Abstract
Lsm1 forms part of a cytoplasmic protein complex, Lsm1-7-Pat1, involved in the degradation of mRNAs. Here, we show that Lsm1 has an important role in promoting genomic stability in Saccharomyces cerevisiae. Budding yeast cells lacking Lsm1 are defective in recovery from replication-fork stalling and show DNA damage sensitivity. Here, we identify histone mRNAs as substrates of the Lsm1-7-Pat1 complex in yeast, and show that abnormally high amounts of histones accumulate in lsm1Δ mutant cells. Importantly, we show that the excess of histones is responsible for the lsm1Δ replication-fork instability phenotype, since sensitivity of lsm1Δ cells to drugs that stall replication forks is significantly suppressed by a reduction in histone gene dosage. Our results demonstrate that improper histone stoichiometry leads to genomic instability and highlight the importance of regulating histone mRNA decay in the tight control of histone levels in yeast.
Keywords: genomic stability, histones, Lsm1, mRNA decay, replication-fork stability
Introduction
The regulation of mRNA turnover is a powerful way to control gene expression. In eukaryotes, mRNAs are primarily degraded by two redundant decay pathways, namely the 3′–5′ and 5′–3′ (Garneau et al, 2007). In Saccharomyces cerevisiae, a required initial step in both pathways is the shortening of the poly(A) tail, or deadenylation, which results in the production of oligoadenylated mRNA from polyadenylated mRNA. After deadenylation, transcripts can be degraded by either the 5′–3′ or the 5′–3′ pathways (Anderson and Parker, 1998; He et al, 2003). In the 3′–5′ pathway, transcripts are further deadenylated and then degraded by a multisubunit 3′–5′ exonuclease complex, known as the exosome (Mitchell et al, 1997), that has been recently shown to have also endonucleolytic activity (Lebreton et al, 2008). In the 5′–3′ decay pathway, poly (A) shortening of the mRNAs triggers the removal of the 5′ cap by the Dcp1/Dcp2 decapping enzyme complex, which then permits the 5′–3′ exonucleolytic degradation of the mRNA by the Xrn1p exonuclease (Larimer et al, 1992; Beelman et al, 1996). Decapping is a critical control point in this pathway, and multiple accessory factors have been described to stimulate decapping (Coller and Parker, 2004). These factors include the Lsm1-7-Pat1 complex (Bouveret et al, 2000; Tharun et al, 2000), the DEAD box helicase Dhh1 (Coller et al, 2001) and the proteins Edc1, Edc2, Edc3 and Scd6 (Dunckley et al, 2001; Decourty et al, 2008). Deletion of any of these factors results in the stabilization of multiple reporter mRNAs, although the relative contribution of each of these proteins to the stability of a particular mRNA is not known.
The Lsm1-7-Pat1 complex is made of seven Sm-like proteins, Lsm1 through Lsm7 and the protein Pat1 (Bonnerot et al, 2000; Bouveret et al, 2000; Tharun et al, 2000). Lsm1 is the key subunit and distinguishes this complex from the related Lsm2–Lsm8 that interacts with U6 snRNAs and functions in pre-mRNA splicing in the nucleus (Mayes et al, 1999; Salgado-Garrido et al, 1999; He and Parker, 2000; Tharun, 2009b). The Lsm1-7-Pat1 complex is highly conserved from yeast to humans and interacts with other factors involved in the 5′–3′ pathway, such as Dhh1, Dcp1 or Xrn1 (Tharun et al, 2000), in cytoplasmic foci known as P-bodies (Sheth and Parker, 2003). In vitro analysis of the Lsm1-7-Pat1 purified from yeast showed that the complex has the intrinsic ability to bind preferentially to the 3′ ends of oligoadenylated mRNAs over polyadenylated mRNAs (Tharun, 2009a). Another interesting observation is that, among the deadenylated RNAs, the complex has a strong binding preference for mRNAs carrying a U-tract over those that do not (Chowdhury et al, 2007), implying that mRNAs with 3′-terminal U-tracts will be further stabilized in the absence of a functional complex. Interestingly, it has been recently shown in human cells that at the end of S-phase, or when DNA replication is inhibited, histone mRNAs acquire 3′-terminal oligo(U) tracts in a process known as oligouridylation (Mullen and Marzluff, 2008). Lsm1-7-Pat1 then recognizes and binds to the oligo (U) tail and possibly leads to the recruitment of decay factors of both 5′–3′ and 3′–5′ pathways. This post-transcriptional modification of histone mRNAs is very important to avoid the toxic effect of an excess of histones in human cells (Marzluff and Duronio, 2002). In S. cerevisiae, mRNA uridylation has not been detected (Rissland and Norbury, 2009), and the lowering of histone levels after inhibition of replication is achieved through a combination of histone gene repression and histone degradation (Gunjan et al, 2005). Rad53, a protein kinase involved in several crucial aspects of the DNA damage response (Gunjan and Verreault, 2003), has been demonstrated to mediate histone degradation in yeast. However, the contribution of Lsm1-7-Pat1 complex to the maintenance of proper histone levels in yeast has not been determined.
Using a genome-wide screen in S. cerevisiae, we identified the lsm1Δ mutant as hypersensitive to DNA-damaging drugs. In this study, we address the role of Lsm1 in preventing DNA damage, and the relationship of this function to its role in mRNA degradation. We demonstrate that Lsm1 promotes the stability of replication forks after stalling induced by DNA alkylation, nucleotide depletion or in natural pause sites in the DNA. Importantly, we show that the stability is maintained by the essential role that Lsm1 exerts in the control of histone mRNA levels. The implications of these findings for genomic stability and cancer are discussed.
Results
Deletion of LSM1 results in hypersensitivity to DNA-damaging drugs and accumulation of double-strand breaks
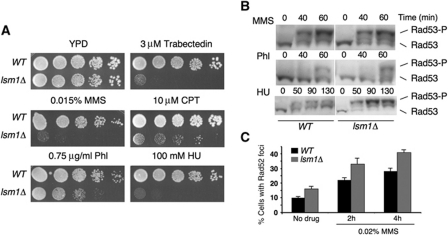
Using a genome-wide screen in S. cerevisiae, we identified the mutant lsm1Δ as hypersensitive to trabectedin, a DNA-binding drug that causes replication-dependent double-strand breaks (DSBs) (Herrero et al, 2006; Guirouilh-Barbat et al, 2008). We then examined the sensitivity of lsm1Δ to other DNA-damaging agents, methyl methanesulfonate (MMS), campthotecin (CPT), phleomycin (Phl) and hydroxyurea (HU), which have been reported to induce DSBs directly or indirectly (Koy et al, 1995; Petermann et al, 2010). lsm1Δ cells were highly sensitive to MMS, CPT and HU and moderately sensitive to Phl (Figure 1A).
Figure 1.
lsm1Δ cells are hypersensitive to DNA damage, proficient to activate the DNA damage checkpoint and accumulate DNA DSBs. (A) Sensitivity of wild-type and lsm1Δ cells to different drugs. (B) Phosphorylation of Rad53 in response to DNA damage. Cells were treated with 0.033% MMS, 2 μg/ml Phl or 0.2 M HU for the indicated time periods. (C) Percentage of cells containing Rad52-YFP foci in wild-type and lsm1Δ strains untreated or treated with 0.02% MMS for 2 and 4 h.
Hypersensitivity to genotoxic drugs could be the result of defects in the activation of DNA damage checkpoints or the accumulation of DNA damage due to a higher incidence, or defects in DNA repair. To test the first possibility, we analysed the phosphorylation state or Rad53, the major DNA damage checkpoint effector kinase of S. cerevisiae (Sanchez et al, 1999), after treatment with different drugs. Rad53 was phosphorylated, and so activated, in both lsm1Δ and wild-type cells following exposure to MMS, Phl or HU (Figure 1B). In fact, compared with the wild-type strain, in the lsm1Δ mutant we observed a further increase in the degree of Rad53 activation after treatment with HU, as revealed by the stronger intensity of the Rad53 phosphorylated form. Our results indicate that Lsm1 is not necessary for the activation of DNA damage checkpoint. In order to analyse the putative accumulation of DNA damage in lsm1Δ cells, we examined the formation of Rad52-YFP foci after treatment with MMS. In normal cells, Rad52 shows a diffuse nuclear localization; when DSBs occur, Rad52 relocalizes and forms discrete foci. Each Rad52 focus represents a centre of recombinational repair capable of processing multiple DNA lesions (Lisby et al, 2001). We found that the number of cells exhibiting Rad52 foci was higher in the lsm1Δ mutant compared with the wild-type strain (Figure 1C), even in the absence of DNA damage, suggesting either a higher incidence of DSBs in lsm1Δ cells or defects in their repair.
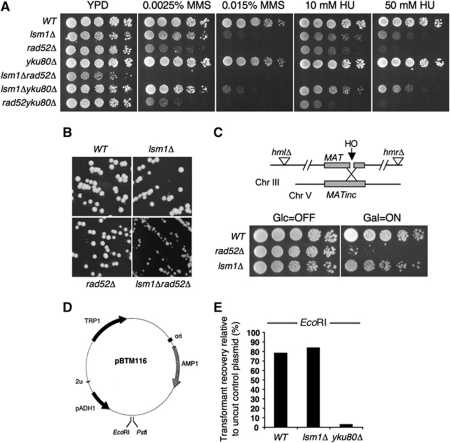
DSBs can be repaired by two mechanisms: homologous recombination (HR) and non-homologous end joining (NHEJ). First, we investigated whether Lsm1 is involved in either pathway by combining the deletion of LSM1 with mutations conferring defects in HR, such as rad52Δ, or NHEJ, such as yku80Δ. Deletion of RAD52 highly increased the sensitivity of lsm1Δ cells to the DNA-damaging drugs MMS, HU (Figure 2A) and Phl (data not shown), suggesting that Lsm1 affects a pathway other than HR. Moreover, we found that lsm1Δ cells required the presence of Rad52 not only to repair exogenous DNA damage, but also for proper growth (Figure 2B), which suggests a higher incidence of DSBs occurring in the lsm1Δ strain. To confirm that Lsm1 is not involved in DSB repair, we took advantage of the HR reporter system shown in Figure 2C (Frank-Vaillant and Marcand, 2002). In this system, a DSB can be induced by the galactose-inducible HO endonuclease. In HR-competent cells, the cleaved MAT locus in chromosome III recombines with the homologous sequence of chromosome V and cells can survive in a galactose-containing medium. In contrast, HR defective strains are unable to form colonies under the same conditions. The lsm1Δ cells were able to grow in the galactose-containing medium, which confirmed that Lsm1 is not involved in DSB repair by HR.
Figure 2.
lsm1Δ cells are competent to repair DSBs by HR and NHEJ. (A) Sensitivity of different mutants to DNA-damaging drugs. (B) Colonies formed by different strains in YPD medium after 2 days of growth at 30°C. (C) Schematic representation of the strain GA2321 used to monitor HR-dependent repair of HO endonuclease-induced DSB. In this strain, both HML and HMR loci are deleted and a non-cleavable copy of the MAT locus is integrated on chromosome V. Expression of HO endonuclease is controlled by the GAL1,10 promoter. Cells were grown overnight in a raffinose-containing medium and then plated on media containing galactose (HO ON) or glucose (HO OFF). (D) Plasmid map of the vector pBTM116 used to monitor NHEJ repair. (E) Percentage of transformant recovery. Each strain was transformed with equal amounts of linear or uncut plasmid, and the number of colonies obtained with the linearized plasmid was normalized to the obtained from uncut plasmid.
To analyse whether lsm1Δ cells are affected in the repair of DSBs by NHEJ, we employed an in vivo plasmid repair assay (Boulton and Jackson, 1996) (Figure 2D). Initially, we assayed the ability of the strains under study to repair a DSB with 5′-overhanging ends produced by digestion with EcoRI. As shown in Figure 2E, transformant recovery in wild-type and lsm1Δ cells was very similar in both cases around 80%. As expected, yku80Δ cells, used as a control, exhibited very low transformant recoveries. Similar results were obtained for the repair of 3′-overhanging ends generated by the enzyme Pst1 (data not shown). Taken together, our results indicate that lsm1Δ cells are competent to repair DSBs by both HR and NHEJ, and, therefore, accumulation of DSBs seemed to be due to a higher incidence of DNA damage.
Lsm1 is necessary to maintain replication-fork stability
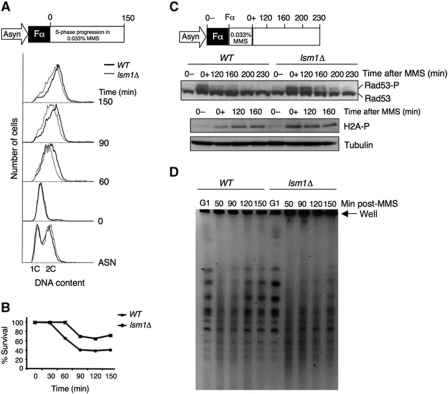
HR is important not only for the repair of DSB, but also for the recovery of stalled or collapsed replication forks (Herzberg et al, 2006; Lambert et al, 2007). The strong requirement of HR for proper growth of lsm1Δ cells, together with their hypersensitivity to MMS and HU, that cause replication-fork slow down or stalling, suggests that Lsm1 may be defective in the stabilization of, or progression through, stalled replication forks. To test this hypothesis, we first monitored S-phase progression in wild-type and lsm1Δ strains in the presence or absence of MMS by flow cytometry (Figure 3A). Cells were synchronized in G1 with α factor and then released into fresh medium, without drug or with 0.033% MMS. At this concentration, MMS reduces the rate of replication-fork progression in wild-type cells (Tercero and Diffley, 2001). In the absence of MMS, both wild-type and lsm1Δ cells reached a 2C DNA content in approximately 1 h (data not shown). However, lsm1Δ was found to be defective in S-phase progression in the presence of MMS (Figure 3A, grey line) compared with the wild-type strain (dark line). This defect resulted in loss of viability as shown in Figure 3B. We then analysed activation and inactivation of Rad53. Wild-type and lsm1Δ cells were synchronized in G1 and released into medium containing 0.033% MMS for 1 h to activate Rad53 (Figure 3C). MMS was then washed out and cells were released into drug-free medium. Rad53 remained activated much longer in lsm1Δ cells than in the wild-type strain. Moreover, we found an increased phosphorylation of H2A in lsm1Δ cells after treatment with MMS, suggesting a higher incidence of DSBs in the lsm1Δ strain (Figure 3C).
Figure 3.
Lsm1 is necessary for the recovery of replication-fork stalling induced by MMS. (A) Schematic representation of the experiment and FACS analysis of the S-phase progression in wild-type and lsm1Δ cells in the presence of 0.033% MMS. (B) Percentage of survival after treatment with 0.033% MMS. (C) Schematic representation of the experiment and Rad53 checkpoint kinase activation and histone H2A phosphorylation upon recovery from MMS damage in wild-type and lsm1Δ cells. (D) Analysis of completion of DNA replication by PFGE. Cells were treated as in (C) and samples were collected at the indicated points, embedded in agarose plugs and analysed by PFGE.
Completion of DNA replication upon treatment with MMS in S-phase was monitored by pulsed field gel electrophoresis (PFGE). This technique allows distinction between linear chromosomal DNA, which enters the gel, from DNA containing replication bubbles, which is trapped inside the agarose plugs and stays in the loading well. Cells were arrested in G1, washed and treated with MMS as described above (Figure 3C). Samples were taken at 50, 90, 120 and 150 min after release from MMS treatment. In both wild-type and lsm1Δ cells, intact chromosomal DNA was separated as individual bands in G1-arrested cells, whereas most of the DNA was retained in the loading well 50 min after drug exposure in S-phase (Figure 3D). Chromosomal DNA re-entered the gel approximately 2 h after drug release in wild-type cells. In contrast, most of the DNA from lsm1Δ cells remained in the well throughout the recovery period, which clearly revealed a defect in the completion of DNA replication after MMS treatment.
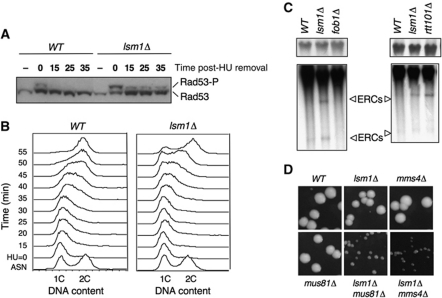
Next, we determined whether Lsm1 was also important for recovery after replication stalling induced by nucleotide depletion. Thus, we assessed the activation status of Rad53 in wild-type and lsm1Δ strains both after 3 h of treatment with HU and following drug removal (Figure 4A). Compared with the wild-type strain, in the lsm1Δ mutant, we observed a further increase in the degree of Rad53 activation after treatment with HU, as revealed by the stronger intensity of the Rad53 phosphorylated form (Figure 4A, time 0). Prolonged Rad53 checkpoint kinase activation upon removal of HU was also evident. This result indicated that the lsm1Δ strain suffered more DNA damage that the wild-type strain after treatment with HU. The persistence of Rad53 phosphorylation also suggested that the lsm1Δ strain exhibits a delay in recovery from replication stress. We, therefore, analysed the DNA content of wild-type and lsm1Δ strains after release from an HU-induced cell-cycle arrest (Figure 4B). As expected, lsm1Δ cells exhibited a clear delay in progression through S-phase as compared with the wild-type strain.
Figure 4.
Lsm1 is required for the recovery of replication-fork stalling induced by nucleotide depletion (HU) and at natural pause sites. (A) Prolonged Rad53 checkpoint kinase activation upon removal of HU in lsm1Δ cells. The phosphorylation state of Rad53 was determined by western blot at the indicated time points. (B) Cell-cycle progression after treatment with HU is delayed in lsm1Δ cells. DNA contents of WT and lsm1Δ cells were determined by FACS analysis of mid-log phase cultures (ASN), cells synchronized in S-phase after treatment with 200 mM HU and at the indicated times after removal of the drug. (C) lsm1Δ cells accumulate ERCs. ERCs were detected by Southern blot hybridization using a radioactive rDNA probe. A shorter exposure of the signal corresponding to the rDNA array is shown in the upper panel. (D) Synthetic fitness interaction between lsm1Δ, mms4Δ and mus81Δ mutants.
Finally, we looked at the effect of LSM1 deletion on the integrity of DNA replication forks arrested at natural pause sites. Fork pausing at the rDNA array induces HR, which results in the accumulation of extra-chromosomal circles (ERCs) (Sinclair and Guarente, 1997). Deletion of FOB1, encoding a protein required for replication-fork blocking, suppresses the elevated pausing, reducing DSB occurrence and ERC formation (Defossez et al, 1999). The consequences of replication-fork instability can thus be measured at the rDNA locus through the quantification of ERCs. Genomic DNA was obtained from wild type, lsm1Δ, fob1Δ and ERCs were detected by Southern blot using an rDNA probe. lsm1Δ cells contained higher levels of ERCs compared with the wild type, which clearly revealed a higher incidence of DSBs occurring at the rDNA locus of the lsm1Δ strain. As expected, ERCs were nearly undetectable in the fob1Δ strain. As a positive control, we used rtt101Δ that has been previously shown to accumulate ERCs (Luke et al, 2006).
These results suggested that Lsm1 is required for the stabilization of replication forks that move through damaged and paused DNA sites, which is supported by the analysis of synthetic genetic interactions of lsm1Δ with mms4Δ and mus81Δ, two genes involved in processing of stalled replication forks (Osman and Whitby, 2007). If deletion of LSM1 results in the accumulation of stalled replication forks, we expected an lsm1Δ strain to require MMS4 or MUS81 for proper growth. Figure 4D shows that lsm1Δ mus81Δ and lsm1Δ mms4Δ double mutants form smaller colonies than single mutants, confirming our expectations.
Inactivation of the 3′–5′ (exosome) pathway in the lsm1Δ mutant enhances hypersensitivity to DNA-damaging drugs
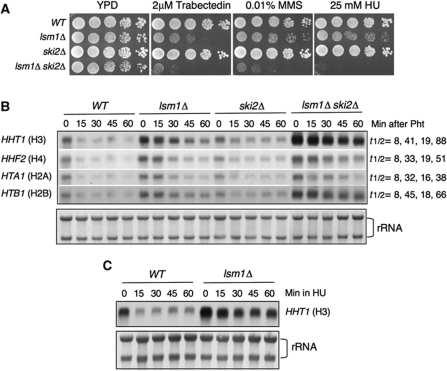
Since Lsm1 is involved in mRNA degradation, we hypothesized that in its absence some proteins that interfere with replication-fork progression would be stabilized leading to the formation of aberrant DNA structures that need to be resolved by HR and replication re-start pathways. If our hypothesis is correct, we expected that a higher stabilization of the putative replication-fork interfering protein(s) would exacerbate the phenotype of lsm1Δ. As mentioned above, there are two general pathways for the degradation of mRNAs, the 5′–3′ and the 3′–5′. Disruption of both pathways leads to loss of viability as revealed by the synthetic lethality of mutants in components of the exosome, ski2Δ or ski3Δ, with the 5′–3′ exonuclease xrn1Δ (Anderson and Parker, 1998). However, since deletion of LSM1 does not cause a complete block of decapping, lsm1Δ is not synthetically lethal with any of the ski mutations (He and Parker, 2001), although the double mutant probably presents stronger defects in mRNA degradation. A double mutant lsm1Δ ski2Δ was, therefore, obtained, and sensitivity to different DNA-damaging drugs analysed. Our results clearly showed that inactivation of the 3′–5′ pathway greatly increased the sensitivity of lsm1Δ cells to DNA damage (Figure 5A).
Figure 5.
Sensitivity to DNA-damaging drugs and histone mRNA levels in mRNA decay mutants. (A) Deletion of SKI2 increases sensitivity of lsm1Δ cells to DNA damage. Five-fold serial dilution of cultures of the indicated mutants were spotted on YPD or YPD containing different drugs at the indicated concentrations. (B) Lsm1 controls histone mRNA levels. Total RNA from the indicated strains were separated on a agarose formaldehyde gel and sequentially hybridized with different histone probes. Histone mRNAs was quantitated by densitometry analysis using Image J. The amount of mRNA was plotted against time to determine the half-lives of histone mRNAs. (C) Histone H3 mRNA levels after treatment with 0.2 M HU. Total rRNA stained with methylene blue was used as loading controls.
Lsm1 controls histone mRNA levels
To uncover the substrates of Lsm1 whose stabilization resulted in the instability of stalled replication forks, we took advantage of the recent findings in human cells indicating that LSM1 is required for histone mRNA degradation at the end of S-phase and following replication stress (Mullen and Marzluff, 2008). We tested the following questions: (i) Are histone mRNAs targets of degradation by the Lsm1 complex in yeast? (ii) Do lsm1Δ cells accumulate free histones? (iii) Is the putative excess of histones in lsm1Δ cells responsible of their replication-fork instability? To answer the first question, we analysed the abundance and decay rates of different histone mRNAs in wild-type, lsm1Δ, ski2Δ and lsm1Δ ski2Δ strains. Cells were collected from the exponential phase of growth and at different times upon inhibition of transcription with 1–10 phenanthroline. We found that deletion of LSM1 increased the stability of all histone mRNAs analysed four- to six-fold (Figure 5B), whereas deletion of SKI2 affected histone mRNA stability to a lesser extent (two-fold). Double mutant lsm1Δ ski2Δ exhibited a strong accumulation of histone mRNAs (6–10-fold). These results indicated that in budding yeast, degradation of histone mRNAs takes place mainly by the 5′–3′ degradation pathway and to a lower extent by the 3′–5′ degradation pathway.
At this point, we were interested in determining how many transcripts were upregulated in the lsm1Δ strain. For this purpose, we performed microarray analysis. Of a total of 5766 genes present in the microarray, 2888 genes were upregulated in the lsm1Δ compared with the wild type (Supplementary Table S3). Consistent with the northern blots, all the histones genes were upregulated.
Maintenance of a short half-life of histone mRNAs must be very important for proper regulation of histone levels when DNA replication is blocked. To test this hypothesis, we measured histone H3 mRNA degradation in wild-type and lsm1Δ cells following HU treatment. As shown in Figure 5C, histone H3 mRNA decay was severely impaired in lsm1Δ compared with the wild-type strain.
lsm1Δ cells are defective in the degradation of overexpressed histones
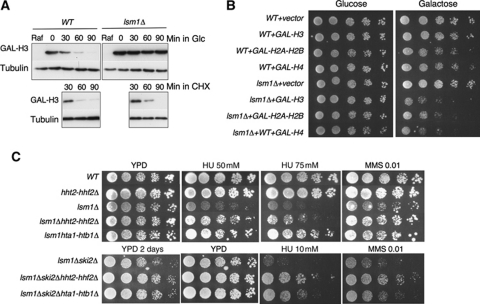
Histones are very stable proteins. However, excess histones that are not packaged into chromatin are degraded very rapidly. In S. cerevisiae, this degradation is dependent upon the catalytic activity of Rad53 (Gunjan and Verreault, 2003). Since lsm1Δ accumulates histone mRNAs, we reasoned that this strain might be accumulating an excess of free histones, even in the presence of a functional Rad53. We, therefore, performed a histone degradation assay as described previously (Gunjan and Verreault, 2003). Strains expressing HA-tagged histone H3 were arrested in G1 to prevent overexpressed histones being packaged into chromatin. Histone H3 overexpression was then induced by addition of galactose. Transcription repression was mediated by switching half of the cultures to a glucose-containing medium. Cycloheximide was added to the other half of the cultures to repress protein synthesis. As shown in Figure 6A, histone degradation is severely delayed in lsm1Δ cells compared with the wild type without cycloheximide. This defect must be due to the translation of stabilized histone mRNAs, since histone protein degradation occurs normally in the lsm1Δ strain treated with cycloheximide.
Figure 6.
lsm1Δ cells accumulate free histones that make replication forks sensitive to drug-induced replication-fork stalling. (A) Degradation of overexpressed histones is impaired in lsm1Δ cells. Wild-type and lsm1Δ strains carrying the 2 μ-URA3-GAL-HA-HHT2 plasmid (pGAL-H3) were grown overnight in minimal medium lacking uracil and with raffinose as a carbon source. Cells were synchronized in G1 with α factor for 2 h and galactose was added to the medium for 1 h. Half of the cultures were then collected and transferred to a glucose-containing medium, and the other half was transferred to the same medium containing galactose, but with the addition of cycloheximide at 35 μg/ml. Samples were taken every 30 min after switching to glucose or cycloheximide. (B) Effect of histone overexpression on the growth of WT, lsm1Δ and lsm1Δ ski2Δ strains. Strains transformed with empty vector or with plasmids expressing histones H3, H4 or H2A–H2B under the control of GAL promoter were grown overnight in minimal medium without uracil and with 2% raffinose as a carbon source. Five-fold serial dilutions of each strain were plated on the same medium, either with glucose or galactose, as carbon source. The plates were incubated for 4 days at 30°C. (C) Deletion of either hht2-hhf2 or hta1htb1 histone gene pairs significantly suppresses the DNA damage sensitivity of the lsm1Δ and lsm1Δ ski2Δ strains.
lsm1Δ cells are hypersensitive to histone overexpression
It has been previously shown that rad53Δ cells, exhibiting defects in histone degradation, are sensitive to histone overexpression (Gunjan and Verreault, 2003). If Lsm1 is indeed important to avoid an excess of free histones in the cell, we expected lsm1Δ cells to be also hypersensitive to histone overexpression. Wild-type, lsm1Δ and lsm1Δ ski2Δ strains were transformed with plasmids encoding histone genes under the control of the GAL1 promoter or with an empty vector. Transformants were then grown in a raffinose-containing medium and serial dilutions of the cell cultures were plated on glucose (promoter OFF) or galactose (promoter ON)-containing media. As shown in Figure 6B, the growth of wild-type cells was not affected by histone H3 overexpression and was minimally affected by overexpression of histones H2A–H2B or H4. In contrast, lsm1Δ cells were found extremely sensitive to overexpression of the four core histones (Figure 6B) as also was lsm1Δ ski2Δ (data not shown).
A reduction in histone H3–H4 or H2A–H2B gene dosage suppresses the sensitivity of lsm1Δ cells to DNA-damaging drugs
The results shown above indicate that Lsm1 exerts an essential role in the maintenance of proper histone levels. We next wondered whether this function was related to the role of Lsm1 in promoting replication-fork stability. If this was the case, we expected lsm1Δ phenotypes to be alleviated by a reduction in histone gene dosage. Each of the four core histones, H3, H4, H2A and H2B, is encoded by a pair of nearly identical genes, HHT1-HHT2, HHF1-HHF2, HTA1-HTA2 and HTB1-HTB2, respectively. It has been previously shown that wild-type cells lacking the gene pair HHT2-HHF2 grow surprisingly well because HHT1-HHF1 alone probably provides sufficient amounts of histones for nucleosome assembly (Gunjan and Verreault, 2003). However, deletion of the HTA1-HTB1 gene pair causes loss of viability (Libuda and Winston, 2006). Recently, Libuda and Winston (2010) performed a genome-wide screen in S. cerevisiae looking for mutations that suppressed the loss of viability caused by HTA1-HTB1 deletion. Interestingly, LSM1, LSM6 and LSM7 were found among the genes that increased viability. Based on these data, we decided to lower the dose of histone H3–H4 in wild-type and lsm1Δ cells and H2A–H2B in lsm1Δ cells, and analyse the effect on the sensitivity to drugs that cause replication-fork stalling. Deletion of HHT2-HHF2 in the wild-type strain had no effect in the sensitivity of the strain to MMS or HU. In contrast, we found that deletion of either HHT2-HHF2 or HTA1-HTB1 histone genes pairs significantly suppressed the DNA damage sensitivity of the lsm1Δ and lsm1Δ ski2Δ strains to both MMS and HU. Moreover, a lower dose of histone H3–H4 or H2A–H2B in the lsm1Δ ski2Δ strain also suppressed its slow growth phenotype. Our results strongly indicate that DNA damage sensitivity of the lsm1Δ mutants is mostly due to the harmful effect of excess histones and that the Lsm1-7-Pat1 complex controls the levels of histone mRNAs in the cell.
Discussion
Histone synthesis and degradation is tightly coupled to DNA synthesis in order to ensure correct chromatin assembly during S-phase. In this study, we have identified Lsm1 as a new factor that contributes to genomic stability in the yeast S. cerevisiae by promoting histone mRNA decay.
In wild-type cells, treatment with MMS activates the DNA damage checkpoint, which results in a slowing down of replication-fork progression and inhibition of origin firing (Tercero and Diffley, 2001). However, these effects are transient and replication continues once the damage is repaired or bypassed. In contrast, lsm1Δ cells prolong the activation of the S-phase DNA damage checkpoint and are defective in completing chromosome replication after treatment with MMS. The same phenotype is observed when replication-fork progression is blocked by treatment with HU. Moreover, replication-fork stalling induced by HU results in an increased DNA damage in the lsm1Δ cells compared with the wild-type strain, as revealed by the stronger intensity of the Rad53 phosphorylated form. These phenotypes indicate that Lsm1 is required to maintain the stability of stalled replication forks.
HR is an important mechanism, not only for the repair of DSBs, but also for the re-start of stalled or collapsed replication forks (Herzberg et al, 2006; Lambert et al, 2007). However, this is a double-edged sword, since unscheduled HR also leads to increased recombination and genomic instability (Tourriere and Pasero, 2007). We found that lsm1Δ cells are fully proficient to repair DNA DSBs, but accumulate this type of lesion, even in the absence of exogenous DNA damage. Moreover, lsm1Δ mutant cells required the HR protein Rad52 for proper growth, exhibited a recombination frequency three times higher than the wild-type strain (Supplementary Figure S1) and had a strong requirement for Mus81–Mms4, a complex involved in the rescue of stalled replication forks (Osman and Whitby, 2007), for proper growth. These observations indicate that Lsm1 has a role in the maintenance of replication forks that stall during normal S-phase. In accordance with this conclusion, deletion of LSM1 also results in the instability of replication forks that encounter natural impediments in the DNA, such as replication-fork barriers in the rDNA locus.
Several lines of evidence suggest that the role of Lsm1 in promoting genomic stability is related to its function in the degradation of mRNAs. First, it has been demonstrated that Lsm1 is mostly a cytoplasmic protein (Tharun et al, 2000). Therefore, the possibility of exerting a direct role in the maintenance of genomic stability in the nucleus is unlikely. Second, some other non-essential members of the Lsm1-7-Pat1 complex were found in our genome-wide screen of mutants hypersensitive to trabectedin, such as lsm6Δ and pat1Δ. Third, the inactivation of the second route of mRNA decay, the 3′–5′ pathway, in the lsm1Δ mutant highly increased its sensitivity to DNA-damaging drugs. Based on these evidences, we hypothesized that in the absence of Lsm1 some proteins that interfere with replication-fork progression would be stabilized, leading to the formation of aberrant DNA structures that need to be resolved by HR and replication re-start pathways.
The Rad53 protein monitors excess histones and targets them for degradation by ubiquitylation-dependent proteolysis (Gunjan and Verreault, 2003; Singh et al, 2009). This mechanism prevents the accumulation of free histones when replication slows down or stops. Excess histones accumulating in rad53Δ result in slow growth, DNA damage sensitivity and chromosome loss. These phenotypes are significantly supressed by a reduction in histone gene dosage. Another way to control histone levels in S. cerevisiae is transcriptional repression mediated by the Hir proteins (Sherwood et al, 1993). In animal cells, regulation of histone synthesis occurs both transcriptionally and post-transcriptionally (Sittman et al, 1983; Ye et al, 2003; Kaygun and Marzluff, 2005; Mullen and Marzluff, 2008). Post-transcriptional regulation consists of rapid degradation of histone mRNAs, both after inhibition of replication and at the end of a normal S-phase. Histone mRNAs are the only mRNAs in mammalian cells that are not polyadenylated. Instead, they end in a stem-loop structure that is recognized by the stem-loop binding protein (SLBP). It has been shown that histone mRNA degradation in human cells requires the protein Upf1, which interacts with the SLBP at the 3′ end of histone mRNAs after treatment with HU (Kaygun and Marzluff, 2005). Recent results obtained by Mullen and Marzluff (2008) postulate that the recruitment of Upf1 results in the recruitment of terminal uridilyl transferases that carry out the oligouridylation of histone mRNAs. Interestingly, these authors show that the Lsm1-7 complex recognizes and binds to the uridylated 3′ end of the mRNAs, possibly triggering the recruitment of decay factors. In S. cerevisiae, histone mRNAs do not terminate in a stem loop and both SLBP and uridylating enzymes are absent (Gunjan et al, 2005). However, we show here that Lsm1 is also very important for the control of histone mRNA levels in yeast. Deletion of LSM1 results in a strong stabilization of histone mRNAs, which leads to the accumulation of abnormally high amounts of histones in the cell. It is possible that the Lsm1-7-Pat1 complex ensures a very short half-life of histone mRNAs by recognizing U-tracts that are present in the 3′ ends of different histone mRNAs, since it has been shown that the Lsm1-7-Pat1 complex has a strong binding preference for mRNAs carrying a U-tract over those that do not (Chowdhury et al, 2007). This short half-life of the histone mRNAs is very important for the proper regulation of histone levels by the known transcriptional (Hir proteins) and post-translational (Rad53) mechanisms. Lsm1 influences yeast histone levels in such a way that deletion of LSM1 leads to the accumulation of histones, even in the presence of a functional Rad53 and intact transcriptional regulation mechanisms. In line with the notion that Lsm1 exerts a strong control over histone levels, we found that the double mutant lsm1Δ rad53Δ is lethal, as are lsm1Δ hir1Δ and lsm1Δ hir2Δ, as revealed by genome-wide synthetic lethal interactions (Pan et al, 2006).
Mammalian histone mRNAs are regulated both at the level of transcription and post-transcriptionally (Sittman et al, 1983). Unlike their mammalian counterparts, yeast histone mRNAs were traditionally thought of being regulated primarily at the level of transcription due to the very short half-lives of these transcripts, particularly in response to replication arrest, while any contribution of post-transcriptional regulation of histone mRNAs was dependent upon the 3′ ends of the histone mRNAs (Lycan et al, 1987). Later, it was suggested that the poly-A polymerases Trf4/5 and the exosome was having a role in post-transcriptional regulation of histone mRNAs in yeast (Reis and Campbell, 2007). More recently, Lsm4 (Mazzoni et al, 2005) and Lsm1 have been implicated in the histone regulation in yeast (Palermo et al, 2010) which, together with our findings, strongly suggest that the traditional view that yeast histone mRNAs are largely regulated at the level of transcription needs to be revised to accommodate the important contribution of post-transcriptional mechanisms in their regulation. This would suggest that despite the significant differences in the structure of yeast and mammalian histone mRNAs, and in the mechanistic details of their regulation, the overall principles governing the regulation of yeast and mammalian histone mRNAs are essentially the same, indicating that yeast is an excellent model for the study of histone regulation and its impact on genomic stability.
It has been demonstrated that accumulation of free histones in rad53Δ cells leads to chromosome loss, a form of genomic instability. Here, we report that lsm1Δ cells accumulate DNA DSBs, known to induce genomic instability, are hyper-recombinant and exhibit instability at the rDNA loci. Since the Lsm1-7-Pat1 complex is highly conserved throughout evolution, we anticipate that defects in this complex will also lead to genomic instability in higher eukaryotes. In this regard, a genome-wide screen recently carried out by Paulsen et al (2009) identified many mRNA processing factors involved in this process. The gene encoding Lsm6, a subunit of the Lsm1-7 complex, was among those identified.
Interestingly, it has been reported that many cancers present alterations in the human LSM1 gene (Takahashi et al, 2002; Fraser et al, 2005; Streicher et al, 2007; Watson et al, 2008). The molecular mechanisms by which alterations of LSM1 lead to neoplastic transformation remain largely unknown. Our results suggest that malignant transformation could be related to the role of Lsm1 in the maintenance of genomic stability through the regulation of histone mRNA levels. Future experiments will be carried out to verify this hypothesis.
Materials and methods
Yeast strains and media
Yeast strains and plasmids used in this study are listed in Supplementary Tables SI and SII. Standard yeast media and growth conditions were used.
Spot assays for analysing sensitivity to DNA-damaging agents
Yeast strains were grown in YP medium, unless otherwise specified, until they reached exponential growth. Cells were harvested by centrifugation and adjusted to an OD600 of 0.2.5 μl of undiluted cell culture and 1/5 serial dilutions of each cell culture were spotted onto plates containing different drugs at the indicated concentration. Plates were incubated at 30°C for 3 days.
Protein extracts and western blots
Protein extracts were obtained by TCA precipitation as described previously (Foiani et al, 1994). For Rad53 detection, protein extracts were run in 7.5% SDS–PAGE gels, transferred to nitrocellulose and incubated with anti-Rad53 antibodies (Santa Cruz Biotechnology, Inc) at 1/500 for 3 h. Equivalent loading in each lane was confirmed by Ponceau staining. Anti-goat antibodies were used as secondary antibodies at 1:3000 dilution and incubated for 1 h at room temperature. For detection of HA-tagged H3, extracts were resolved in 12% SDS–PAGE gels and probed with mouse anti-HA 12CA5 antibodies (Roche Applied Sciences) at 1/3000. For detection of tubulin, mouse anti-tubulin antibodies (a gift of Dr Keith Gull) were used in a 1/3000 dilution. The immunoblots were developed using ECL™ western blotting detection reagents (Amersham, UK).
Detection of Rad52-YFP
Rad52-YFP foci from exponentially growing cells transformed with plasmid pWJ1213 were visualized by fluorescence microscopy using a Leica DM6000B microscope controlled by the MetaMorph software. Three independent experiments counting 300 cells each time were performed for each strain and condition.
NHEJ assays
The plasmid repair assay for NHEJ was performed as previously described (Boulton and Jackson, 1996). Briefly, plasmid pBTM116 was digested with the appropriate restriction enzyme and then heat inactivated by treatment at 65°C for 20 min. The same amount of uncut or linearized plasmid DNA was used to transform each strain with the lithium acetate method. Diluted samples were plated on minimal media lacking the appropriate amino acids, and colonies were counted after incubation at 30°C for 4–5 days. Percentage of transformants relative to uncut plasmid determined the efficiency of NHEJ.
Frequency of recombination
The frequency of HR was determined by using the plasmidic inverted repeat system SU. This system is based on two truncated leu2 alleles that share a 0.6-kb internal fragment. HR between the repeats leads to the inversion of the intervening sequence and the formation of an LEU2 wild-type copy (Prado and Aguilera, 1995). Spontaneous recombination frequencies were obtained by fluctuation test as the median value of six independent colonies as described previously (Prado and Aguilera, 1995).
Fluorescence-activated cell sorting analysis
Samples of 2 × 107 cells were collected by centrifugation, fixed in 70% ethanol and processed for flow cytometry as described previously (Hutter and Eipel, 1979). A Becton Dickinson (Mountain View, CA) FACSCalibur was used to determine the DNA content.
Pulse-field gel electrophoresis
For PFGE analysis, 6 × 108 cells were washed with 1 ml of 10 mM Tris, 50 mM EDTA and 0.1% sodium azide, pH 8. Agarose plugs containing chromosomal DNA from 5 × 107 cells were prepared as described previously (Lengronne et al, 2001). Electrophoresis was performed for 24 h at 6 V/cm with a switch time of 60–120 s in 0.5 × TBE at 14°C. The gels were stained with 0.5 g/ml ethidium bromide for 20 min, destained in deionized water for 20 min and then photographed.
Extra-chromosomal circles
For the detection of ERCs, genomic DNA was extracted and purified using the DNAeasy Blood and tissue kit from Quiagen. Equal amounts of DNA (1.5 μg) were separated on a 0.7% agarose gel at 1 V/cm for 24 h. rDNA species were detected by Southern blot using a radioactive rDNA fragment, RDN25.
RNA extraction and northern blots
Total RNA from cells was obtained following the recommendations of the RNAeasy Mini kit from Qiagen. A total of 5 μg of each sample were separated on a agarose formaldehyde gel and northern blotting was carried out following the protocol of the ExpressHybTM Hybridization Solution (BD Biosciences). Probes corresponding to different mRNAs were obtained by PCR and labelled with [α-32P]dCTP. The Rediprime II Random Prime labelling System kit (Amersham, UK) was used.
Supplementary Material
Acknowledgments
We thank Drs Stanley Fields, Susan Gasser, Akash Gunjan, Felix Prado and Alain Verrault for plasmids and strains, José Antonio Tercero for suggestions in experimental design and Chris Norbury for his careful and critical reading of the manuscript. ABH is supported by a postdoctoral grant from the Fundación Científica de la Asociación Española Contra el Cáncer. Work in our group is supported by Grants BFU2008-01808, Consolider CSD2007-00015 and Junta de Castilla y León Grupo de Excelencia GR 265.
Author contributions: ABH and SM conceived and designed the experiments. ABH performed the experiments and analysed the data. ABH and SM wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson JS, Parker RP (1998) The 3′–5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′–5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 [DOI] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B (2000) The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol 20: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24: 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B (2000) A Sm-like protein complex that participates in mRNA degradation. EMBO J 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S (2007) The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R (2004) Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R (2001) The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, Jacquier A (2008) Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci USA 105: 5821–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3: 447–455 [DOI] [PubMed] [Google Scholar]
- Dunckley T, Tucker M, Parker R (2001) Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Marini F, Gamba D, Lucchini G, Plevani P (1994) The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol 14: 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell 10: 1189–1199 [DOI] [PubMed] [Google Scholar]
- Fraser MM, Watson PM, Fraig MM, Kelley JR, Nelson PS, Boylan AM, Cole DJ, Watson DK (2005) CaSm-mediated cellular transformation is associated with altered gene expression and messenger RNA stability. Cancer Res 65: 6228–6236 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Redon C, Pommier Y (2008) Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11-Rad50-Nbs1 complex. Mol Biol Cell 19: 3969–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Paik J, Verreault A (2005) Regulation of histone synthesis and nucleosome assembly. Biochimie 87: 625–635 [DOI] [PubMed] [Google Scholar]
- Gunjan A, Verreault A (2003) A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115: 537–549 [DOI] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′–3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- He W, Parker R (2000) Functions of Lsm proteins in mRNA degradation and splicing. Curr Opin Cell Biol 12: 346–350 [DOI] [PubMed] [Google Scholar]
- He W, Parker R (2001) The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158: 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero AB, Martin-Castellanos C, Marco E, Gago F, Moreno S (2006) Cross-talk between nucleotide excision and homologous recombination DNA repair pathways in the mechanism of action of antitumor trabectedin. Cancer Res 66: 8155–8162 [DOI] [PubMed] [Google Scholar]
- Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, Anderson S, Bashkirova EV, Yates JR III, Heyer WD (2006) Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol 26: 8396–8409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter KJ, Eipel HE (1979) Microbial determinations by flow cytometry. J Gen Microbiol 113: 369–375 [DOI] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF (2005) Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Koy JF, Pleninger P, Wall L, Pramanik A, Martinez M, Moore CW (1995) Genetic changes and bioassays in bleomycin- and phleomycin-treated cells, and their relationship to chromosomal breaks. Mutat Res 336: 19–27 [DOI] [PubMed] [Google Scholar]
- Lambert S, Froget B, Carr AM (2007) Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 6: 1042–1061 [DOI] [PubMed] [Google Scholar]
- Larimer FW, Hsu CL, Maupin MK, Stevens A (1992) Characterization of the XRN1 gene encoding a 5′-->3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120: 51–57 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B (2008) Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E (2001) Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res 29: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Winston F (2006) Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443: 1003–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Winston F (2010) Alterations in DNA replication and histone levels promote histone gene amplification in Saccharomyces cerevisiae. Genetics 184: 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M (2006) The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol 16: 786–792 [DOI] [PubMed] [Google Scholar]
- Lycan DE, Osley MA, Hereford LM (1987) Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol 7: 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol 14: 692–699 [DOI] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD (1999) Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18: 4321–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni C, Palermo V, Torella M, Falcone C (2005) HIR1, the co-repressor of histone gene transcription of Saccharomyces cerevisiae, acts as a multicopy suppressor of the apoptotic phenotypes of the LSM4 mRNA degradation mutant. FEMS Yeast Res 5: 1229–1235 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF (2008) Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′–3′ and 3′–5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Whitby MC (2007) Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 6: 1004–1017 [DOI] [PubMed] [Google Scholar]
- Palermo V, Cundari E, Mangiapelo E, Falcone C, Mazzoni C (2010) Yeast lsm pro-apoptotic mutants show defects in S-phase entry and progression. Cell Cycle 9: 3991–3996 [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD (2006) A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA (2009) A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (1995) Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics 139: 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CC, Campbell JL (2007) Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics 175: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ (2009) Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J 18: 3451–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ (1999) Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286: 1166–1171 [DOI] [PubMed] [Google Scholar]
- Sherwood PW, Tsang SV, Osley MA (1993) Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 13: 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Singh RK, Kabbaj MH, Paik J, Gunjan A (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol 11: 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman DB, Graves RA, Marzluff WF (1983) Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci USA 80: 1849–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher KL, Yang ZQ, Draghici S, Ethier SP (2007) Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene 26: 2104–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Suzuki S, Inaguma S, Cho YM, Ikeda Y, Hayashi N, Inoue T, Sugimura Y, Nishiyama N, Fujita T, Ushijima T, Shirai T (2002) Down-regulation of Lsm1 is involved in human prostate cancer progression. Br J Cancer 86: 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tharun S (2009a) Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol 6: 228–232 [DOI] [PubMed] [Google Scholar]
- Tharun S (2009b) Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol 272: 149–189 [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Watson PM, Miller SW, Fraig M, Cole DJ, Watson DK, Boylan AM (2008) CaSm (LSm-1) overexpression in lung cancer and mesothelioma is required for transformed phenotypes. Am J Respir Cell Mol Biol 38: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD (2003) Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell 11: 341–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.