Abstract
Type 2 T helper (TH2) cells are critical for the development of allergic immune responses; however, the molecular mechanism controlling their effector function is still largely unclear. Here, we report that the transcription factor NFIL3/E4BP4 regulates cytokine production and effector function by TH2 cells. NFIL3 is highly expressed in TH2 cells but much less in TH1 cells. Production of interleukin (IL)-13 and IL-5 is significantly increased in Nfil3−/− TH2 cells and is decreased by expression of NFIL3 in wild-type TH2 cells. NFIL3 directly binds to and negatively regulates the Il13 gene. In contrast, IL-4 production is decreased in Nfil3−/− TH2 cells. Increased IL-13 and IL-5 together with decreased IL-4 production by antigen-stimulated splenocytes from the immunized Nfil3−/− mice was also observed. The ability of NFIL3 to alter TH2 cytokine production is a T-cell intrinsic effect. Taken together, these data indicate that NFIL3 is a key regulator of TH2 responses.
Keywords: helper T-cell differentiation, IL-13, IL-4, transcriptional regulation
Introduction
Immune responses to certain parasites or those underlying atopy are primarily driven by type 2 helper T (TH2) cells through the production of specific cytokines including interleukin (IL)-4, IL-5, and IL-13. These cytokines have separate but overlapping functions. IL-4 has a role in the regulation of immunoglobulin (Ig) class switching to IgE in B cells, in the activation and proliferation of B cells, and also in promoting TH2 responses. Furthermore, IL-4 upregulates MHC class II on B cells and IgE receptors on inflammatory cells, resulting in the enhancement of antigen presenting ability to T cells and IgE-mediated secretion of inflammatory mediators, respectively (Nelms et al, 1999). IL-13 induces IgE class switching in human B cells and allergic inflammation (Wills-Karp et al, 1998). IL-5 has a central role in eosinophilia by chemoattracting eosinophils and regulating the activation, expansion, and differentiation of eosinophils. IL-5 is produced not only by TH2 cells but also by eosinophils themselves, which serves to autoregulate and augment their accumulation in the airways. Other TH2 cytokines including IL-9 and IL-25 (IL-17E) are also thought to be involved in the pathogenesis of atopic immune responses (Barrett and Austen, 2009; Hamid and Tulic, 2009).
The production of TH2 cytokines is controlled by several transcription factors including STAT6, GATA-3, NFAT, NF-κB, c-Maf, and AP-1 (Li-Weber and Krammer, 2003; Gilmour and Lavender, 2008). STAT6 directly mediates IL-4 signalling leading to the transcriptional activation of target genes (Nelms et al, 1999). Expression of GATA-3 is very low in naive CD4+ T cells but is upregulated during TH2 differentiation (Ho et al, 2009). Studies utilizing mice lacking genes encoding these factors have demonstrated their importance in TH2 cytokine production (Cousins et al, 2008). Recently, Dec2 was also reported as critical for the commitment and promotion of TH2 differentiation (Liu et al, 2009; Yang et al, 2009). These transcription factors are believed to act as positive regulators for TH2 differentiation and TH2 cytokine production. The transcriptional repressor Mina has been shown to repress Il4 gene expression by directly binding to its promoter, thereby, controlling TH2 bias (Okamoto et al, 2009). Under TH1 conditions, IRF1 and IRF2 have been shown to repress Il4 promoter activity (Elser et al, 2002). However, the mechanism of negative regulation by which TH2 differentiation and thus TH2-driven pathologies are prevented is still largely unknown.
To elucidate the regulation of helper T-cell differentiation, many studies have focused on the identification of genes specifically expressed or repressed during the differentiation process. Microarray studies have shown that Nfil3 (nuclear factor, IL-3 regulated, also called E4bp4) expression increases during TH2 differentiation (Chen et al, 2003; Lund et al, 2003, 2005; Lu et al, 2004), suggesting that NFIL3 may be involved in the regulation of TH2 differentiation and effector function. NFIL3 is a basic leucine zipper transcription factor that was identified as a transcriptional activator for the human IL-3 promoter and a transcriptional repressor for the adenovirus E4 promoter (Cowell et al, 1992; Zhang et al, 1995). NFIL3 has structural similarity in its basic region and extended region to PAR (proline and acidic residue-rich) family proteins, and NFIL3 and PAR family protein bind to similar DNA sequences (Cowell, 2002). To date, most studies of NFIL3 have focused on its role in the regulation of circadian rhythm due to its oscillatory pattern of expression in both the suprachiasmatic nucleus and peripheral tissues as well as its association with the regulation of Per2 gene expression (Mitsui et al, 2001; Cowell, 2002; Ueda et al, 2005; Akashi et al, 2006; Ohno et al, 2007). Nfil3 expression is very low in many cell types and is induced in response to cytokines and hormones (Ikushima et al, 1997; Altura et al, 1998; Lang et al, 2002; Chen et al, 2003; Ozkurt and Tetradis, 2003; Sartipy and Loskutoff, 2003; Ramsborg and Papoutsakis, 2007). Nfil3 expression is minimal in immune cells including B cells and T cells, but is strongly induced by IL-4 stimulation in these cells (Chu and Paul, 1998; Schroder et al, 2002; Chen et al, 2003; Kashiwada et al, 2010). Recently, we demonstrated that Nfil3−/− mice showed impaired IgE class switching in B cells, suggesting a role for NFIL3 in IL-4-induced IgE class switching (Kashiwada et al, 2010). Nfil3 is highly expressed in natural killer (NK) cell and NKT cells (Gascoyne et al, 2009; Kamizono et al, 2009). Interestingly, Nfil3−/− mice lack NK cells, suggesting that NFIL3 is critical for NK cell development (Gascoyne et al, 2009; Kamizono et al, 2009; Kashiwada et al, 2010). These observations suggest that NFIL3 has several important roles in the immune system.
To explore the role of NFIL3 in TH2 differentiation and TH2 cytokine production, we examined TH2 cytokine production in Nfil3−/− TH2 cells. We found that Nfil3−/− TH2 cells produced significantly more IL-13 and IL-5, but less IL-4, compared with wild-type (WT) TH2 cells. In addition, we found that NFIL3 directly binds and negatively regulates Il13 gene expression. In contrast, NFIL3 indirectly regulates Il4 gene expression. Immunization of Nfil3−/− mice demonstrated that NFIL3 modulated TH2 responses in vivo. These findings demonstrate that NFIL3 is a critical regulator of TH2 cytokine production by CD4+ T cells.
Results
Reduced IL-4-producing TH2 cells in the absence of NFIL3 in vitro
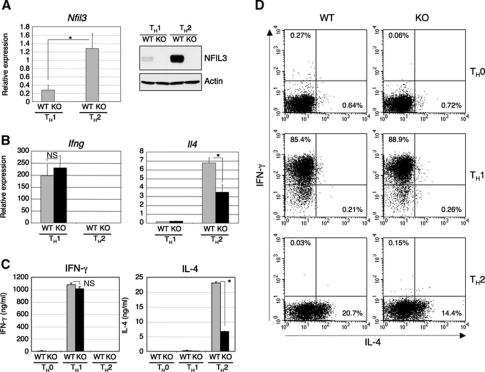
Microarray studies have demonstrated that several sets of genes are differentially expressed during TH1 and TH2 cell differentiation. Expression of Nfil3 gene is induced in both TH1 and TH2 cultures although its expression level in TH2-skewed cells is significantly higher than that in TH1-skewed cells (Chen et al, 2003; Lund et al, 2003, 2005; Lu et al, 2004). This preferential expression of Nfil3 mRNA in TH2 cells suggests a role for NFIL3 in TH2 differentiation and/or TH2 effector function. In order to further evaluate this, we examined the expression of Nfil3 mRNA and NFIL3 protein in TH1- and TH2-skewed cells. Naive CD4+ T cells from the spleen of WT and Nfil3−/− mice were cultured for 7 days under TH1 or TH2 polarizing conditions, and the levels of Nfil3 mRNA and NFIL3 protein were determined by real-time RT–PCR and western blotting, respectively. As expected from the microarray data, both mRNA and protein for NFIL3 were highly expressed in TH2-skewed cells (Figure 1A). Apparent but lower expression of Nfil3 mRNA was also detected in TH9 cells compared with TH2 cells, which also require IL-4 for their differentiation (Dardalhon et al, 2008; Veldhoen et al, 2008), and Nfil3 expression was much lower in other helper T-cell lineages (Supplementary Figure S1A). To assess the role of NFIL3 in TH1/TH2 cell differentiation, we examined TH1/TH2 differentiation of naive CD4+ T cells from Nfil3−/− mice by monitoring the production of IFN-γ and IL-4. Under TH1-skewing conditions, Nfil3−/− T cells expressed levels of Ifng mRNA comparable to WT T cells whereas Il4 mRNA expression in TH2-skewed Nfil3−/− T cells was significantly reduced (Figure 1B). Consistent with these observations, levels of the expression of transcription factors critical for TH1 differentiation were similar between Nfil3−/− and WT T cells (Supplementary Figure S2). IL-4 secretion by TH2-skewed Nfil3−/− T cells was also significantly reduced compared with WT T cells although the secretion of IFN-γ by TH1-skewed cells from WT and Nfil3−/− mice was comparable (Figure 1C). Intracellular cytokine staining also confirmed that TH1 differentiation is not affected by the loss of expression of NFIL3, whereas under TH2-skewing conditions the number of IL-4-producing Nfil3−/− T cells was reduced compared with that of WT T cells (Figure 1D). These results indicate that NFIL3 has a role in the regulation of IL-4 production during TH2 differentiation in vitro.
Figure 1.
Altered IL-4-producing TH2 differentiation in the absence of NFIL3. (A) Expression of Nfil3 mRNA and NFIL3 protein in TH1 and TH2 cells differentiated in vitro. Expression of Nfil3 mRNA was determined by quantitative real-time RT–PCR and data shown are the mean and s.d. from four experiments (left, *P<0.01). Expression of NFIL3 protein was determined by western blot analysis and data are representative of four experiments with similar results (right). (B) Quantitative real-time RT–PCR analysis of Ifng and Il4 mRNA in TH1 and TH2 cells differentiated in vitro. Data show the mean and s.d. from four experiments (n.s., not significant; *P<0.001). (C) Secretion of IFN-γ and IL-4 by TH1 and TH2 cells restimulated with anti-CD3/CD28 for 24 h. Cytokine concentration was determined by ELISA and data show the mean and s.d. from four experiments (n.s., not significant; *P<0.0001). (D) Intracellular staining for IFN-γ and IL-4 of TH0, TH1, and TH2 cells differentiated in vitro. Restimulated cells were stained and analysed by flow cytometry. Data are representative of four experiments with similar results.
T-cell intrinsic NFIL3 regulates TH2 cytokine production in vitro
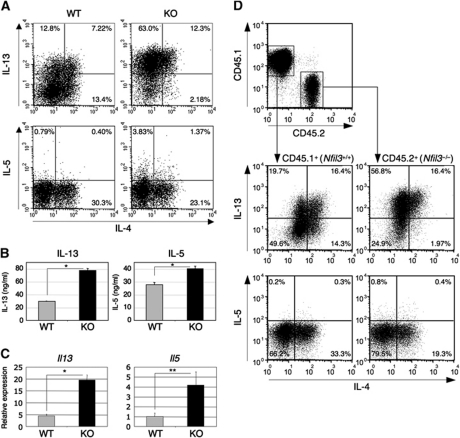
In the absence of NFIL3 expression, IL-4 production was significantly reduced in TH2-skewed cells. The genes encoding the TH2 cytokines, IL-4, IL-13, and IL-5 are located in the same chromosomal region called the TH2 cytokine locus and their expression is coordinately regulated (Wilson et al, 2009). Given the decreased levels of IL-4 expression in the absence of NFIL3, we determined whether the production of IL-13 and IL-5 was similarly affected. Interestingly, the number of IL-13 and IL-5-producing Nfil3−/− cells was significantly increased compared with that of WT cells (Figure 2A). Additionally, the mean fluorescence intensity (MFI) of IL-13+ Nfil3−/− cells was significantly higher than that of WT cells (MFI: 129.2 ± 5.7 for WT cells and 224.8 ± 20.1 for Nfil3−/− cells). The secretion of these cytokines by Nfil3−/− T cells was also increased (Figure 2B), as was the expression of both Il13 and Il5 mRNA (Figure 2C). Interestingly, expression of Il13 in TH1 and TH9 cells was also significantly increased in the absence of Nfil3 expression although Il13 expression in these cells was very low compared with that of TH2 cells. This suggests that NFIL3 regulates Il13 expression in these helper cell subsets (Supplementary Figure S1B). Additionally, under TH2 conditions Nfil3−/− T cells had increased IL-9 production and Il9 mRNA compared with WT T cells, although these levels were extremely low (Supplementary Figure S3A and B). However, when cultured under TH9 conditions, no differences in the production of IL-9 and Il9 mRNA expression were observed between WT and Nfil3−/− cells. Moreover, while IL-10 and IL-3 production was significantly decreased in Nfil3−/− TH2 cells, no difference in IL-10 production was observed between WT and Nfil3−/− TH1 cells (Supplementary Figure S3C–H).
Figure 2.
Altered TH2 cytokine production in Nfil3−/− TH2 cells. (A) Increased IL-13 and IL-5 production in Nfil3−/− TH2 cells. Cytokine production of TH2 cells restimulated with PMA/ionomycin was determined by intracellular staining and analysed by flow cytometry. Data are representative of four experiments with similar results. (B) Increased secretion of IL-13 and IL-5 by Nfil3−/− TH2 cells restimulated with anti-CD3/CD28 for 24 h. Cytokine concentration was determined by ELISA and data show the mean and s.d. from four experiments (*P<0.001). (C) Increased expression of Il13 and Il5 mRNA in Nfil3−/− TH2 cells. Expression of mRNA was determined by quantitative real-time RT–PCR. Data show the mean and s.d. from four experiments (*P<0.0001, **P<0.005). (D) Altered cytokine production in Nfil3−/− TH2 cells is cell intrinsic. Naive CD4+ T cells from WT mice (CD45.1+) and Nfil3−/− mice (CD45.2+) were co-cultured under TH2 conditions for 7 days and cytokine production by cells restimulated with PMA/ionomycin was determined by intracellular staining and analysed by flow cytometry. Data are representative of three experiments with similar results.
Because perturbed IL-4 production might affect IL-13 and IL-5 production in the absence of NFIL3, we determined whether the altered TH2 cytokine production in Nfil3−/− cells was T-cell intrinsic. Naive CD4+ T cells from WT mice (CD45.1) and Nfil3−/− mice (CD45.2) were co-cultured under TH2 condition for 7 days and were examined for TH2 cytokine production. The number of IL-13 and IL-5-producing cells from CD45.2+ Nfil3−/− mice was significantly higher when compared with the cells from CD45.1+ WT mice (Figure 2D). Similarly, the number of IL-4-producing cells was lower from CD45.2+ Nfil3−/− mice compared with the cells from CD45.1+ WT mice. These data suggest that the level of NFIL3 within the T cell regulates TH2 cytokine production. Taken together, these results suggest that NFIL3 specifically regulates cytokine production by TH2 cells in vitro.
Normal IL-4 signalling and proliferation of Nfil3−/− T cells
In the presence of exogenous IL-4 under TH2-skewing condition, Nfil3−/− T cells produced fewer cells that were capable of expressing IL-4. This may be due to altered IL-4 signalling in Nfil3−/− T cells, and thus we examined IL-4 signalling in Nfil3−/− T cells. Splenic CD4+ T cells were stimulated with IL-4, and the activation of STAT6 was examined by flow cytometry. STAT6 activation by IL-4 stimulation occurred in Nfil3−/− T cells at a level comparable to WT cells, suggesting IL-4 signalling in CD4+ T cells is not affected in the absence of NFIL3 expression (Supplementary Figure S4A). We also examined IL-4Rα expression in cells cultured under TH0 and TH2 conditions. Expression of Il4ra gene was similar between Nfil3−/− cells and WT cells subjected to either condition (Supplementary Figure S4B). Thus, Nfil3 deficiency did not alter IL-4 signalling.
IL-4 production of TH2 cells is dependent on cellular proliferation (Gett and Hodgkin, 1998). Thus, the impaired production of IL-4 by Nfil3−/− T cells could be secondary to defective proliferation. Therefore, we next examined the proliferative response of Nfil3−/− CD4+ T cells. In response to CD3/CD28 stimulation, thymidine incorporation of Nfil3−/− CD4+ T cells was comparable to those of WT CD4+ T cells (Supplementary Figure S4C). The addition of IL-4 to this culture showed similar enhancement of proliferation of Nfil3−/− and WT T cells. These results showing normal proliferation suggest the altered TH2 cytokine production of Nfil3−/− CD4+ T cells is not due to a proliferative defect.
We also asked whether IL-4 negatively regulate Il13 and Il5 expression by effector TH2 cells by upregulating the expression of Nfil3. Although the addition of neutralizing anti-IL-4 antibody in the TH2 cell culture reduced Nfil3 expression, expression of Il13 and Il5 genes was not affected (Supplementary Figure S5). This suggests that Il13 and Il5 expression is not altered by IL-4 stimulation of differentiated TH2 cells.
NFIL3 is required for control of Il13 and Il5 at an early stage of TH2 differentiation
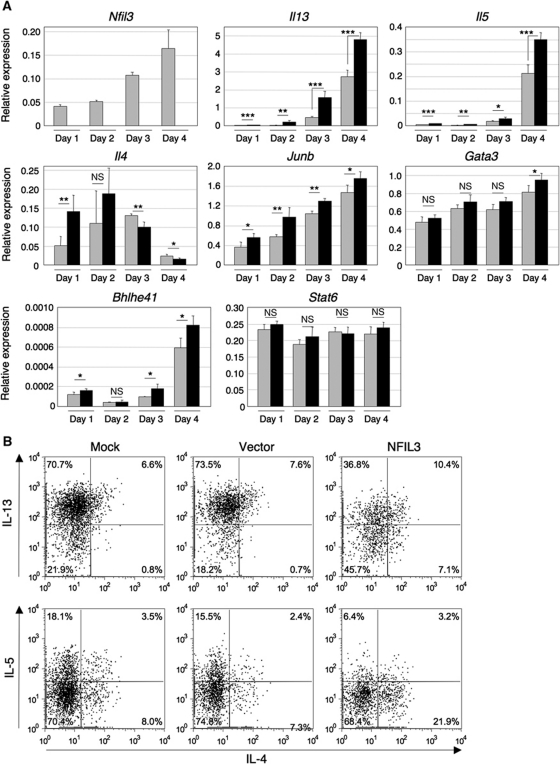
We next assessed whether the abnormalities displayed by TH2 effector cells extended to the earlier stages in TH2 differentiation by examining the expression of cytokine genes and transcription factors essential for TH2 differentiation. Naive CD4+ T cells from Nfil3−/− and WT mice were cultured under TH2 condition for 1–4 days, and gene expression was analysed by real-time RT–PCR. Nfil3 expression increased with time during TH2 differentiation as did the expression of the Il13 and Il5 genes (Figure 3A). Notably, expression of these cytokine genes was significantly higher in Nfil3−/− cells compared with WT cells at all time points, suggesting that NFIL3 negatively regulates these genes from the very early stages of TH2 differentiation (Figure 3A). Interestingly, early expression of the Il4 gene was higher in Nfil3−/− T cells than that of WT T cells. Consistent with this observation, Nfil3−/− cells cultured under neutral conditions also expressed more Il4 mRNA than WT cells (Supplementary Figure S6). Gene expression of the TH2 transcription factors Gata3, Junb, Bhlhe41 (which encodes Dec2), and Stat6, was comparable between WT and Nfil3−/− T cells (Figure 3A). GATA-3 is the master transcription factor for TH2 differentiation. Therefore, we examined whether GATA-3 is required for the expression of Nfil3 gene in response to IL-4. Efficient knockdown of GATA-3 expression did not alter Nfil3 expression in response to IL-4, indicating that GATA-3 is not required for the expression of IL-4-induced Nfil3 gene (Supplementary Figure S7). Taken together, these results suggest that NFIL3 is involved in the regulation of cytokine production but not in the regulation of transcription factors induced during the early stages of TH2 differentiation.
Figure 3.
Role of NFIL3 expression at an early stage of TH2 differentiation. (A) Required expression of Nfil3 at an early stage of TH2 differentiation for the normal expression of Il13 and Il5 genes. Naive CD4+ T cells from WT (grey bar) and Nfil3−/− (closed bar) mice were cultured under TH2 conditions up to 4 days and RNA was prepared for quantitative RT–PCR. Relative expression of each gene was normalized by the expression of Hprt1 mRNA. Data show the mean and s.d. from four experiments (NS, not significant; *P<0.05, **P<0.01, ***P<0.001) (B) Introduction of NFIL3 into Nfil3−/− T cells at an early time point of TH2 differentiation restores the impairment of TH2 cytokine production. CD4+ T cells under TH2 conditions for 30 h were infected with retroviruses carrying NFIL3 or vector (pMiT). Infected cells were cultured for an additional 6 days under TH2 conditions and then restimulated with PMA/ionomycin in the presence of Brefeldin A to examine cytokine production by intracellular staining. Data are representative of three experiments with similar results.
Next, we asked whether the restoration of NFIL3 expression in Nfil3−/− T cells during TH2 differentiation is sufficient to induce normal TH2 cytokine production. We expressed NFIL3 in Nfil3−/− naive CD4+ T cells cultured under TH2 condition using retroviral transduction. Six days after infection, transduced cells were analysed for TH2 cytokine production by flow cytometry. Transduction of NFIL3 into Nfil3−/− CD4+ T cells resulted in increased IL-4 production and decreased IL-13 and IL-5 production in comparison to Nfil3−/− CD4+ T cells transduced by empty vector (Figure 3B). These results indicate that the induced NFIL3 expression seen at early stages of TH2 differentiation is required for the normal cytokine production during TH2 differentiation.
We also expressed NFIL3 during the later stage of TH2 differentiation. Similar to the transduction into the early stage of TH2 differentiation, transduction of NFIL3 resulted in decreased IL-13 production, but the effect of NFIL3 transduction on IL-5 production was less clear (Supplementary Figure S8A). Surprisingly, IL-4 production was also decreased by NFIL3 transduction in contrast to NFIL3 transduction at the early stage. Furthermore, we examined the effect of knockdown of NFIL3 expression in polarized TH2 cells. Consistent with NFIL3 transduction, knockdown of NFIL3 expression resulted in increased IL-13 and IL-4 production (Supplementary Figure S8B). These results suggest that altered expression of NFIL3 in polarized TH2 cells may affect IL-4 production in a different manner to that in the early stage of TH2 differentiation.
NFIL3 regulates TH2 response in vivo
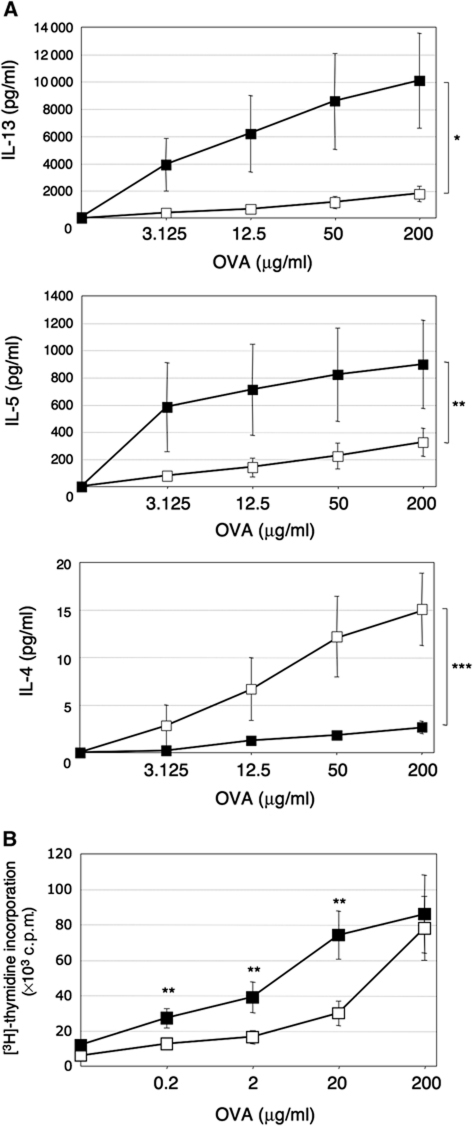
In order to determine whether NFIL3 regulates TH2 responses in vivo, mice were immunized with ovalbumin (OVA) adsorbed to alum, and cytokine production by splenic T cells after restimulation with OVA was examined by ELISA. In response to OVA restimulation, IL-13 and IL-5 production by Nfil3−/− cells was significantly increased compared with WT cells (Figure 4A). In contrast, IL-4 production was decreased in Nfil3−/− mice. These observations are consistent with cytokine production observed by TH2 cells differentiated in vitro (Figures 1 and 2). Thus, these results suggest that NFIL3 regulates TH2 cytokine production in vivo.
Figure 4.
NFIL3 regulates TH2 response in vivo. Mice (n=4 for each genotypes, WT mice: open square, Nfil3−/− mice: closed square) were immunized with OVA plus alum. After 6 days, splenocytes were prepared and cultured in the presence of OVA for 3 days. (A) Increased IL-13 and IL-5 but decreased IL-4 production in response to OVA in Nfil3−/− mice. Supernatants were harvested and secreted TH2 cytokines were examined by ELISA (*P=0.078, **P=0.16, ***P<0.05). (B) Increased OVA-specific cell proliferation in Nfil3−/− mice. Proliferation was measured during the last 16 h by measurement of thymidine incorporation in triplicate. Data show the mean and s.e. from four mice in each group and representative of two independent experiments (**P<0.05).
We also examined OVA-specific proliferation of the cells from OVA-immunized mice. Nfil3−/− cells were hyperproliferative in response to OVA at lower concentration in comparison to WT cells (Figure 4B). However, freshly isolated CD4+ T cells from unimmunized Nfil3−/− and WT mice showed comparable proliferation in response to CD3 stimulation (Supplementary Figure S4C). These results may implicate the role of Nfil3 in the antigen-specific cell expansion leading to the efficient desired effector function such as cytokine production.
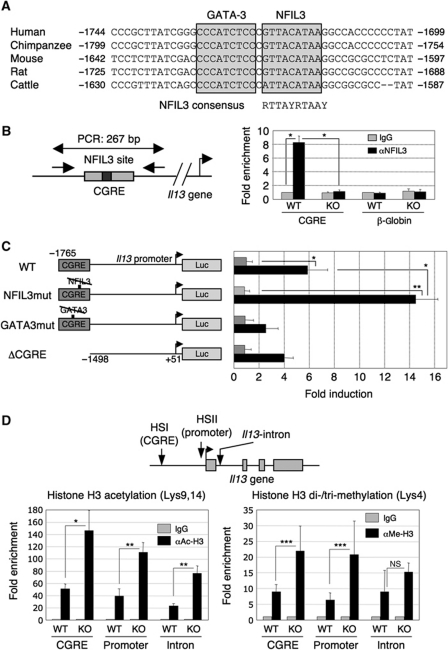
NFIL3 directly binds to CGRE region and regulates IL-13 gene expression
In TH2 cells, both Il13 mRNA expression and IL-13 production were increased in the absence of NFIL3 (Figure 2), suggesting that NFIL3 may directly regulate the Il13 gene. Therefore, we searched the mammalian Il13 gene locus for NFIL3-binding sites using the TRANSFAC program (Matys et al, 2003). We found an evolutionarily conserved consensus sequence for NFIL3 binding 1.6–1.7 kb upstream of the first exon of the Il13 gene (Figure 5A). Interestingly, this putative NFIL3-binding sequence is located next to the GATA-3-binding site in the CGRE/HS1/CS1 region, which was identified as a DNase I hypersensitivity site and corresponds to the 5′ border of the histone acetylation region in TH2 cells (Agarwal and Rao, 1998; Kishikawa et al, 2001; Yamashita et al, 2002).
Figure 5.
NFIL3 binds to CGRE and suppresses Il13 gene expression. (A) Sequence alignment around the putative NFIL3-binding site in the CGRE region. GATA-3 and NFIL3-binding sequences are boxed and the consensus NFIL3-binding sequence is shown. The numbers of positions relative to the transcriptional start site (human, mouse, and cattle) or the translational start site (chimpanzee and rat) are indicated, respectively. (B) NFIL3 binds to CGRE in vivo. The region amplified by PCR and the primers are indicated (left). Splenic CD4 T cells from WT and Nfil3−/− mice cultured under TH2 conditions for 7 days were crosslinked and soluble chromatin complexes were immunoprecipitated by anti-NFIL3 antibody or control IgG. The region including CGRE in the co-precipitated DNA was amplified by PCR. The β-globin gene was used as a negative control. The average and s.d. of enrichment from four experiments were indicated (right). *P<0.0001. (C) Negative regulation of Il13 gene transcription by NFIL3. The reporter constructs of WT, NFIL3-binding mutant, GATA-3-binding mutant, and CGRE-deletion mutant used are shown (left). The TH2 cell line, D10.G4.1, was transfected with the indicated reporter constructs and stimulated with anti-CD3/CD28 antibodies for 48 h. Cell lysates were subjected to luciferase assay. Five experiments were performed with similar results. *P<0.01, **P<0.001. (D) Chromatin modification in the Il13 gene locus. The region amplified by PCR indicated (top). Splenic CD4 T cells from WT and Nfil3−/− mice cultured under TH2 conditions for 7 days were crosslinked and soluble chromatin complexes were immunoprecipitated by anti-acetyl Histone H3, anti-di + tri-methyl Histone H3, or control IgG. The region including the CGRE, promoter region, and first intron in the co-precipitated DNA were amplified by PCR. The average and s.d. of enrichment from four experiments are indicated (*P<0.005, **P<0.001, ***P<0.05; NS, not significant).
We first determined whether NFIL3 binds to this sequence in vivo. We performed chromatin immunoprecipitation (ChIP) analysis by amplifying the CGRE region of WT and Nfil3−/− TH2 cells. ChIP analysis of WT but not Nfil3−/− TH2 cells clearly demonstrated a significant enrichment of NFIL3 binding to the CGRE region upstream of Il13 gene (Figure 5B). No enrichment of NFIL3 binding to β-globin gene was observed as a negative control. Therefore, NFIL3 specifically binds to the CGRE region in vivo. Because the NFIL3-binding site and GATA-3-binding site are very close it is possible that binding of NFIL3 to the CGRE could interfere with GATA-3 binding to the CGRE. However, the level of GATA-3 binding to the CGRE was not affected in Nfil3−/− TH2 cells compared with WT cells (Supplementary Figure S9).
Next, we asked whether NFIL3 binding to the CGRE region is functionally relevant for Il13 gene transcription. D10.G4.1 cells were transfected with a CGRE-containing 1.8 kb promoter-luciferase construct (WT), NFIL3-binding site mutant construct (NFIL3mut) (Ozkurt and Tetradis, 2003), GATA-3-binding site mutant construct (GATA3mut) (Yamashita et al, 2002) or CGRE-deletion mutant construct (ΔCGRE), followed by CD3/CD28 stimulation for 48 h, after which cell lysates were subjected to luciferase assay (Figure 5C, left). CD3/CD28 stimulation induced six-fold transcriptional activation of the WT-reporter gene (Figure 5C, right). Notably, mutation of the NFIL3-binding site significantly enhanced transcriptional activity (14-fold). On the other hand, mutation in the GATA-3-binding site and CGRE deletion diminished transcriptional activity (two-fold and four-fold, respectively). These results suggest that the NFIL3-binding site in the CGRE region functions to regulate Il13 transcription.
Previous studies have demonstrated that epigenetic changes are observed in the Il13 gene locus during TH2 differentiation (Yamashita et al, 2002; Baguet and Bix, 2004). Therefore, we examined histone modification at the Il13 locus in the absence of NFIL3 expression by ChIP analysis. Interestingly, Nfil3−/− TH2 cells showed increased histone H3 acetylation in the Il13 gene locus including the CGRE region, promoter region, and the first intron sequence compared with WT TH2 cells (Figure 5D, left). In addition, Nfil3−/− TH2 cells also showed increased histone H3 methylation at lysine 4 in the Il13 locus compared with WT TH2 cells (Figure 5D, right). These changes of chromatin modification are strongly correlated with active transcription of the affected genes (Fischle et al, 2003). Thus, NFIL3 may be involved in the regulation of epigenetic changes in the Il13 locus leading to altered transcription of the Il13 gene.
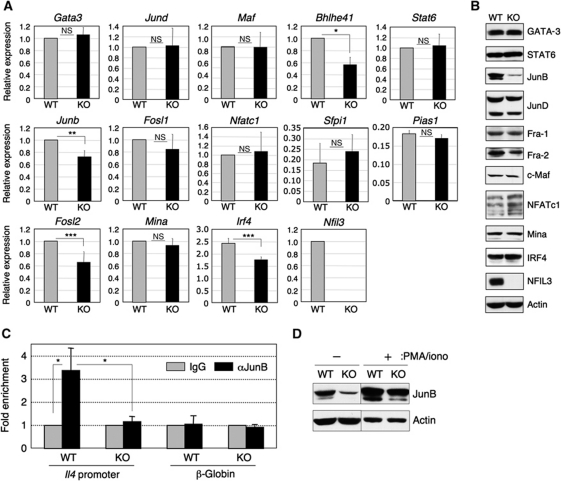
NFIL3 regulates the levels of AP-1
As we could not identify a likely NFIL3-binding site within the Il4 gene locus, we hypothesized that NFIL3 regulation of Il4 expression might be indirect. In order to assess this possibility, we examined the expression levels of transcription factors involved in the regulation of Il4 gene expression. Among these, expression of the Junb, Fosl2, Bmhle41 genes were slightly reduced in Nfil3−/− TH2 cells (Figure 6A). Both JunB and Fra-2 are components of the AP-1 complex in TH2 cells (Rooney et al, 1995; Li et al, 1999). At the protein level, JunB was markedly reduced in Nfil3−/− TH2 cells (Figure 6B). Expression of Fra-2 protein was also slightly reduced. These data suggest that diminished upregulation of JunB and Fra-2 might be the mechanism by which NFIL3 deficiency causes reduced Il4 expression in TH2 cells. Next, we tested this hypothesis through assessment of the level of JunB protein bound to the Il4 promoter by ChIP analysis. As predicted, ChIP analysis demonstrated significantly reduced JunB binding to the Il4 promoter region in Nfil3−/− TH2 cells compared with WT TH2 cells following stimulation with PMA/ionomycin (Figure 6C). A small level of JunB binding to the Il4 promoter in WT but not Nfil3−/− TH1 cells was observed (Supplementary Figure S10) likely because small amounts of JunB protein is also detectable in TH1 cells (Li et al, 1999). We confirmed the lower JunB protein level in both unstimulated and PMA/ionomycin-stimulated Nfil3−/− TH2 cells in comparison to the levels in WT TH2 cells (Figure 6D). We also asked whether the reduced JunB binding to the Il4 promoter in the absence of NFIL3 alters histone modification. The levels of both histone H3 acetylation and methylation were similar between WT TH2 cells and Nfil3−/− TH2 cells, indicating that JunB reduction in the absence of NFIL3 results in downregulation of Il4 transcription independent of histone modification (Supplementary Figure S11). Last, we examined whether transduction of JunB into Nfil3−/− TH2 cells restores reduced IL-4 production. Interestingly, transduction of JunB into Nfil3−/− TH2 cells could not rescue the impairment of IL-4 production (Supplementary Figure S12). This observation suggests that transduction of JunB alone is not sufficient to rescue IL-4 production.
Figure 6.
Indirect regulation of Il4 gene expression by NFIL3. (A) Gene expression levels of transcription factors involved in the regulation of Il4 gene expression. Real-time RT–PCR analysis for the genes listed in Nfil3−/− and WT cells cultured under TH2 condition for 7 days was performed. Relative expression of each gene was normalized to the expression of Hprt1 mRNA and WT value was set as 1. Data show the mean and s.d. from four experiments (*P<0.001, **P<0.002, ***P<0.01). (B) Protein expression levels of transcription factors listed in (A). Cell lysates from the cells above were subjected to western blot analysis. At least four experiments were performed with similar results. (C) Reduced JunB binding to the Il4 promoter in Nfil3−/− TH2 cells (left). Nfil3−/− and WT TH2 cells were restimulated with PMA/ionomycin for 4 h and were crosslinked and soluble chromatin complexes were immunoprecipitated by anti-JunB antibody or control IgG. Il4 promoter region in the co-precipitated DNA were amplified by PCR. The β-globin gene was used as a negative control. The average and s.d. of enrichment from three experiments are indicated (*P<0.05). (D) Reduced JunB expression in Nfil3−/− cells both before and after PMA/ionomycin restimulation. Cells were unstimulated or stimulated with PMA/ionomycin for 4 h and prepared cell lysates for western blot analysis. Data are representative from more than five independent experiments and two independent experiments for the unstimulated and PMA/ionomycin-stimulated samples, respectively. The lanes are from the same blot.
Discussion
There have been extensive studies over the past two decades focused on elucidating the mechanisms that regulate helper T-cell differentiation and effector function. Specific cytokines direct naive CD4+ T cells to induce the required lineage-specific transcription factors at the appropriate stage of lineage specification in the context of T-cell receptor (TCR) signalling. For TH2 differentiation, IL-4 is critical for initiating, as well as maintaining, TH2 phenotypes. Indeed, TCR-stimulated naive CD4+ T cells produce IL-4, which stimulates these T cells by positive autocrine feedback. IL-4 signalling induces STAT6 activation, which is constitutively expressed, leading to GATA-3 expression. GATA-3 has a critical role in the chromatin remodelling of the TH2 cytokine locus, driving the commitment to TH2 differentiation (Ansel et al, 2006; Lee et al, 2006). The collaboration of GATA-3 with other transcription factors including JunB, c-Maf, NFATc, and AP-1 facilitates TH2 differentiation and TH2 cytokine production. However, dysregulated or inappropriate production of these TH2 cytokines leads to pathogenic allergic immune responses. Our data described here suggest that NFIL3 may be a key regulator of normal cytokine production.
NFIL3 expression in TH2 cells is much higher when compared with TH1 cells, implicating a role for NFIL3 in TH2-mediated responses, such as allergic inflammation. We demonstrated that NFIL3 functions as a negative regulator of IL-13 and IL-5 production and as a positive regulator of IL-4 production under TH2 condition in vitro and in vivo. A previous report describing a putative NFIL3-binding site in the proximal promoter of Ifng gene suggests a potential role for NFIL3 in TH1 cytokine production (Zhang et al, 1995). Despite this implication TH1 differentiation of Nfil3−/− T cells appeared normal as determined by the expression and production of IFN-γ and transcription factors. However, Il13 expression in Nfil3−/− cells was slightly increased compared with WT cells under TH1 conditions (Supplementary Figure S1B), suggesting that NFIL3 may also contribute to negative regulation of the Il13 gene in TH1 cells. Moreover, expression of Nfil3 in TH9 cells, which is induced by IL-4 plus TGF-β from naive CD4 T cells, was higher than TH1 cells, and Nfil3−/− TH9 cells highly expressed Il13 gene compared with WT cells (Supplementary Figure S1A and B). These observations suggest that NFIL3 regulates expression of the Il13 gene not only in TH2 cells but also TH1 and TH9 cells.
How does NFIL3 regulate Il13 gene expression? Our analysis demonstrated that NFIL3 directly bound to the CGRE region and negatively regulated Il13 gene expression. Enhanced Il13 expression in Nfil3−/− TH2 cells was observed not only in the differentiated TH2 cells but also at the very early stages of TH2 differentiation. Interestingly, the CGRE region corresponds to the 5′ border of the histone acetylation region in TH2 cells (Yamashita et al, 2002). The NFIL3-binding site is very close to the GATA-3-binding site in the CGRE region. However, the level of GATA-3 binding to the CGRE was similar between WT and Nfil3−/− TH2 cells, suggesting that NFIL3 regulated Il13 gene expression without affecting GATA-3 activity in TH2 cells. Moreover, epigenetic analysis revealed increased levels of histone acetylation and methylation leading to transcriptional activation in Nfil3−/− TH2 cells compared with WT TH2 cells. Therefore, NFIL3 may contribute to the chromatin remodelling on the Il13 gene locus.
Similarly to IL-13 production, we also showed increased IL-5 production in Nfil3−/− TH2 cells and OVA-immunized mice. Although we could not find putative NFIL3-binding sites in the Il5 gene locus, preliminary experiments showed increased acetylation and methylation of histone H3 in the Il5 gene locus in Nfil3−/− TH2 cells compared with WT TH2 cells (data not shown). These chromatin modifications were correlated with the transcription level of the Il5 gene in TH2 cells from Nfil3−/− and WT mice. Thus, NFIL3 may regulate Il5 gene expression either indirectly through controlling other transcription factors involved in the regulation of Il5 gene transcription, or directly through intrachromosomal interaction between the Il13 promoter region containing CGRE and the Il5 gene. Indeed, the Il5 promoter region has been shown to co-localize with the Il13 promoter region in TH2 cells (Spilianakis and Flavell, 2004; Cai et al, 2006), and the CGRE functions as an enhancer if fused to the Il5 promoter (Yamashita et al, 2002). By elucidating the mechanism of Il13 and Il5 regulation by NFIL3, we could identify putative targets for therapeutic intervention to control allergic immune responses. Although a coordinate regulation of the TH2 cytokine locus to express Il4, Il5, and Il13 genes is well documented, discordant expression of these cytokine genes has also been demonstrated (Kishikawa et al, 2001; Wilson et al, 2009). Our data indicate that the IL-13+IL-4− population is dramatically increased in the absence of NFIL3, and that NFIL3 regulates Il13 and Il5 gene expression but not Il4 gene expression at very early stages of TH2 differentiation, suggesting that NFIL3 may differentially regulate the expression of these cytokines independent of the coordinated TH2 cytokine locus accessibility. These observations may explain why there is heterogeneity in the expression of other TH2 cytokine genes in the individual TH2 cells or clones (Bucy et al, 1995; Kelso et al, 1999; Kishikawa et al, 2001). It has been shown that expression of PU.1, IRF4, and Pias1 contributes to TH2 heterogeneity (Chang et al, 2005; Zhao et al, 2007; Ahyi et al, 2009). However, we could not find a potential role for NFIL3 in regulating these genes, suggesting that NFIL3 may not be involved in the regulation of these transcriptional regulators. How NFIL3 contributes to TH2 heterogeneity needs to be elucidated.
In contrast to the differentiated TH2 cells, early expression of IL-4 by Nfil3−/− T cells under TH2 condition and neutral condition were higher than that of WT T cells. Initial expression of the Il4 gene is induced by TCR stimulation, and this induction is independent on GATA-3 (Ansel et al, 2006). Under these conditions, expression of Junb and Gata3 in Nfil3−/− T cells was similar to that of WT T cells, suggesting that NFIL3 may regulate Il4 expression in the different mechanisms between the early stage of TH2 differentiation and TH2-polarized cells.
We have shown a correlation between reduced Il4 expression and a significant reduction of JunB protein in Nfil3−/− T cells. This marked decrease in JunB protein was found in the context of only a modest decrease in Junb expression. These observations have led us to speculate that NFIL3 may regulate both Junb expression and perhaps JunB protein stability. Importantly, JunB transduction into Nfil3−/− TH2 cells did not restore reduced IL-4 production (Supplementary Figure S12). Therefore, JunB alone may be insufficient to rescue IL-4 production. Alternately, JunB protein may be unstable in the absence of NFIL3. It has been demonstrated that JunB protein stability was controlled by Itch, an E3 ubiquitin ligase, whose activity is regulated by Ndfip1 (Fang et al, 2002; Oliver et al, 2006). It is also known that transcriptional activity of JunB is regulated by SUMOylation in T cells (Garaude et al, 2008). SUMOylation is one of the post-translational modifications with SUMO protein that, unlike ubiquitin, modifies the target protein's function but dose not cause protein degradation. However, how JunB stability is regulated is still unknown. Therefore, the mechanism by which NFIL3 modifies the JunB protein level needs to be elucidated.
Similar to the increased Il13 and Il5 expression, Il9 expression was also increased in Nfil3−/− TH2 cells. In contrast, reduced Il10 and Il3 expression (along with Il4) was observed in Nfil3−/− TH2 cells. NFIL3 was previously identified as a transcriptional activator for human IL3 promoter (Zhang et al, 1995). Our data show that Nfil3−/− TH2 cells produced decreased Il3 expression and IL-3 production, suggesting that NFIL3 directly regulates Il3 transcription. Currently, the molecular mechanisms by which NFIL3 regulates the expression of Il9 and Il10 genes are unclear.
In summary, we demonstrated that NFIL3 has a critical role in the TH2 cytokine gene expression. Previously, we have also demonstrated that NFIL3 controls IgE class switching. In addition, NFIL3 controls NK cell development. Therefore, NFIL3 is a pleiotropic transcriptional regulator in both acquired immunity and innate immunity.
Materials and methods
Mice and cell line
Generation of Nfil3−/− mice was previously described (Kashiwada et al, 2010). Nfil3−/− mice backcrossed with C57BL/6 for at least 10 generations were used for all experiments. CD45.1+ C57BL/6 mice were obtained from the Jackson Laboratory. All mice were bred and maintained under specific pathogen-free conditions. All experimental mouse protocols were adhered to Institutional Animal Care and Use Committee guidelines and were approved by the IACUC of University of Iowa. The TH2 cell line, D10.G4.1, was maintained in complete RPMI1640 medium containing sodium pyruvate, non-essential amino acids and IL-2 (5 ng/ml). D10.G4.1 were stimulated with conalbumin every 2 weeks and used 2 weeks after stimulation.
In vitro helper T-cell differentiation
Naive CD4+ T cells (CD4+CD62L+CD25−) purified with CD4+CD62L+ T-Cell Isolation Kit II (Miltenyi) were cultured with plate-bound anti-CD3 (10 μg/ml) and soluble anti-CD28 (5 μg/ml) in the presence of the following cytokines and antibodies; IL-12 (20 ng/ml), anti-IL-4 (5 ng/ml), IL-2 (20 ng/ml) for TH1 differentiation; IL-4 (20 ng/ml), anti-IFN-γ (5 ng/ml), IL-2 (20 ng/ml) for TH2 differentiation; anti-IFN-γ (5 ng/ml), anti-IL-4 (5 ng/ml), IL-2 (20 ng/ml) for TH0 condition; IL-4 (20 ng/ml), TGF-β (5 ng/ml), anti-IFN-γ (5 ng/ml), IL-2 (20 ng/ml) for TH9 differentiation; TGF-β (5 ng/ml), anti-IFN-γ (5 ng/ml), anti-IL-4 (5 ng/ml) for iTreg differentiation; IL-6 (10 ng/ml), TGF-β (5 ng/ml), anti-IFN-γ (5 ng/ml), anti-IL-4 (5 ng/ml) for TH17 differentiation; 1α, 25-dihydroxyvitamin D3 (4 × 10−8 M; Sigma), dexamethasone (5 × 10−8M; Sigma) for Tr1 differentiation. All recombinant cytokines except IL-4 (BD Bioscience) were purchased from R&D Systems. All antibodies for helper T-cell differentiation were purchased from eBioscience.
Western blotting
Preparation of cell lysates and western blot analysis was performed as described previously (Kashiwada et al, 2010). Briefly, cell lysates prepared in lysis buffer (50 mM Tris–HCl, pH 8.0, 1% Nonidet P-40, 150 mM NaCl with protease inhibitor cocktail obtained from Roche) were subjected to SDS–PAGE and transferred onto Immobilon™ PVDF membrane (Millipore). Membranes were probed with the antibodies indicated, and detected with ECL detection system (Amersham). Antibodies used are anti-NFIL3, STAT6, NFATc1, IRF4 (Santa Cruz), GATA-3 (eBioscience), actin (Sigma), JunB (Cell Signaling Technology), JunD (R&D Systems), Mina (Proteintech Group), Fra-1 (Aviva Systems Biology), Fra-2 (Bioworld Technology), and c-Maf (Novus Biologicals).
Real-time RT–PCR
RNA was prepared from unstimulated cells or cells stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml) for 4 h, and cDNA was synthesized using the SuperScript® First-Strand Synthesis System (Invitrogen). PCR analysis of mRNA expression was performed using SYBR® GREEN PCR Master Mix on the 7900HT Fast Real-Time PCR System (Applied Biosystems). The expression levels of each gene were normalized to the expression of Hprt1. The primers used are listed in Supplementary Table 1.
Detection of cytokine by ELISA
After a 7-day culture for differentiation, cells (1 × 106 cells) were restimulated with plate-bound anti-CD3 (10 μg/ml) and soluble anti-CD28 (5 μg/ml) for 24 h, and then the culture supernatants were harvested. The concentration of cytokines was measured by ELISA. All cytokine antibodies were obtained from eBioscience, and alkaline phosphatase-conjugated avidin was obtained from BD Biosciences.
Intracellular staining
After a 7-day culture for differentiation, cells were restimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml) for 4 h in the presence of Brefeldin A (eBioscience). After stimulation, cells were stained with anti-CD4, CD45.1, CD45.2, and anti-Thy1.1 for virus-infected cells, fixed, permeabilized, and then stained with antibodies for the cytokines indicated. For detection of phospho-STAT6, cells were stimulated with IL-4 for 20 min, then fixed, permeabilized, and then stained with anti-phospho-STAT6 antibody (BD Biosciences). The stained cells were analysed with LSR II and CellQuest (BD Biosciences). All antibodies for cytokine detection were obtained from eBioscience except the anti-IL-9 antibody (Biolegend).
Viral infection
Retroviral construct for NFIL3 in pMiT was described previously (Kashiwada et al, 2010). The lentiviral construct for JunB was generated by cloning JunB cDNA into pCDH-Thy1.1, which was generated by inserting an IRES-Thy1.1 fragment from pMiT into pCDH-CMV-MCS-EF1-copGFP (System Biosciences). The shRNA constructs for the Nfil3 gene were generated using pLKO.1-Thy1.1 constructed by replacing Puror gene in pLKO.1 (Addgene) with a Thy1.1 fragment. The scramble shRNA construct was obtained from Addgene. For infection at the early stage of TH2 differentiation, naive CD4+ T cells were cultured under TH2 conditions overnight, and then activated cells were spin infected with the viruses produced by transient transfection of viral constructs into 293T cells. After infection, cells were grown under TH2 conditions for another 6 days before being subjected to assays. For infection at the late stage of TH2 differentiation, Nfil3−/− CD4+ T cells cultured for 6 days under TH2 condition were spin infected with viruses and cultured for 2 days. For Nfil3 knockdown experiments, WT CD4+ T cells cultured for 6 days under TH2 condition were spin infected with shRNA viruses, and cultured for 3 days. Infected cells were identified as Thy1.1+ cells by flow cytometry. For Gata3 knockdown experiments, D10.G4.1 cells were spin infected with lentivirus produced by transient transfection of viral constructs into 293T cells. The shRNA constructs for the Gata3 gene were kindly provided by Dr Soumen Paul (University of Kansas Medical Center, KS) (Home et al, 2009).
Luciferase assay
Luciferase-reporter plasmids were generated with pGL2-basic (Promega) and mutations were introduced using QuikChange Site-Directed Mutagenesis Kit (Stratagene) with the primers as follows: for the NFIL3-binding mutant, 5′-CATCTCCCGTTACATTAGGCCGCGCCTCTAT-3′ and 5′-ATAGAGGCGCGGCCTAATGTAACGGGAGATG-3′, for the GATA-3-binding mutant, 5′-TCCTCTTATCGACCCAAAATCCCGTTACATAAGG-3′ and 5′-CCTTATGTAACGGGATTTTGGGTCGATAAGAGGA-3′. D10.G4.1 cells were transiently transfected by electroporation with the reporter gene constructs in pGL2 and pRL-TK (Promega) as an internal control. Cells were cultured with or without plate-bound anti-CD3 (10 μg/ml) and soluble anti-CD28 (5 μg/ml). After 48 h of transfection, the Dual-Reporter Luciferase Assay® system (Promega) was used for the preparation of cell lysates and subsequent assays.
Proliferation assay
Naive CD4+ T cells were seeded at a density of 2 × 105 cells per well in 96-well plates in complete RPMI 1640 with varying final concentrations of plate-bound anti-CD3 (0.3, 1, and 3 μg/ml), soluble anti-CD28 (5 μg/ml), mIL-4 (20 ng/ml) as indicated in triplicate. One μCi of [3H]-thymidine (Perkin-Elmer) was added 40 h after stimulation followed by incubation for 8 h before analysis. For OVA-specific proliferation, splenocytes were cultured in the presence of OVA at the indicated concentration for 56 h followed by incubation with 1 μCi of [3H]-thymidine for 16 h before analysis. [3H]-thymidine incorporation was measured with a liquid scintillation counter.
Chromatin immunoprecipitation
ChIP assay was performed as described previously (Kashiwada et al, 2010). Briefly, cells were crosslinked with formaldehyde and chromatin was fragmented by sonication. Chromatin was immunoprecipitated with the antibodies indicated or control IgG, and then purified co-precipitated DNA was quantified by real-time PCR as above. Data are normalized to input Ct values and indicated as fold enrichment relative to the values for control IgG. The primers used are listed in Supplementary Table 1. Antibodies used for IP are anti-NFIL3, JunB, GATA-3 (Santa Cruz), anti-acetyl Histone H3 (Millipore), and anti-di+tri-methyl Histone H3 (Abcam).
Statistical analysis
Statistical significance was determined using a two-tailed Student's t-test.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant R01 AI54821 (to PBR). Author contributions: MK and SLC performed the experiments; MK and JDC analysed the data; MK, JDC, and PBR designed the experiments; and MK and PBR wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agarwal S, Rao A (1998) Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9: 765–775 [DOI] [PubMed] [Google Scholar]
- Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH (2009) IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol 183: 1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Ichise T, Mamine T, Takumi T (2006) Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol Biol Cell 17: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altura RA, Inukai T, Ashmun RA, Zambetti GP, Roussel MF, Look AT (1998) The chimeric E2A-HLF transcription factor abrogates p53-induced apoptosis in myeloid leukemia cells. Blood 92: 1397–1405 [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24: 607–656 [DOI] [PubMed] [Google Scholar]
- Baguet A, Bix M (2004) Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc Natl Acad Sci USA 101: 11410–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett NA, Austen KF (2009) Innate cells and T helper 2 cell immunity in airway inflammation. Immunity 31: 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucy RP, Karr L, Huang GQ, Li J, Carter D, Honjo K, Lemons JA, Murphy KM, Weaver CT (1995) Single cell analysis of cytokine gene coexpression during CD4+ T-cell phenotype development. Proc Natl Acad Sci USA 92: 7565–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH (2005) PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity 22: 693–703 [DOI] [PubMed] [Google Scholar]
- Chen Z, Lund R, Aittokallio T, Kosonen M, Nevalainen O, Lahesmaa R (2003) Identification of novel IL-4/Stat6-regulated genes in T lymphocytes. J Immunol 171: 3627–3635 [DOI] [PubMed] [Google Scholar]
- Chu CC, Paul WE (1998) Expressed genes in interleukin-4 treated B cells identified by cDNA representational difference analysis. Mol Immunol 35: 487–502 [DOI] [PubMed] [Google Scholar]
- Cousins DJ, McDonald J, Lee TH (2008) Therapeutic approaches for control of transcription factors in allergic disease. J Allergy Clin Immunol 121: 803–809; quiz 810–801 [DOI] [PubMed] [Google Scholar]
- Cowell IG (2002) E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Bioessays 24: 1023–1029 [DOI] [PubMed] [Google Scholar]
- Cowell IG, Skinner A, Hurst HC (1992) Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol 12: 3070–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, Khoury S, Oukka M, Kuchroo VK (2008) IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat Immunol 9: 1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser B, Lohoff M, Kock S, Giaisi M, Kirchhoff S, Krammer PH, Li-Weber M (2002) IFN-gamma represses IL-4 expression via IRF-1 and IRF-2. Immunity 17: 703–712 [DOI] [PubMed] [Google Scholar]
- Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, Hunter T, Copeland N, Jenkins N, Liu YC (2002) Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol 3: 281–287 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003) Histone and chromatin cross-talk. Curr Opin Cell Biol 15: 172–183 [DOI] [PubMed] [Google Scholar]
- Garaude J, Farras R, Bossis G, Charni S, Piechaczyk M, Hipskind RA, Villalba M (2008) SUMOylation regulates the transcriptional activity of JunB in T lymphocytes. J Immunol 180: 5983–5990 [DOI] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ (2009) The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol 10: 1118–1124 [DOI] [PubMed] [Google Scholar]
- Gett AV, Hodgkin PD (1998) Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc Natl Acad Sci USA 95: 9488–9493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour J, Lavender P (2008) Control of IL-4 expression in T helper 1 and 2 cells. Immunology 124: 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid Q, Tulic M (2009) Immunobiology of asthma. Annu Rev Physiol 71: 489–507 [DOI] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY (2009) GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol 9: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S (2009) GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 284: 28729–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima S, Inukai T, Inaba T, Nimer SD, Cleveland JL, Look AT (1997) Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci USA 94: 2609–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW (2009) Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med 206: 2977–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M, Levy DM, McKeag L, Murray K, Schroder AJ, Canfield SM, Traver G, Rothman PB (2010) IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc Natl Acad Sci USA 107: 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso A, Groves P, Ramm L, Doyle AG (1999) Single-cell analysis by RT–PCR reveals differential expression of multiple type 1 and 2 cytokine genes among cells within polarized CD4+ T cell populations. Int Immunol 11: 617–621 [DOI] [PubMed] [Google Scholar]
- Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC (2001) The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol 167: 4414–4420 [DOI] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ (2002) Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol 169: 2253–2263 [DOI] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA (2006) T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24: 369–379 [DOI] [PubMed] [Google Scholar]
- Li B, Tournier C, Davis RJ, Flavell RA (1999) Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J 18: 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Weber M, Krammer PH (2003) Regulation of IL4 gene expression by T cells and therapeutic perspectives. Nat Rev Immunol 3: 534–543 [DOI] [PubMed] [Google Scholar]
- Liu Z, Li Z, Mao K, Zou J, Wang Y, Tao Z, Lin G, Tian L, Ji Y, Wu X, Zhu X, Sun S, Chen W, Xiang C, Sun B (2009) Dec2 promotes Th2 cell differentiation by enhancing IL-2R signaling. J Immunol 183: 6320–6329 [DOI] [PubMed] [Google Scholar]
- Lu B, Zagouras P, Fischer JE, Lu J, Li B, Flavell RA (2004) Kinetic analysis of genomewide gene expression reveals molecule circuitries that control T cell activation and Th1/2 differentiation. Proc Natl Acad Sci USA 101: 3023–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund R, Ahlfors H, Kainonen E, Lahesmaa AM, Dixon C, Lahesmaa R (2005) Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur J Immunol 35: 3307–3319 [DOI] [PubMed] [Google Scholar]
- Lund R, Aittokallio T, Nevalainen O, Lahesmaa R (2003) Identification of novel genes regulated by IL-12, IL-4, or TGF-beta during the early polarization of CD4+ lymphocytes. J Immunol 171: 5328–5336 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H (2001) Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev 15: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE (1999) The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol 17: 701–738 [DOI] [PubMed] [Google Scholar]
- Ohno T, Onishi Y, Ishida N (2007) A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res 35: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Van Stry M, Chung L, Koyanagi M, Sun X, Suzuki Y, Ohara O, Kitamura H, Hijikata A, Kubo M, Bix M (2009) Mina, an Il4 repressor, controls T helper type 2 bias. Nat Immunol 10: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PM, Cao X, Worthen GS, Shi P, Briones N, MacLeod M, White J, Kirby P, Kappler J, Marrack P, Yang B (2006) Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25: 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkurt IC, Tetradis S (2003) Parathyroid hormone-induced E4BP4/NFIL3 down-regulates transcription in osteoblasts. J Biol Chem 278: 26803–26809 [DOI] [PubMed] [Google Scholar]
- Ramsborg CG, Papoutsakis ET (2007) Global transcriptional analysis delineates the differential inflammatory response interleukin-15 elicits from cultured human T cells. Exp Hematol 35: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney JW, Hoey T, Glimcher LH (1995) Coordinate and cooperative roles for NF-AT and AP-1 in the regulation of the murine IL-4 gene. Immunity 2: 473–483 [DOI] [PubMed] [Google Scholar]
- Sartipy P, Loskutoff DJ (2003) Expression profiling identifies genes that continue to respond to insulin in adipocytes made insulin-resistant by treatment with tumor necrosis factor-alpha. J Biol Chem 278: 52298–52306 [DOI] [PubMed] [Google Scholar]
- Schroder AJ, Pavlidis P, Arimura A, Capece D, Rothman PB (2002) Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL-4-stimulated B lymphocytes. J Immunol 168: 996–1000 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37: 187–192 [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B (2008) Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol 9: 1341–1346 [DOI] [PubMed] [Google Scholar]
- Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD (1998) Interleukin-13: central mediator of allergic asthma. Science 282: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M (2009) Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol 9: 91–105 [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T (2002) Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem 277: 42399–42408 [DOI] [PubMed] [Google Scholar]
- Yang XO, Angkasekwinai P, Zhu J, Peng J, Liu Z, Nurieva R, Liu X, Chung Y, Chang SH, Sun B, Dong C (2009) Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol 10: 1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer SD (1995) Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol 15: 6055–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zheng B, Huang Y, Yang D, Katzman S, Chang C, Fowell D, Zeng WP (2007) Interaction between GATA-3 and the transcriptional coregulator Pias1 is important for the regulation of Th2 immune responses. J Immunol 179: 8297–8304 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.