position in the phylogeny of the genus [8]. This suggests that most or all Alternaria species may have had sexual ancestors as has been suggested for other asexual ascomycetes [14]. A formal study to determine the mating system of A. alternata has not been conducted and we are currently unable to rule out the possibility that Alternaria genomes may regularly recombine through cryptic meiotic or parasexual processes. Results of Peever et al. [15] found two genetically distinct clusters of A. alternata infecting citrus within a small area (250 m2) of a single citrus grove. The strongly clonal population structure within each cluster was suggestive of asexual reproduction [15], although small sample sizes and lack of genetic linkage analyses of the markers precluded critical estimation of the mating system.
Molecular evolution analyses can be used to uncover patterns in codon usage to help reveal functionality, expression levels, and mechanisms of natural selection acting on a gene [16]–[17]. Codon usage has been shown to be highly biased [18], reflecting a balance among the forces of mutation, selection, and random genetic drift [19]. This bias appears to maximize the efficiency of translation [20], and differs among species due to changes in the complement of tRNAs or life history [21]–[23]. Comparing rates of synonymous and non-synonymous substitutions across a gene can provide powerful inferences about gene evolution [24]. Synonymous substitutions, codon changes that do not result in amino acid changes, are thought to be largely invisible to natural selection whereas non-synonymous substitution changes that lead to changes in amino acid may be under strong selective pressures. Purifying selection occurs when non-synonymous changes are suppressed and is thought to be a common force in maintaining gene function [25]. Several examples of fungal genes under purifying selection include ubiquitin, a protein that plays a major role in cellular stress response and protein degradation in eurkaryotes, was found to be conserved among 28 species of fungi, plants and animals [26], and the telomere-linked helicase gene family of Magnaporthe oryzae [25]. Positive selection, where non-synonymous changes are favored, has been identified in fungal genes related to defense-related genes and toxin protein genes [27]–[29] such as the phytotoxic protein-encoding genes (NEP1 and NEP2) from Botrytis [30] and the host-specific toxin gene (SnToxA) from Phaeosphaeria nodorum [31].
The objective of this study was to infer the evolutionary processes acting on the mating type-locus, MAT1, in a filamentous fungus with no known sexual state. Sequence data from the Alternaria MAT1 locus were used to estimate the direction and strength of selection acting on MAT1 genes and to compare codon usage patterns to other genes in the A. alternata genome and MAT1 genes in related species. Diversity, neutrality and codon usage patterns of MAT1 of Alternaria were compared to MAT1 of a closely related dothideomycete genus Pleospora (Anamorph Stemphylium) to test the hypothesis that MAT1 in these closely related genera have similar signals of molecular evolution.
Materials and Methods
Fungal culture and DNA extraction
Twenty-one isolates of Alternaria spp. were used in this study (Table S1). MAT1 sequence data from 16 closely related, heterothallic, and putatively sexually reproducing Pleospora spp. (Table S1) were downloaded from GenBank and TreeBase. These taxa were previously described as having typical heterothallic MAT1 locus organization by Inderbitzin et al. [32] but were not verified as heterothallic in laboratory matings. For simplicity, these outgroup taxa are referred to using their anamorphic name, Stemphylium. Sequence data for 11 housekeeping genes in A. alternata and A. brassicicola were downloaded from GenBank and the DOE Joint Genome Institute (Alternaria brassicicola genome sequence) for comparison with Alternaria MAT1 sequences to obtain baseline codon usage patterns for Alternaria spp.
For DNA extraction, fungi were cultivated in liquid 2YEG medium (2 g yeast extract, 10 g glucose per liter) for 1 week at room temperature on an orbital shaker at 150 rpm. Genomic DNA was extracted from powdered, lyophilized mycelium following the methods of Peever et al. [15], quantified using a spectrophotometer, and used as template for PCR. Isolates were maintained in long-term storage on sterilized filter paper at 4°C [15].
PCR Amplification of MAT1 sequences from Alternaria species
Mating type of each isolate was determined using mating type-specific PCR with primers designed to the MAT1-1 (GenBank accession AB009451) and MAT1-2 (GenBank accession AB009452) idiomorphs of the Japanese pear pathotype of A. alternata [7]. The full length MAT1 gene (1.5 kb for MAT1-1-1 or 2.4 kb for MAT1-2-1) was amplified from A. alternata using AAM1-11+AAM1-12 or AAM2-1+AAM2-2 (7) (Table 1), respectively. The full length MAT1-1-1 gene was amplified from isolates SH-MIL-11s, SH-MIL-22s, SH-MIL-34s and SH-MIL-38s and full length MAT1-2-1 was amplified in isolates SH-MIL-1s, SH-MIL-13s, SH-MIL-14s and SH-MIL-19s (Table S1). Twenty microliter PCR reaction mixtures contained 20 ng genomic DNA, 1×PCR buffer (New England Biolabs (NEB), Ipswich, MA), 4 nmol of each dNTP (NEB), 50 pmol primer, and 1 U of Taq polymerase (NEB). Cycling conditions consisted of denaturation at 94°C for 4 min; 44 cycles of 94°C for 1 min, 58 or 55°C for 30 sec, and 72°C for 2 min; final extension was at 72°C for 7 min depending on the optimal conditions for each primer set.
Table 1. Primers used to amplify MAT1 locus from Alternaria spp.
| Primer Name | Sequence (5′ to 3′) | Position | Description | References |
| AAM1-11 | CATCATGATCATTGTTGT | 252–2691 | A. alternata full length MAT1-1-1 | this study |
| AAM1-12 | GCACACCTCAAGTGATCA | 2650–26331 | A. alternata full length MAT1-1-1 | this study |
| AAM2-1 | TAGCGTTTGTCCGTACCGA | 750–7682 | A. alternata full length MAT1-2-1 | this study |
| AAM2-2 | GTAACGAGCATGAACATT | 2211–22282 | A. alternata full length MAT1-2-1 | this study |
| ASML-1 | GGGTTGTTGTGGTCAAGGTT | 144–1631 | Alternaria spp. full length MAT1-1-1 or MAT1-2-1 | this study |
| ASMR-1 | GTCATGATCAAGCAAGGGCA | 2584–25651 | Alternaria spp. full length MAT1-1-1 or MAT1-2-1 | this study |
| AaM1-8 | GGTCGTGAGTCGTGATCG | 2257–22401 | Alternaria spp. full length MAT1-1-1 or MAT1-2-1 | this study |
| ASML-2 | GGACGCATCGCAGATTGGAA | 170–1891 | Alternaria spp. full length MAT1-1-1 or MAT1-2-1 (nested PCR) | this study |
Several approaches were used to amplify the entire MAT1 idiomorph from other Alternaria species because primers designed for A. alternata did not yield amplicons of the expected size for A. brassicicola, A. brassicae and A. solani. Full length MAT1 genes (2.4 kb for MAT1-1-1 or 2. 2 kb for MAT1-2-1) from A. brassicicola isolates 01-1c-s, 01-2a-s, 01-9c-s, 01-23a-s and 01-41a-s were amplified using primers ASML-1+ASMR-1 or AsM1-8+ASML-2, respectively (Table 1). Nested PCR was used to amplify MAT1 from A. brassicae and A. solani isolates 01-8a, 01-8b, 21ss, 39ss, IdahoA and EGS 44-098 (Table S1). Primers ASML-1 and ASMR-1 amplified MAT1 with a primary PCR reaction. This reaction yielded product containing several amplicons. PCR products were diluted 50-fold and were used as template for a second PCR reaction using primers ASML-2 and AaM1-8 to target a fragment containing full length MAT1 gene only. Primary and secondary PCR conditions consisted of denaturation at 94°C for 4 min; 44 cycles of 94°C for 1 min, 58 or 55°C for 30 sec, and 72°C for 2 min; final extension was at 72°C for 7 min.
Amplified DNA fragments were sequenced directly on both strands following treatment with EXOSAP-IT (USB, Cleveland, OH) using the Big Dye terminator kit (Applied Biosystems, Foster City, CA). Sequence reads were performed on a PE Biosystems model 3700 automated DNA Sequencer by the Laboratory for Biotechnology and Bioinformatics at Washington State University, Pullman, WA. Sequences of MAT genes have been deposited under GenBank accession numbers GU735410–GU735428 (Table 1).
Evolutionary analyses of MAT1 genes in Alternaria spp. and closely related species
MAT1-1-1 and MAT1-2-1 ORFs from A. alternata, A. brassicicola, A. brassicae, and A. solani were predicted using two A. alternata reference isolates, 15A and O-276 carrying MAT1-1-1 and MAT1-2-1, respectively [4]. Sequences were aligned manually and using DIALIGN [33]. Comparisons were made between MAT1-1-1 and MAT1-2-1 of Alternaria spp. and MAT1 of putatively asexual Alternaria spp. and putatively sexual, heterothallic Stemphylium spp. (Table S1). Sequence diversity (S), the number of segregating sites, number of haplotypes (Nhap) and haplotype diversity (Hd) were estimated using DnaSP v4.5 [34]. Sequence diversity was also quantified using Watterson's θ parameter [35]. To determine if the patterns of polymorphisms and divergence within and among groups deviated from neutral evolution predictions [36], Tajima's D, DT [37] and Fu and Li's D, DFL [38] tests were performed. DT compares the number of segregating sites to the average number of pairwise nucleotide differences. DFL compares the number of recent (external branches) and ancestral (internal branches) mutations on a phylogenetic tree. Under a neutral evolution model, numbers of mutations on internal and external branches are expected to be equal. Increased number of external branch mutations indicates purifying selection whereas increased number of mutations on internal branches indicates balancing selection [39]. The significance of DT and DFL was tested using coalescent simulations where a neutral coalescent process was used to simulate 1,000 replicates with the number of segregating sites set to the observed data. When positive selection is acting, DT tends to be positive, whereas DT is negative in cases of purifying selection [40].
Signatures of purifying or positive selection acting on MAT1 were tested at the codon level using codon-based likelihood analyses. A maximum likelihood implementation was used to fit codon substitution models to the data using the CODEML program within PAML v 4.2 [41]. Three random site models were used to describe the variation of ω ( = dN/dS) among codon sites within each MAT1 gene. Random site models M0 (one ratio), M7 (beta) and M8 (beta&ω) [42]–[44] were used to describe the variation of ω (ω = dN/dS) among codon sites within each MAT1 gene. M0 is the simplest model, assuming one ω for all codons in a dataset, which can be used to check parameter estimates in the other more complex models. M7 is a flexible null model in which a ω ratio for each codon is randomly selected from a beta distribution between 0 and 1. M8 adds one additional site class to M7 allowing for positive selection (ω>1). A test for positive selection was implemented using likelihood ratio tests (LRT) that compare models M7 and M8. The Bayes Empirical Bayes (BEB) approach [44] was used, within CODEML, to estimate ω using model M8 for each codon site for MAT1-1-1 and MAT1-2-1. BEB also calculates the posterior probability (pp) that a codon site is from a positive-selection site class (ω>1), determining which codon sites are under positive selection, or from a purifying selection class (ω<0.25) [44]. Codon sites with ω>1 and ω<0.25 and pp values greater than 80% were considered to be under positive or purifying selection, respectively [44]. The tree file used as an input file for CODEML was produced using the parsimony search criterion in PAUP.
Codon bias, where certain codons are used preferentially, has been described in many organisms [45]–[46] and can give insight into translation efficiency and levels of protein expression [47]. Three measures of codon usage were used to estimate codon bias at MAT1 of Alternaria spp. using CodonW 1.4.2 [48], the frequency of G+C at the third synonymous variable codon position (GC3), a measure of the effective number of different codons used in a gene (ENC) [49], and codon adaptation index (CAI) which is a univariate measure of synonymous codon usage used as an indicator of gene expressivity. CAI is the geometric mean of relative synonymous codon usage (RSCU) values, which is the observed frequency of a codon divided by the frequency expected under the assumption of equal usage of synonymous codons for an amino acid [50]. To obtain baseline codon usage patterns for Alternaria spp., 11 housekeeping genes from Alternaria spp. were compared (Table S1). GC3 and ENC were plotted and compared to ENC*, which is the null hypothesis that GC bias at the third position is solely due to mutation rather than selection [49], [51]. Genes lacking codon bias are expected to have an ENC score of 61, where all possible codons are used [49]. The reference species used for CAI analyses was Saccharomyces cerevisae [52]. Using this comparison, highly expressed genes that use the same codon set as Saccharomyces cerevisae would have a CAI value of 1. Lower values of CAI indicate smaller levels of expression [50]. The Mann-Whitney U test [53], implemented in R (v2.8.1) was used to determine if observed differences in ENC, GC3 and CAI between MAT1-1-1, MAT1-2-1, and housekeeping genes were significant.
Results
Evolution and patterns of purifying selection on MAT1 of Alternaria
Analyses of nucleotide variation between Alternaria MAT1-1-1 (2.4 kb) and MAT1-2-1 (2.2 kb) without regard to codon structure revealed no statistically significant differences in diversity (Table 2). MAT1-1-1 and MAT1-2-1 had similar diversity (within one standard deviation of each other) for variable sites (S) and nucleotide diversity (Watterson's θ). We failed to reject the null hypothesis of neutrality using Tajima's D (DT), which tests for neutrality using the average number of pairwise nucleotide differences. This indicated that nucleotide diversity at MAT1 appeared not to deviate from neutrality when nucleotides across the entire gene were compared (Table 2). However, analyses of Fu and Li's D (DFL), which examines the distribution of mutations on internal or external branches, found that Alternaria MAT1-1-1 was significantly greater than 1 (DFL = 1.6, P<0.05), indicating that more mutations were found on internal branches, suggestive of positive selection.
Table 2. Genetic diversity and results of neutrality test of putatively asexual Alternaria spp. MAT1 and closely related putatively heterothallic sexual Stemphylium spp.
| MAT 1-1-1 | MAT 1-2-1 | |||||
| Summary statistic | Alternaria spp. | Stemphylium spp. | All isolates | Alternaria spp. | Stemphylium spp. | All isolates |
| N | 11 | 10 | 21 | 10 | 6 | 16 |
| L | 1173 | 1173 | 1173 | 1039 | 1039 | 1039 |
| H | 7 | 9 | 16 | 7 | 6 | 13 |
| Hd | 0.927 | 0.978 | 0.976 | 0.933 | 1.00 | 0.97 |
| S | 267 | 301 | 599 | 202 | 181 | 483 |
| Π | 0.095 | 0.094 | 0.21 | 0.086 | 0.085 | 0.208 |
| 0.078 | 0.093 | 0.146 | 0.069 | 0.076 | 0.143 | |
| DT | 1.06 | −0.39 | 0.73 | 0.98 | −0.52 | 0.99 |
| DFL | 1.61P | −0.47 | 0.84 | 1.29 | −0.64 | 1.24 |
Signature of selection at Alternaria MAT1
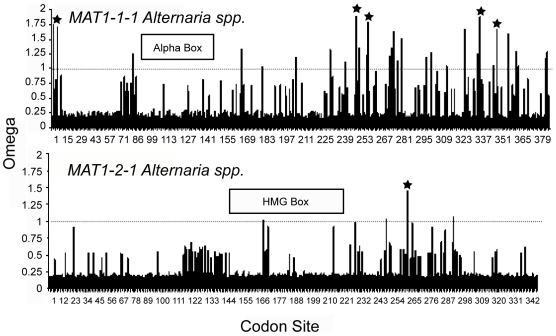
Using the site-branch model in PAML, we failed to reject the null hypothesis of neutrality using a likelihood ratio test comparing the M7 (null) and M8 (positive selection) site models across the entire genes of MAT1-1-1 and MAT1-2-1. However, tests for selection using a codon-site model (BEB approach), which focuses on selection at individual codons, revealed an overall signal purifying selection acting on Alternaria MAT1 with potential signatures of positive selection at few specific codon sites within MAT1-1-1 and MAT1-2-1 (pp greater than 80%). For MAT1-1-1, five of 387 codon sites had ω values greater than one with a pp over 80%, indicating possible positive selection. Two hundred and twenty five sites had a ω<0.25 at a pp over 90%, indicating purifying selection and of these 110 sites had a pp over 98% suggesting strong purifying selection (Figure 1). For MAT1-2-1, only one site had a ω over 1 with a pp greater than 80%, indicating possible positive selection. Of 342 codon sites, 124 had a ω<0.25 at a pp over 90% indicating purifying selection. When the pp stringency was decreased to 80% for MAT1-2-1, 256 of 342 sites were shown to be under purifying selection, indicating that 75% of the codons within MAT1-2-1 are possibly under purifying selection. ω values for each of the conserved protein regions within each idiomorph, alpha box of MAT1-1-1 (aa. 70–127) and HMG box of MAT1-2-1 (aa 129–205) also provided evidence for purifying selection (Figure 1).
Figure 1. Non-synonymous to synonymous substitution ratio (ω) for MAT1-1-1 (upper) and MAT1-2-1 (lower) in Alternaria estimated in CODEML in PAML.
The graph shows approximate posterior means of ω calculated as an average of ω's weighted by their posterior probabilities of the 11 site classes using in model 8a. The 11 ω ratios for MAT1-1-1 are 0.08896, 0.10488, 0.11513, 0.12372, 0.13175, 0.13979, 0.14838, 0.15827, 0.17113, 0.19391, and ωs = 1.46207. The 11 ω ratios for MAT1-2-1 are 0.08585, 0.12609, 0.15474, 0.18009, 0.20463, 0.22990, 0.25745, 0.28968, 0.33196, 0.40659, and ωs = 2.22313. Sites with low mean ω are inferred to be under purifying selection. Asterisks indicate sites with posterior probabilities more than 0.80 for ωs>1.
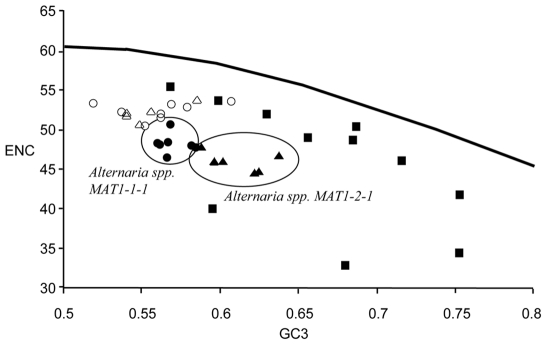
Observed differences in effective codon usage (ENC), GC (GC3) content at the third position, and codon adaption index (CAI) between Alternaria MAT1-1-1 and MAT1-2-1 indicated divergent codon usage patterns. Significant differences for GC3 and ENC (P<0.001) were detected between MAT1-1-1 and MAT1-2-1 within Alternaria. Means for each were 0.57 and 48 for MAT1-1-1 and 0.62 and 46 for MAT1-2-1. Overall, MAT1-2-1 of Alternaria had larger GC3 values and lower ENC values, similar to housekeeping genes where no significant differences were observed (Figure 2). GC3 and ENC values plotted for all MAT1 genes were significantly different from those of ENC*, indicating that selection was likely driving biased codon usage (Figure 2).
Figure 2. Effective number of codons (ENC) used in a gene plotted against the G+C content at the synonymously variable third position (GC3), for 21 MAT1-1 genes, 16 MAT1-2 genes, and 11 highly conserved genes (Table S1).
Circles indicate MAT1-1-1 gene of Alternaria (black), and Stemphylium (white). Triangles indicate MAT1-2-1 genes of Alternaria (black) and Stemphylium (white). Squares indicate highly conserved genes of Alternaria spp. and A. alternata (black). The solid line is the expected ENC* curve, representing the null hypothesis that GC bias at the third position is solely due to mutation rather than selection.
Comparisons of codon adaptation index (CAI) mean values of Alternaria MAT1 and housekeeping genes indicated that MAT1 had reduced levels of expression compared to housekeeping genes. Mean CAI values for MAT1-1-1 and MAT 1-2-1 were 0.107 and 0.095, respectively (Table 3). These values were lower than CAI values observed for Alternaria housekeeping genes which had a mean CAI of 0.151. MAT1-2-1 had significantly smaller CAI values than MAT1-1-1, indicating lower expression. MAT1-1-1 was not significantly different than housekeeping genes, though this was most likely the result of the high variance in CAI values of the housekeeping genes (Table 3). High variance within Alternaria housekeeping genes can be attributed to the diversity of genes incorporated in the analyses, which included an endoxylanase, exoglucanse, chitin synthase, kinase and G protein (Table 3).
Table 3. Mean codon adaptation index (CAI) values for Alternaria MAT1 (Am1 and Am2), housekeeping genes (Ahk), and Stemphylium MAT1 (Sm1 and Sm2).
| Group | NA | Mean±STD | Pairwise comparisons | ||||
| Am1 | Am2 | Ahk | Sm1 | Sm2 | |||
| Am1 | 11 | 0.107±0.004 | - | P = 0.0005* | P = 0.262 | P = 0.001* | P = 0.116 |
| Am2 | 10 | 0.095±0.005 | - | P = 0.001* | P = 0.0002* | P = 0.001* | |
| Ahk | 11 | 0.151±0.069 | - | P = 0.716 | P = 0.925 | ||
| Sm1 | 10 | 0.117±0.007 | - | P = 0.080 | |||
| Sm2 | 6 | 0.111±0.003 | - | ||||
*Significant pairwise comparisons.
Number of genes included in each group.
Comparisons of nucleotide variation and codon usage at MAT1 in Alternaria and Stemphylium
Alternaria and Stemphylium MAT1 had similar numbers of polymorphic nucleotide sites and levels of diversity. All values were within one standard deviation of each other. Results examining nucleotide differences and diversity between Stemphlium MAT1-1-1 and MAT1-2-1 also yielded no significant differences results, as observed with Alternaria.
Codon usage patterns (ENC, GC3 and CAI) between Alternaria and Stemphylium MAT1 were significantly different. MAT1 of Stemphylium had significantly smaller GC3 (P<0.01), larger ENC (P<0.01), and higher CAI values (Table 3) compared to Alternaria MAT1-2-1. Comparisons of Stemphylium MAT1 and Alternaria MAT1-1-1 yielded no significant differences for GC3, but Stemphlium MAT1 had significantly larger ENC values. Results of mean CAI value comparisons showed mixed results; Stemphylium MAT1-1-1, but not MAT1-2-1, CAI means were significantly larger than Alternaria MAT1-1-1.
Overall, codon usage between Stemphylium MAT1-1-1 and MAT1-2-1 did not vary. Mean values for ENC and GC3 were 52.16 and 0.561 for MAT1-1-1 and 52.38 and 0.558 for MAT1-2-1. Mean CAI values for Stemphylium MAT1 were also similar, 0.117 for MAT1-1-1 and 0.113 for MAT1-2-1. Though not significantly different, Stemphylium MAT1 had lower values than Alternaria housekeeping genes, indicating lower expression levels.
Discussion
Almost all known Alternaria species are considered to be asexual and only a few Alternaria anamorphs have been connected to a teleomorphic stage. A sexual stage for A. alternata has never been observed in nature and attempts to produce a sexual stage in the laboratory have failed [8]. Although a sexual stage has never been described for A. alternata, this species carries functional genes at the MAT1 locus [7] and both mating types are routinely recovered from populations of this fungus [T.L. Peever, unpublished]. Our codon-site analyses rejected the neutrality hypothesis and indicated that these loci are not evolving neutrally but rather are subject to purifying (negative) selection. Over half the codons (387 aa and 342 aa for MAT1-1-1 and MAT1-2-1, respectively) were under purifying selection. Further, patterns of selection differed between the idiomorphs with MAT1-1-1 under strong purifying selection with many sites with high posterior probability values and MAT1-2-1 under weaker purifying selection. Purifying selection might be the result of a cryptic contemporary sexual cycle, a recent sexual past or the involvement of MAT1 in other critical cellular functions. Aspergillus fumigatus, a fungus long thought to be asexual, was recently shown to have a sexual stage [55], which might also be possible for Alternaria. Moreover, if the MAT1 locus in Alternaria controls cellular functions in addition to mating, the differences in codon usage between MAT1-1-1 and MAT1-2-1 may indicate that genes encoded by each idiomorph have different roles in the cell.
In sexual species, MAT1-1-1 and MAT1-2-1 are known to play different roles in the sexual cycle and possibly in other cellular processes [1]. In an elegant study using gene deletions of MAT1 and MAT2 in Aspergillus nidulans, Paoletti et al. [55] found that mutations affected components of the sexual cycle differentially, particularly the development of thick-walled Hülle cells. The MAT1-1 alpha box and the MAT1-2 HMG DNA-binding box motifs are thought to regulate different classes of sex pheromones and their receptors [56] so a differential role in the cell is not unexpected. Another putative role for the MAT1 locus may be the control of virulence in plant-pathogenic fungi. In a population study, Zhan et al. [57] found that MAT1-1 strains of the sexual wheat pathogen, Mycosphaerella graminicola, were more virulent than MAT1-2 strains. Limitations in the experimental design did not allow the authors to determine if these associations were due to pleiotropic effects of MAT1 or other loci tightly linked to MAT1 [57]. Several studies have suggested that mating-type genes are involved in additional cellular processes such as cell wall maintenance, cellular resistance to DNA damage, and MAT pheromones have been shown to induce G proteins which are linked to protein kinase cascades [58]–[60].
Similar to the results presented here, O'Donnell et al. [61] showed that MAT1 genes in Fusarium graminearum were subject to purifying selection, and likewise Rau et al. [62] found amino acid conservation (non-synonymous substitution ratio<synonymous substitution ratio) in MAT1 genes of Pyrenophora teres. Turgeon [4] observed a paucity of silent substitutions in MAT1 of C. heterostrophus, leading to speculation that mating-type genes may be under strong diversifying selection to prevent interspecific mating [63]–[65]. Using likelihood ratio tests of codon site models with and without positive selection, we saw no evidence of overall positive selection at MAT1 of Alternaria, although several positively selected sites within each idiomorph were identified. Several studies have demonstrated heterologous complementation of MAT-deficient mutants [7], [14], [66], suggesting that function is retained across genera and this result is not consistent with strong diversifying selection as suggested by Turgeon [4].
Wik et al. [67] found higher levels of positive selection on MAT1 in heterothallic Neurospora species compared to homothallic species. Comparing the results Wik et al. [67] with those presented here for with Alternaria MAT1, we find similar numbers of positively selected codons, albeit at different codon sites. Wik et al. [67] found 2, 7 and 1 positively selected codons in Neurospora mat-a-1, mat-A-1, and mat A-3, respectively. Similarly, we found that MAT1-1-1 and MAT1-2-1 had 5 and 1 positively selected codon sites, respectively. Rare signatures of positive selection scattered within an overall strong signal of purifying selection at MAT1 in Alternaria may not be due to asexuality, but rather rapid divergence in the heterothallic mating system due to adaptive evolution or a lack of selective constraint in the mating type genes [67].
The strength of codon bias can be used to make predictions about expression levels in a gene, where a smaller ENC value is an indicator of the overall codon bias which is then correlated with higher expression levels (higher CAI values) [55]. ENC and CAI values for Alternaria MAT1 did not follow this trend. Alternaria MAT1-2-1 showed more codon bias (lower ENC) but also low expression levels (smaller CAI), whereas MAT1-1-1 had decreased levels of codon bias, but higher expressions levels. The reasons for these differences in codon usage and expression patterns between Atlernaria MAT1 are unknown. It may signal variation in the type and strength of selective forces. A positive correlation between codon bias and expression level can be attributed to translational efficiency [68]. Codon Adaptation Index (CAI) measures the difference between observed codon frequencies from the null expectation that each amino acid has an equal chance of being encoded by all possible codons, and the difference can be correlated to translational efficiency because as fewer codons are used to encoded amino acids, increasing bias [69]. Alternaria housekeeping genes follow this ENC and CAI trend with significant linear correlation (R2 = 0.62, P = 0.003), whereas for MAT1 in Alternaria, the correlation was weaker and was statistically insignificant (R2 = 0.14, P = 0.09). We might speculate that lack of correlation between ENC and CAI indicates that translational selection is a weak force in shaping Alternaria MAT1. The combined results of GC3, ENC and CAI highlight the possible complexity of selective forces acting on MAT1 in Alternaria.
Our results demonstrating differential codon bias between MAT1 idiomorphs in Alternaria suggest that MAT1 is conserved. Critical determination of the mating system of any Alternaria spp. has not been performed to date but our results may suggest that this genus has a sexual cycle or recent sexual past. The differences detected between MAT1-1-1 and MAT1-2-1 may also indicate that the MAT1 alleles are involved in different biological functions, which may lead to differential fitness between mating types. Future comparisons of Alternaria MAT1 with MAT1 from sexual species may help to deduce if the selection and codon bias differences observed in MAT1-1-1 and MAT1-2-1 are due to differences in gene function or if they signify divergent histories.
Supporting Information
Isolates used in evolutionary analyses.
(DOCX)
Acknowledgments
We thank Dr. Lindsey DuToit for isolates of A. brasssicicola and Drs. Lori Carris and Eric Roalson for editing earlier versions of this manuscript. PPNS No. 0532, Department of Plant Pathology, College of Agriculture, Human, and Natural Resource Sciences, Agricultural Research Center, Project No. WNP03061-0300, Washington State University, Pullman, WA 99164-6430, USA.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: ZA is partially funded by NIH P20RR16448. JS received partial financial support from the ARCS Foundation (www.arcsfoundation.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding was received for this study.
References
- 1.Debuchy R, Turgeon BG. Mating-type structure, evolution, and function in Euascomycetes. In: Kües U, Fisher R, editors. The Mycota: Growth, Differentiation and Sexuality. Berlin: Springer-Verlag; 2006. pp. 293–323. [Google Scholar]
- 2.Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora. Bioassay. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 3.Turgeon BG, Yoder OC. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol. 2000;31:1–5. doi: 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- 4.Turgeon BG. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol. 1998;36:115–137. doi: 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- 5.Herskowitz I. A Regulatory Hierarchy for Cell Specialization in Yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 6.Sharon A, Yamaguchi K, Christiansen S, Horwitz BA, Yoder OC, et al. An asexual fungus has the potential for sexual development. Mol Gen Genet. 1996;251:60–68. doi: 10.1007/BF02174345. [DOI] [PubMed] [Google Scholar]
- 7.Arie T, Kaneko I, Yoshida T, Nouchi M, Nomura Y, et al. Mating-type gene from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol Plant Microbe Interact. 2000;13:1330–1339. doi: 10.1094/MPMI.2000.13.12.1330. [DOI] [PubMed] [Google Scholar]
- 8.Berbee ML, Payne BP, Zhang G, Roberts RG, Turgeon BG. Shared ITS DNA substitutions in isolates of opposite mating type reveal a recombining history for three presumed asexual species in the filamentous ascomycete genus Alternaria. Mycol Res. 2003;107:169–182. doi: 10.1017/s0953756203007263. [DOI] [PubMed] [Google Scholar]
- 9.Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, et al. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol. 2005;15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 10.Groenewald M, Groenewald JZ, Harrington TC, Abeln ECA, Crous PW. Mating type gene analysis in apparently asexual Cercospora species is suggestive of cryptic sex. Fungal Genet Biol. 2006;43:813–825. doi: 10.1016/j.fgb.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Taylor JW, Jacobson DJ, Fisher MC. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 12.Burt A, Carter DA, Koenig GL, White TJ, Taylor JW. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmons EG. Alternaria themes and variations (22–26). Mycotaxon. 1986;25:287–308. [Google Scholar]
- 14.Yun SH, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life-style from self-sterile ancestors. Proc Natl Acad Sci USA. 1999;96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peever TL, Canihos Y, Olsen L, Ibanez A, Liu YC, et al. Population genetics structure and host specificity of Alternara species causing brown spot of Minneola tangelo and rough lemon in Florida. Phytopathology. 1999;89:851–860. doi: 10.1094/PHYTO.1999.89.10.851. [DOI] [PubMed] [Google Scholar]
- 16.Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 17.Duret L. Evolution of synonymous codon usage in metazoan. Curr Opin Genet Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 18.Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grocock RJ, Sharp PM. Synonymous codon usage in Pseudomonas aeruginosa PA01. Gene. 2002;289:131–139. doi: 10.1016/s0378-1119(02)00503-6. [DOI] [PubMed] [Google Scholar]
- 20.Dong HJ, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 21.Andersson SGE, Sharp PM. Codon usage in the Mycobacterium tuberculosis complex. Microbiology. 1996;142:915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- 22.Kanaya S, Yamada Y, Kudo Y, Ikemura T. Study of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- 23.Lafay B, Atherton JC, Sharp PM. Absence of translationally selected synonymous codon usage bias in Helicobacter pylori. Microbiology. 2000;146:851–860. doi: 10.1099/00221287-146-4-851. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolution models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 25.Rehmeyer CJ, Weixi L, Kusaba M, Farman ML. The telomere-linked helicase (THL) gene family in Magnaporthe oryzae: revised gene structure reveals a novel TLH-specific protein motif. Current Genetics. 2009;55:253–262. doi: 10.1007/s00294-009-0240-3. [DOI] [PubMed] [Google Scholar]
- 26.Nei M, Rogozin IB, Piontkivska H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc Natl Acad Sci USA. 2000;97:10888–71. doi: 10.1073/pnas.97.20.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen R. Molecular signatures of natural selection. Ann Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 28.Duda TF, Palumbi SR. Evolutionary diversification of multigene families: allelic selection of toxins in predatory cone snails. Mol Biol Evol. 2000;17:1286–1293. doi: 10.1093/oxfordjournals.molbev.a026412. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Bos JIB, Armstrong M, Whisson SC, da Cunha L, et al. Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol Biol Evol. 2005;22:659–672. doi: 10.1093/molbev/msi049. [DOI] [PubMed] [Google Scholar]
- 30.Staats M, van Baarlen P, Schouten A, van Kan JAL, Bakker FT. Positive selection in phytotoxic protein-encoding genes of Botrytis species. Fungal Gen Bio. 2007;44:52–63. doi: 10.1016/j.fgb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Stukenbrock EH, McDonald BA. Geographic variation and positive diversifying selection in the host specific toxin SnToxA. Mol Plant Pathol. 2007;8:321–332. doi: 10.1111/j.1364-3703.2007.00396.x. [DOI] [PubMed] [Google Scholar]
- 32.Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. Lateral transfer of mating system in Stemphlium. Proc Natl Acad Sci USA. 2005;102:11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgenstern B. DIALIGN: Multiple DNA and Protein Sequence Alignment at BiBiServ. Nucleic Acids Res. 2004;32:33–36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozas J, Sanchez-DelBarrio JC, Messequer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 35.Watterson G. On the number of segregating site in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 36.Graur D, Li W-H. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- 37.Tajima F. Statistical methods for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Y, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;155:1405–1413. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhauser C. Mathematical models in population genetics. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of Statistical Genetics, 2nd edition. West Sussex, UK: Wiley; 2003. pp. 577–599. [Google Scholar]
- 40.Innan H, Stephan W. The coalescent in an exponentially growing metapopulation and its application to Arabidopsis thaliana. Genetics. 2000;155:2015–2019. doi: 10.1093/genetics/155.4.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and application to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Nielsen R, Goldman N, Pedersen A-MK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 45.Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 46.Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucl Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higgs PG, Ran W. Coevolution of codon usage and tRNA genes leads to alternative stable states of codon usage. Mol Biol Evol. 2008;25:2279–2291. doi: 10.1093/molbev/msn173. [DOI] [PubMed] [Google Scholar]
- 48.Peden JF. Analysis of codon usage. 1999. Ph.D. Thesis, University of Nottingham.
- 49.Wright F. The effective number of codon used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 50.Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucl Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shackelton LA, Parrish CR, Holmes EC. Evolutionary bias of codon usage and nucleotide composition bias in vertebrate DNA virus. J Mol Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 52.Sharp PM, Cowe E. Synonymous codon usage in Saccaromyces cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- 53.Hollander M, Wolfe DA. Nonparametric Statistical Methods, 2nd Edition. Wiley, New York,; 1999. 1999. [Google Scholar]
- 54.O'Gorman CM, Hubert TF, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 55.Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, et al. Mating Type and the Genetic Basis of Self-Fertility in the Model Fungus Aspergillus nidulans. Cur Biol. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Debuchy R. Internuclear recognition: a possible connection between euascomycetes and homobasidiomycetes. Fungal Genet Biol. 1999;27:218–223. doi: 10.1006/fgbi.1999.1142. [DOI] [PubMed] [Google Scholar]
- 57.Zhan J, Torriana SFF, McDonald BA. Significant difference in pathogenicity between MAT1-1 and MAT1-2 isolates in the wheat pathogen Mycosphaerella graminicola. Fungal Genet Biol. 2007;44:339–346. doi: 10.1016/j.fgb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbour L, Xiao W. Mating type regulation of cellular tolerance to DNA damage is specific to the DNA postreplication repair and mutagenesis pathway. Mol Microbiol. 2006;59:637–650. doi: 10.1111/j.1365-2958.2005.04965.x. [DOI] [PubMed] [Google Scholar]
- 60.Yamagishi D, Otani H, Kodama M. G protein signaling mediates developmental processes and pathogenesis of Alternaria alternata. Mol Plant Microb Interac. 2006;19:1280–1288. doi: 10.1094/MPMI-19-1280. [DOI] [PubMed] [Google Scholar]
- 61.O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within Fusarium graminearum clade. Fungal Genet Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Rau D, Attene G, Brown AHD, Nanni L, Maier FJ, et al. Phylogeny and evolution of mating-type genes from Pyrenophora teres, the causal agent of barley “net blotch” disease. Curr Genet. 2007;51:377–392. doi: 10.1007/s00294-007-0126-1. [DOI] [PubMed] [Google Scholar]
- 63.Swanson WJ, Vacquier VD. Extraordinary divergence and positive Darwinian selection in a fusagenic protein coating acrosomal process of abalone spermatozoa. Proc Natl Acad Sci USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metz EC, Palumbi SR. Positive selection and rearrangements generate extensive polymorphism in the gamete recognition protein binding. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 65.Civetta A, Rajakumar SA, Brouwers B, Bacik JP. Rapid evolution and gene-specific patterns of selection for three genes of spermatogenesis in Drosophila. Mol Biol Evol. 2005;23:655–662. doi: 10.1093/molbev/msj074. [DOI] [PubMed] [Google Scholar]
- 66.Wirsel S, Horwitz B, Yamaguchi K, Yoder OC, Turgeon BG. Both mating and fertility are controlled by single mating type specific genes at the Cochliobolus heterostrophus MAT locus. Mol Gen Genet. 1998;259:272–281. doi: 10.1007/s004380050813. [DOI] [PubMed] [Google Scholar]
- 67.Wik L, Karlsson M, Johannesson H. The evolutionary trajectory of the mating-type genes in Neurospora relates to reproductive behavior of taxa. BMC Evolutionary Biology. 2008;8:109–119. doi: 10.1186/1471-2148-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duret L, Mouchiroud D. Determininat of substation rates in mannalian genes: expression pattern affects selection intensity butnot mutation rate. Mol Biol Evol. 1999;17:68–70. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- 69.Genereux D. Evolution of genomic GC variation. Genome Biology. 2002;3:58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolates used in evolutionary analyses.
(DOCX)