Abstract
MicroRNAs have well-established roles in eukaryotic host responses to viruses and extracellular bacterial pathogens. In contrast, microRNA responses to invasive bacteria have remained unknown. Here, we report cell type-dependent microRNA regulations upon infection of mammalian cells with the enteroinvasive pathogen, Salmonella Typhimurium. Murine macrophages strongly upregulate NF-κB associated microRNAs; strikingly, these regulations which are induced by bacterial lipopolysaccharide (LPS) occur and persist regardless of successful host invasion and/or replication, or whether an inflammatory response is mounted, suggesting that microRNAs belong to the first line of anti-bacterial defence. However, a suppression of the global immune regulator miR-155 in endotoxin-tolerant macrophages revealed that microRNA responses also depend on the status of infected cells. This study identifies the let-7 family as the common denominator of Salmonella-regulated microRNAs in macrophages and epithelial cells, and suggests that repression of let-7 relieves cytokine IL-6 and IL-10 mRNAs from negative post-transcriptional control. Our results establish a paradigm of microRNA-mediated feed-forward activation of inflammatory factors when mammalian cells are targeted by bacterial pathogens.
Keywords: IL-10, let-7, miR-155, miRNA, Salmonella
Introduction
MicroRNAs are a class of genome-encoded small RNAs, of ∼22 nt in length, that govern post-transcriptional repression of target mRNAs in a wide range of biological processes including development, cellular differentiation, apoptosis, fat metabolism and growth control. They also have important roles in human disorders including cancer, neurodegenerative and cardiovascular diseases (Filipowicz et al, 2008; Stefani and Slack, 2008; Bartel, 2009). Regarding the host response to pathogens, microRNAs have well-established roles in viral infections (Ding and Voinnet, 2007; Umbach and Cullen, 2009). Several host microRNAs such as miR-29a and miR-32 exert direct anti-viral activity by downregulating viral mRNAs (Lecellier et al, 2005; Pedersen et al, 2007; Nathans et al, 2009). Conversely, some DNA viruses encode microRNAs and utilize the host machinery to regulate either host or viral mRNAs for their own benefit (Pfeffer et al, 2004; Gottwein et al, 2007; Umbach and Cullen, 2009).
A role of microRNAs in bacterial infections was first discovered in plants where Arabidopsis miR-393 contributed to resistance against the extracellular pathogen Pseudomonas syringae, presumably by repressing auxin signalling (Navarro et al, 2006). In addition, P. syringae was shown to secret effector proteins (Navarro et al, 2008) that similarly to virus-encoded suppressor proteins (Burgyan, 2008; de Vries and Berkhout, 2008) antagonize the small RNA-directed basal immunity of the host.
Before the work in plants, mammalian microRNAs were implicated in bacterial infections because of associations with immunity and inflammation (Baltimore et al, 2008; Lindsay, 2008). That is, recognition of pathogen-associated molecular patterns (PAMPs) by Toll-like receptors (TLRs) was found to differentially regulate several microRNAs, for example, TLR4-mediated sensing of bacterial lipopolysaccharide (LPS) and downstream NF-κB activity induced miR-146a/b and miR-155 (Taganov et al, 2006; O’Connell et al, 2007; Tili et al, 2007; Androulidaki et al, 2009; Ceppi et al, 2009; Liu et al, 2009). Moreover, mice deficient in miR-155 display an altered immune response, and fail to be immunized by an attenuated bacterial pathogen (Salmonella) (Rodriguez et al, 2007). Subsequently, miR-155 was also shown to be induced by Helicobacter pylori (Xiao et al, 2009; Fassi Fehri et al, 2010), a pathogen of the human stomach.
Whereas P. syringae and H. pylori remain extracellular during infection, many bacterial pathogens actively invade host cells or become intracellular after ingestion by phagocytic immune cells such as macrophages. During their multistage infection, intracellular pathogens extensively manipulate the signalling and gene expression cascades of the host for survival and replication (Diacovich and Gorvel, 2010). Whether and how host microRNAs are regulated by invasive and intracellular bacteria remained unknown.
Salmonella enterica serovar Typhimurium (henceforth, Salmonella) is an intensely investigated intracellular bacterial pathogen that causes gastroenteritis in humans and lethal typhoid fever in mice (Cossart and Sansonetti, 2004; Mastroeni et al, 2009). Both host-cell invasion and intracellular replication by Salmonella can be recapitulated in defined cell lines in vitro (Valdez et al, 2009). Pathogenesis is mediated by secreted effector proteins that Salmonella injects into eukaryotic cells via the two type 3 secretion systems (T3SS) that are encoded by the major pathogenicity islands, SPI-1 (invasion) or SPI-2 (intracellular survival). These effectors act in concert to subvert the host cell cytoskeleton, signal transduction pathways, membrane trafficking and pro-inflammatory responses, by directly interacting with host proteins (Galan, 2009; McGhie et al, 2009).
This paper reports microRNA responses of phagocytic (RAW 264.7) and non-phagocytic (HeLa) cells during the course of infection with Salmonella. Using defined bacterial mutants impaired in invasion, intracellular survival or both, we show that bacterial LPS through the innate immune response induces long-lasting microRNA programs that prevail during the course of infection. While these programmes vary considerably depending on cell type and status, downregulation of the let-7 microRNA family appears to be a generic and consistent response, even in endotoxin-tolerant macrophages with mute TLR4-based innate immunity. We provide evidence suggesting that let-7 family members post-transcriptionally repress production of the major cytokine, interleukin-10 (IL-10), via multiple pairing sites in the 3′ UTR of IL-10 mRNA. Collectively, our data suggest that differential expression of microRNAs caused by bacterial infection results in cell type-dependent feed-forward activation of cytokine expression.
Results
Salmonella infection regulates host microRNAs
We identified differentially regulated microRNAs by comparative deep sequencing of a total of 14 cDNA libraries prepared from the small RNA population (19–34 nt) of host cells before or after Salmonella infection, or in mock-treated cells. Experiments were performed in two of the most commonly used cell lines for in vitro study of Salmonella infections, that is, murine RAW 264.7 cells due to their macrophage-like characteristics and human HeLa cells as a common epithelial infection model. In addition to infecting both cell types with wild-type bacteria (Figure 1), we determined microRNA changes in RAW 264.7 cells after challenge with Salmonella mutants deleted for the SPI-1 or SPI-2 major virulence regions (Supplementary Figure S1). On average, ∼50 000 cDNAs were sequenced per library, and typically >70% of all cDNAs in a library matched the known or predicted microRNAs (Supplementary Table S1) compiled by mirBASE version 14 (Griffiths-Jones et al, 2008).
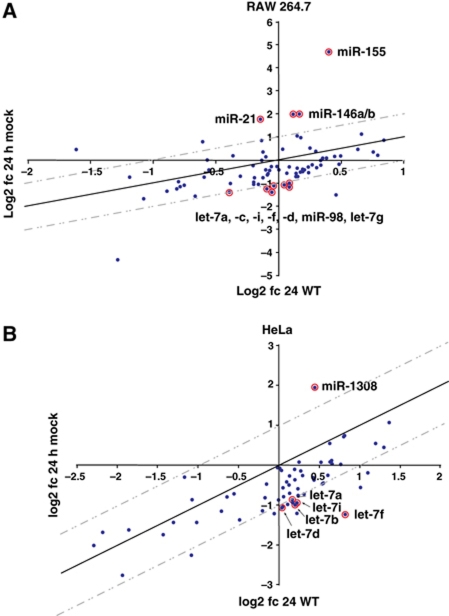
Figure 1.
Salmonella infection regulates host microRNA expression in (A) RAW 264.7 and (B) HeLa cells. Small RNA libraries of uninfected cells at 0 h, cells infected with wild-type Salmonella at 24 h p.i. and mock-treated cells at 24 h p.i., were analysed by 454 sequencing, and microRNA expression changes were calculated by comparison of cDNA hits in the libraries (24 h infection versus 0 h and 24 h mock versus 0 h). The graphs show log2 fold changes in infected cells (y axis) versus log2 fold changes in mock-treated cells (x axis).
Calculation of microRNA expression changes in RAW 264.7 cells as detected by 454 sequencing at 24 h p.i. (post infection) with wild-type Salmonella versus mock treatment (Figure 1A; Supplementary Table S2) readily detected the expected upregulation of miR-21 (3.4-fold), miR-146a/b (4.0-fold) and miR-155 (25.7-fold), all of which are known to accumulate following stimulation of macrophages with bacterial LPS (Taganov et al, 2006). Intriguingly, we observed significant downregulation of several let-7 family members, namely let-7a/c/d/f/g/i and miR-98. All of the above regulations were validated in independent northern blot (Figure 2A) and quantitative real-time PCR (qRT–PCR; Figure 2B) assays. Analysis of additional time points (4 and 8 h p.i.) by both cDNA sequencing and qRT–PCR showed that the upregulation or downregulation of miR-155 and the let-7 family, respectively, is a fast response and most dramatic within the first 4 h of infection (Supplementary Figure S2).
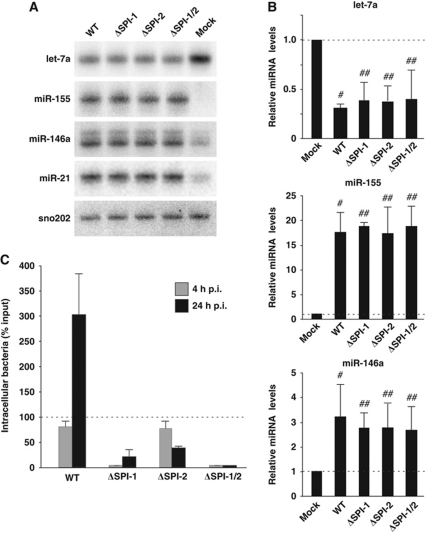
Figure 2.
Expression of let-7 microRNAs is downregulated upon Salmonella infection. (A) Northern blot analysis of let-7a, miR-155, miR-146a and miR-21 expression in RAW 264.7 cells after 24 h of mock treatment or infection with Salmonella wild type, or with mutants defective in invasion (ΔSPI-1), intracellular replication (ΔSPI-2) or both (ΔSPI-1/2). (B) qRT–PCR analysis of expression changes of let-7a, miR-155 and miR-146a in RAW 264.7 cells as above. Mean values±s.d. from three independent experiments are shown. (C) Invasion/infection assays performed in RAW 264.7 cells with Salmonella wild type and the three virulence mutants. Bacteria were enumerated at 4 and 24 h p.i. Mean values±s.d. from three independent experiments are shown. #Significant difference between wild-type infection and mock control (P-value <0.05). ##No significant difference compared with wild-type infection (P-value >0.05).
In HeLa cells, a significant upregulation of microRNAs by Salmonella was limited to miR-1308 (Figure 1B; Supplementary Table S3), a candidate microRNA of unknown function that derives from tRNA (tRF-5005; Lee et al, 2009). In contrast, miR-21, miR-146a/b or miR-155 remained unaffected, which is in accord with a lack of expression of the LPS sensing components TLR2 or the TLR4 co-factor MD-2 in this epithelial cell type (Wyllie et al, 2000; Taganov et al, 2006; O’Connell et al, 2007). Intriguingly, our sequencing data suggested that downregulation of let-7 microRNAs also occurred in HeLa cells (Figure 1B; Supplementary Table S3), and this was further validated by qRT–PCR and northern blot analyses (Supplementary Figure S3B and C). Taken together, these results suggest that Salmonella impacts on microRNA expression in a cell type-dependent manner and generically promotes downregulation of the let-7 family in both phagocytic and non-phagocytic cells.
Extracellular stimulus, rather than invasion, drives the microRNA response to Salmonella
Previous studies of mammalian microRNA alterations induced by bacterial pathogens were limited to extracellular stimulation of TLR4 by treatment with purified LPS or by non-invasive H. pylori (Baltimore et al, 2008; Zhang et al, 2008; Xiao et al, 2009). To evaluate the contributions of extracellular versus intracellular bacteria, we sequenced small RNA populations of RAW 264.7 cells infected with Salmonella mutants defective in invasion (ΔSPI-1), intracellular replication (ΔSPI-2) or both (ΔSPI-1/2). Invasion/infection assays confirmed the expected behaviour in RAW 264.7 cells (Figure 2C): ΔSPI-1 bacteria were poorly internalized yet then replicated at the same rate as the wild type (compare 4 and 24 h p.i.); the ΔSPI-2 strain showed normal uptake at 4 h p.i. followed by intracellular clearance as evident from reduced bacterial load at 24 h p.i.; only few intracellular bacteria were recovered for the ΔSPI-1/2 strain at either infection time point. These infection defects notwithstanding, the three mutants induced alterations to the levels of miR-21, miR-146, miR-155 and the let-7 family similar to those induced by wild-type Salmonella (Figure 2A and B). Importantly, downregulation of the let-7 microRNAs proceeds at the same speed with the wild-type or the ΔSPI-1/2 strain as determined by qRT–PCR at 4 h p.i. (Supplementary Figure S2E).
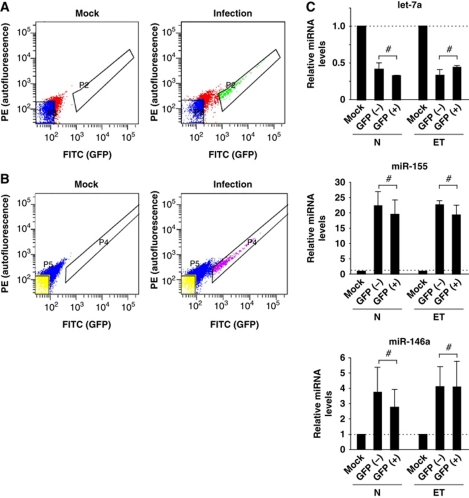
To further distinguish the effects of intracellular and extracellular bacterial stimuli, RAW 264.7 cells were infected with Salmonella expressing green fluorescent protein (GFP), and the fraction of cells which had internalized bacteria were separated from the uninfected fraction by fluorescence-activated cell sorting (FACS). Using an MOI of 1, ∼10% of the host cells contained Salmonella at 24 h p.i. (Figure 3A). Comparison of let-7, miR-146 and miR-155 levels between the two fractions revealed no difference in the regulation of these microRNAs (Figure 3C).
Figure 3.
An extracellular Salmonella stimulus is sufficient to induce a microRNA response in the host cells. (A) Naive (N) and (B) endotoxin-tolerant (ET) RAW 264.7 cells were infected with Salmonella expressing GFP. At 24 h p.i., the fraction of cells with internalized bacteria (GFP+) were sorted from the uninfected cells (GFP−) by FACS. (C) qRT–PCR analysis of expression changes of let-7a, miR-155 and miR-146a in the infected (GFP+) and uninfected (GFP−) RAW 264.7 cells. Mean values±s.d. from three independent experiments are shown. #No significant difference between GFP+ and GFP− fraction (P-value>0.05).
Salmonella may induce apoptosis and cytotoxicity in macrophages by the activity of its secreted SipB protein (Hersh et al, 1999). The congruent patterns of microRNA changes by Salmonella strains that do (wild type, ΔSPI-2) or do not encode SipB (ΔSPI-1 and ΔSPI-1/2) argued, however, against a significant contribution of apoptosis or cytotoxicity to the altered microRNA levels observed upon Salmonella infection. To assess this further, we quantified the apoptosis and cytotoxicity levels by Annexin V and propidium iodide (PI) staining, and by measurement of LDH release. At 4 and 24 h p.i., the invasive wild-type and ΔSPI-2 strains caused a mild increase in the fraction of apoptotic cells and in cytotoxicity, while no significant apoptosis or cytotoxicity was observed in infections with ΔSPI-1 and ΔSPI-1/2 bacteria (Supplementary Figure S4). Moreover, as shown below, non-pathogenic Escherichia coli K12 bacteria impact on miR-155 and let-7 microRNAs similar to Salmonella, which further argues that apoptosis or cytotoxicity effects have little if any role in the regulations reported here. Thus, the sensing of extracellular bacterial antigens sets the microRNA response to Salmonella, at least for the first 24 h of infection.
Repression of let-7 in macrophages is triggered by LPS
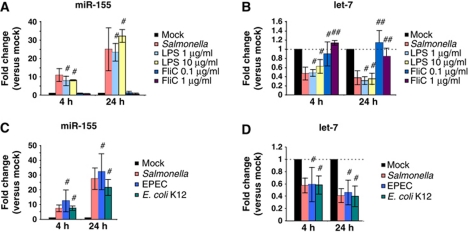
Whereas the induction of miR-21, miR-146 and miR-155 through TLR signalling had been well established, the signal to downregulate let-7 was unknown. Since extracellular bacteria were sufficient, we hypothesized that TLR sensing of main surface antigens of bacteria might as well mediate the repression of let-7. To test this hypothesis, RAW 264.7 cells were exposed to purified LPS or FliC flagellin—sensed by TLR4 or TLR5, respectively—for 4 or 24 h, and miRNA expression was analysed by qRT–PCR. In line with previous reports (O’Connell et al, 2007), LPS strongly induced miR-155 expression in RAW 264.7 cells (Figure 4A). Intriguingly, LPS also fully recapitulated the let-7 repression observed with Salmonella (Figure 4B). By contrast, the TLR5 ligand FliC impacted neither on miR-155 nor let-7 (Figure 4A and B). To ascertain that TLR4 signalling mediates let-7 repression, we treated bone marrow-derived macrophages from TLR4+/+ and TLR4−/− mice with heat-killed Salmonella; as predicted, the downregulation of let-7 was only observed in TLR4+/+ cells (Supplementary Figure S5).
Figure 4.
Regulation of miR-155 and let-7 can be recapitulated with purified Salmonella LPS and pathogenic/non-pathogenic Escherichia coli. Expression changes of (A) miR-155 and (B) let-7a upon stimulation of RAW 264.7 cells with the indicated concentrations of LPS and FliC or wild-type Salmonella as determined by qRT–PCR. Expression changes of (C) miR-155 and (D) let-7a upon challenge of RAW 264.7 cells with wild-type Salmonella, enteropathogenic E. coli (EPEC) or non-pathogenic E. coli K12 as determined by qRT–PCR. Mean values±s.d. from three independent experiments are shown. #No significant difference compared with Salmonella infection (P-value>0.05). ##Significant difference compared with Salmonella infection (P-value <0.05).
Taken together, our experimental data suggest the reduction of let-7 expression during Salmonella infection occurs by TLR4 sensing of bacterial LPS. Importantly, we determined that this regulation is not limited to Salmonella but is also observed after exposure of RAW 264.7 cells to two other gram-negative species, non-pathogenic E. coli K12 and enteropathogenic E. coli (EPEC) (Figure 4C and D).
Endotoxin-tolerant macrophages also exhibit a robust microRNA response
Under physiological conditions, macrophages can become less susceptible to extracellular bacterial stimuli, for example, after pre-exposure to LPS; this desensitized state protecting against septic shock is referred to as endotoxin tolerant and results from a loss of MyD88 recruitment to TLR4 and thus NF-κB activity (Biswas and Lopez-Collazo, 2009).
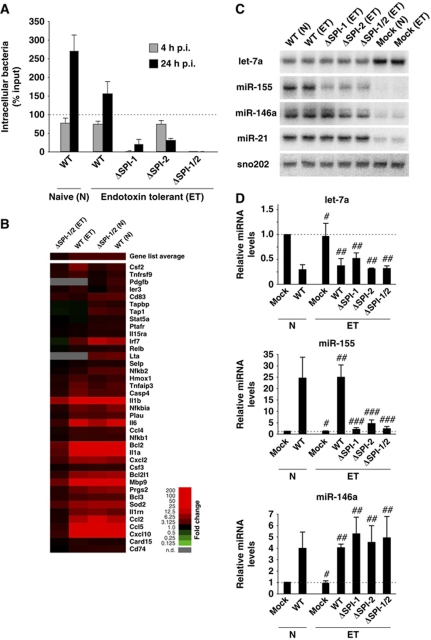
We evaluated the robustness of the microRNA response by infection of endotoxin-tolerant RAW 264.7 macrophages. Wild-type Salmonella invaded naïve and endotoxin-tolerant cells at comparable rates (with a slight drop in replication in the latter), and the ΔSPI-1, ΔSPI-2 and ΔSPI-1/2 showed the same phenotypes as in naïve macrophages (compare Figure 5A versus Figure 2C). Nonetheless, global microarray analysis confirmed that the endotoxin tolerance altered the strength of NF-κB activation by extracellular microbial stimulation (Figure 5B). That is, whereas naïve macrophages responded with the activation of well-established NF-κB-dependent mRNA genes to wild-type and ΔSPI-1/2 bacteria alike, endotoxin-tolerant macrophages mounted a full NF-κB response only when infected with wild-type Salmonella. In contrast, the strictly extracellular ΔSPI-1/2 strain only marginally activated NF-κB-dependent genes in endotoxin-tolerant macrophages. In other words, when TLR4 signal transduction was impaired, an upregulation of NF-κB-dependent mRNAs relies upon the sensing of intracellular Salmonella.
Figure 5.
Salmonella infection induces host microRNA responses in naive (N) and endotoxin-tolerant (ET) macrophages. (A) Invasion/infection assays performed in ET RAW 264.7 cells with Salmonella wild type and the ΔSPI-1, ΔSPI-2, ΔSPI-1/2 mutants. Infection rates were determined at 4 and 24 h post infection. The result of the corresponding assays for naive macrophages infected with wild-type bacteria is shown for comparison. (B) Microarray expression analysis of selected NF-κB-dependent genes in naive and ET RAW 264.7 cells infected with Salmonella wild type and the ΔSPI-1/2 mutant compared with the respective mock-infected cells. The relative expression level of each mRNA is colour-coded, as indicated to the right. (C) Northern blot analysis of let-7a, miR-155, miR-146a and miR-21 expression in ET RAW 264.7 cells infected for 24 h with the bacteria indicated. (D) Corresponding qRT–PCR analysis of expression changes of let-7a, miR-155 and miR-146a. Mean values±s.d. from three independent experiments are shown. #No significant difference between naive and ET mock controls (P-value>0.05); ##no significant difference compared with naive wild-type infection (P-value>0.05); ###significant difference compared with naive wild-type infection (P-value <0.05).
By contrast, the NF-κB-dependent microRNAs still responded to extracellular stimuli. That is, miR-21, miR-146 and let-7 levels were invariably altered in endotoxin-tolerant macrophages by wild-type Salmonella and any of the three mutants (northern blot and qRT–PCR results in Figure 5C and D; additional data with sorted macrophage populations for wild-type Salmonella in Figure 3B and C). The exception was miR-155, which showed a similar pattern as the NF-κB-dependent mRNAs, being poorly induced by the three Salmonella mutants in endotoxin-tolerant cells. The induction levels of miR-155 also indicated a requirement for intracellular sensing such that the invasive ΔSPI-2 mutant caused higher (∼5-fold) upregulation of miR-155 than the non-invasive ΔSPI-1 or ΔSPI-1/2 mutants (∼2-fold). Regardless of the deviant behaviour of miR-155, the macrophage desensitization experiment shows that there is a core set of microRNAs that respond to Salmonella in a very robust manner; this set includes the let-7 family.
let-7 targets the major immune-modulatory cytokines IL-6 and IL-10
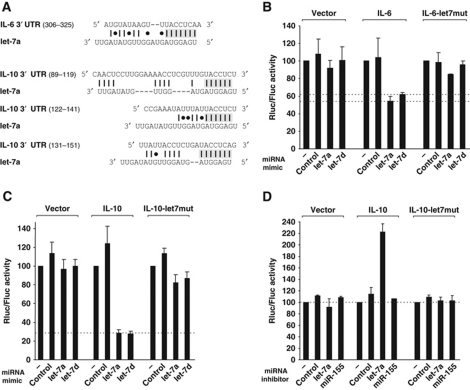
Macrophages are central players in the regulation of the immune response to systemic bacterial infection, part of which is achieved by the secretion of interleukins for intercellular communication. To obtain evidence of a physiological relevance of let-7 downregulation in this process, we focused on potential targeting of cytokine mRNAs by this microRNA family. Bioinformatic analysis had predicted the let-7 family members to repress several important immune-related genes (Krek et al, 2005; Lewis et al, 2005; Asirvatham et al, 2008) including IL-6 and IL-10, two major cytokines of the acute innate immune response. To test whether let-7 post-transcriptionally regulates the mouse IL-6 and IL-10 genes via predicted RNA–RNA complementarity (Figure 6A), the corresponding full-length 3′ UTRs were cloned downstream of the Renilla luciferase ORF in a reporter vector that also expresses firefly luciferase (our internal reference). The reporters were introduced into mouse embryonic fibroblast (MEF) cells, a highly transfectable murine fibroblast line. Co-transfection of synthetic oligo-ribonucleotides mimicking let-7a or let-7d caused 1.8-fold (IL-6) or 3.5-fold (IL-10) downregulation of the reporters, as compared with transfections with no or control microRNA (Figure 6B and C). Regulations were considered specific since neither microRNA regulated Renilla luciferase expression in the parental vector (Figure 6B and C).
Figure 6.
let-7 microRNA family targets the cytokines IL-6 and IL-10. (A) Schematic representation of let-7a complementarity to the 3′ UTRs of IL-6 or IL-10. Shaded regions denote the IL-6 or IL-10 sequences that were deleted or mutated in the reporter vector below. (B, C) MEF cells were transfected with a mixture containing the psicheck2 plasmid (vector) or the Renilla luciferase reporter plasmids carrying the 3′ UTRs of IL-6 or IL-10, or mutants thereof (indicated above the graphs), in combination with microRNA mimics. (D) Same experiment as in (C) but using microRNA inhibitors instead of mimics. Renilla luciferase activity is normalized to that of the Firefly luciferase and set to 100 in cells with no microRNA mimic or inhibitor. Mean values±s.d. from three independent experiments are shown.
IL-6 and IL-10 mRNAs contain let-7 binding sites in their 3′ UTRs
The 3′ UTR of the mouse IL-6 gene was predicted to contain one binding site (positions 318–324) complementary to the seed sequence (7 nt) of all let-7 family members; the interaction would be stabilized by an additional seven base-pair complementarity with the 3′ region of let-7 microRNAs (Figure 6A). Deletion of or point mutations in the anti-seed region in the IL-6 reporter abrogated repression by let-7a and let-7d (Figure 6B and data not shown), supporting the prediction that let-7 directly targets the IL-6 mRNA.
In contrast to IL-6, deletion of the single let-7 binding site initially predicted in the IL-10 reporter (3′ UTR positions 144–150) only slightly reduced repression by let-7a or let-7d, to ∼2-fold as compared with 3.5-fold of the parental IL-10 reporter (Supplementary Figure S6A). Since some microRNAs were shown to act at multiple sites in the same mRNA, we used the RNAhybrid algorithm (Rehmsmeier et al, 2004) to predict additional let-7 binding sites. Using this approach, we identified two potential let-7 binding sites that are complementary to 6 nt of the seed sequence of the let-7 family members (positions 113–118 and 135–140) in the 3′ UTR of IL-10 (Figure 6A). Similar to the first binding site, individual deletions of these two additional sites reduced but did not abolish repression of the IL-10 reporter by let-7; however, simultaneous deletion or mutation of all three sites rendered it refractory to the action of let-7a as well as let-7d (Figure 6C; Supplementary Figure S6A), suggesting that the targeting of all three binding sites is essential for post-transcriptional repression of IL-10. The identified let-7 binding sites in the IL-6 and IL-10 mRNAs are conserved in sequence throughout mammalian genomes, suggesting an evolutionary conserved targeting mechanism (Supplementary Figure S6B and C).
The post-transcriptional control of IL-10 was further confirmed with a let-7 anti-sense LNA (anti-let-7), which acts to repress the endogenous let-7 microRNAs. MEF cells showed an ∼2-fold increase in IL-10 reporter activity when co-transfected with the anti-let-7 as compared with a control inhibitor or anti-miR-155 (Figure 6D), supporting our model that a depletion of the endogenous let-7 pool alleviates IL-10 repression. Again, this effect was specific as the anti-let-7 did not impact on the mutant IL-10 reporter in which the three let-7 binding sites are mutated.
Salmonella infection post-transcriptionally activates cytokine expression
To address whether the observed regulations in the 3′ UTR reflected regulation of the native target mRNAs, we measured let-7-dependent changes in both 3′ UTR reporter activity and IL-6/IL-10 levels in RAW 264.7 macrophages, inducing cytokine production by infection with wild-type Salmonella (Supplementary Figure S7). Increasing concentrations of let-7a or let-7d RNA mimics (Supplementary Figure S7A) caused a concomitant, up to two-fold reduction of 3′ UTR reporter activity and the level of the respective secreted cytokine within a 2.5–20 nM range of co-transfected microRNA mimic (Supplementary Figure S7B–E); above this range, the regulation of both 3′ UTR reporter and secreted cytokine was equally saturated. Conversely, we antagonized let-7 in RAW 264.7 cells with anti-sense LNA oligonucleotides before infection with Salmonella, expecting to see an increase in both the activity of co-transfected 3′ UTR reporters, and the corresponding secreted cytokines. Again, the observed ∼1.4-fold increase in 3′ UTR reporter activity correlated well with a determined increase in secreted IL-6 and IL-10 (Supplementary Figure S7F–H). In other words, the let-7-mediated regulation of the 3′ UTR reporter is representative of regulation of the endogenous target mRNA.
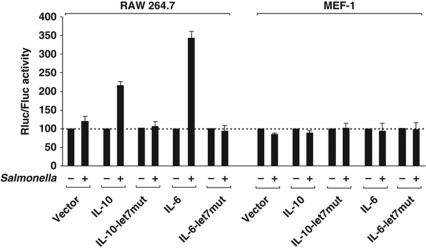
To determine the degree of IL-6 and IL-10 regulation that can be achieved with the decrease of let-7 levels as a consequence of Salmonella infection, RAW 264.7 cells transfected with the 3′ UTR reporters of IL-6 and IL-10 were infected with Salmonella for 24 h. The bacteria triggered a 3.5-fold (IL-6) or 2.2-fold (IL-10) increase in reporter activity, as compared with 1–1.2-fold regulation of the reporters in which the let-7 binding sites were mutated or the empty vector (Figure 7).
Figure 7.
Salmonella infection post-transcriptionally activates expression of IL-6 and IL-10. RAW 264.7 and MEF cells were transfected with the empty psicheck2 plasmid (vector) or luciferase reporters carrying the IL-6 or IL-10 3′ UTRs, or mutants. After 24 h, the cells were mock infected or infected for 24 h with Salmonella wild type before reporter activity was assayed. Renilla and Firefly luciferase activity were analysed as described in Figure 6.
The same experiment was performed in non-responsive MEF cells in which let-7 and miR-155 levels remained constant even after prolonged (24 h p.i.) infection with Salmonella (Supplementary Figure S8). In agreement with the unchanged let-7 levels, the Salmonella infection also failed to impact on IL-6 or IL-10 reporter activity in this cell line (Figure 7). Collectively, the sum of our data suggests that let-7 serves as a post-transcriptional brake of IL-6 and IL-10 synthesis that is released upon the recognition of a pathogen.
Discussion
The present study is the first to address the role of microRNAs in the eukaryotic response to an invasive and intracellular bacterium, and the question whether Salmonella can manipulate host microRNAs similarly to its well-established interference with protein-based signalling cascades by. Our results show that the initial sensing of LPS of extracellular Salmonella induces cell type-dependent microRNA expression programs that predominate during the subsequent intracellular phase of infection; there is a core set of microRNAs that act in the first line of anti-bacterial defence and their expression is refractory to reprogramming by Salmonella's arsenal of effectors. In addition, experiments with endotoxin-tolerant macrophages show that the microRNA response is dramatically influenced by the physiological state of the cells, which along with the cell type specificity has implications for the potential use of microRNAs as gene expression markers in infected tissues or animals. Finally, our study is the first to implicate the widespread let-7 family in anti-bacterial defence, and we provide molecular evidence of how the downregulation of these microRNAs promotes the expression of the key cytokines, IL-6 and IL-10, during Salmonella infection. The results establish a paradigm of how microRNAs facilitate post-transcriptional feed-forward activation of inflammatory factors when mammalian cells are targeted by invasive bacteria.
MicroRNA responses to bacterial infection
Pioneering work on plant–microbe interactions showed that host microRNAs are both passively and actively regulated by bacterial pathogens (Ruiz-Ferrer and Voinnet, 2009). Specifically, Arabidopsis miR-393 is induced at the level of transcription by innate immune sensing of the flagellin-derived PAMP peptide of P. syringae (Navarro et al, 2006). This incurs repression of the main receptor (TIR1) of the hormone auxin, which is a negative regulator of defence responses. Thus, a main scheme in potentiating the basal defence responses is to activate a repressor of a negative regulator of defence. In mammal, the PAMP LPS reduces the levels of let-7 microRNAs by a yet to be discovered mechanism, and this alleviates post-transcriptional repression of IL-6 synthesis; here, the PAMP sensing promotes defence by repressing a repressor of a positive regulator of defence.
Another difference between animals and plants with respect to the employment of the microRNA machinery during host pathogen interplay might be the involvement of secreted bacterial effectors. The effector protein HOPT1-1 of P. syringae was found to antagonize host microRNA activity at the level of Ago1, at least when expressed as a transgene in plants (Navarro et al, 2008). No homologues of HOPT1-1 or other microRNA suppressor proteins have been reported in Salmonella or other bacterial pathogens of mammals so far. Moreover, it appears that the animal microRNA response to infection is persistent and robust to the activity of the >30 translocated effector proteins of Salmonella. Specifically, the SPI-1 and/or SPI-2 mutants elicited the same microRNA response as did wild-type Salmonella or purified LPS. This robustness is remarkable given that numerous Salmonella effectors target NF-κB signalling which also controls microRNA transcription (McGhie et al, 2009). As a note of caution, increased sequencing depth and better cell fractionation might still discover microRNAs that are differentially regulated by extracellular versus intracellular bacteria; similarly, Salmonella effectors might alter microRNA activity rather than the steady-state levels profiled here. It also remains to be seen whether the infection studies so far employed the right conditions for host microRNAs to have the highest impact and bacterial effectors to sufficiently accumulate in the host for measurable interference with microRNA-mediated regulations.
Our deep sequencing results identify miR-21, miR-146 and miR-155 as the most highly regulated microRNAs in macrophages, irrespective of whether infective or attenuated Salmonella are used, and thus provide the first experimental evidence for predicted key roles of LPS and NF-κB in the microRNA response to bacterial infections (Taganov et al, 2006; Chen et al, 2007; Tili et al, 2007). However, host cells must rely on additional stimuli for the sensing of pathogens, especially so if pre-exposure to LPS renders macrophages endotoxin tolerant (Biswas and Lopez-Collazo, 2009). We have discovered highly similar response patterns of NF-κB-dependent microRNAs in naïve versus endotoxin-tolerant macrophages, whereas NF-κB-dependent mRNAs were induced fully by Salmonella in naïve macrophages only. Moreover, miR-155 differed from the above as it failed to be induced by the ΔSPI-1/2 strain in endotoxin-tolerant cells. These differential expressions identify microRNAs as novel candidates for the emerging plasticity of NF-κB-dependent gene regulation. For example, NF-κB initiates cytokine IL-6 expression in primary macrophages upon LPS stimulation, but this response is then sustained by NF-κB-induced C/EBPδ and later attenuated by ATF3, allowing the cells to differentially respond to transient or recurring stimulation of TLR4 (Litvak et al, 2009). In our infection experiments, the limited LPS stimulus by ΔSPI-1/2 bacteria might be below the threshold for miR-155 induction, yet still permit alterations to the levels of the other microRNAs in endotoxin-tolerant cells. Although ΔSPI-1/2 bacteria are both non-invasive and rapidly cleared from macrophages after minor phagocytosis, they might still induce weak NF-κB activity. Thus, the differential expression of miR-155 in endotoxin-tolerant cells constitutes an example of how cells discriminate between short-term and long-term challenges with bacterial pathogens, and argues further that control by NF-κB is a plastic rather than an all-or-nothing event. Importantly, our data also show that let-7, miR-21 and miR-146 belong to those inflammation-related genes that respond to even minute stimulation of NF-κB activity. In contrast, miR-155 as a global regulator of the immune response with many targets (Selbach et al, 2008) is controlled much more stringently during infection.
Profiling epithelial (HeLa) cells revealed yet another uncoupled expression of NF-κB-dependent microRNAs: miR-21, miR-146 and miR-155 did not respond to Salmonella, while let-7 family members were still downregulated. Altogether, the cell types tested here exhibit different microRNA response patterns to infection by one and the same bacterium.
let-7 family regulates cytokines IL-6 and IL-10
We have identified the let-7 family as new microRNAs associated with bacterial infection and shown that its downregulation promotes cytokine production upon attack by Salmonella. MicroRNAs of the let-7 family are highly conserved across species in sequence and function, and associated with cell differentiation and development of cell-based diseases such as cancer (Roush and Slack, 2008). Recently, let-7 repression was also observed in human cholangiocytes by LPS and the protozoan Cryptosporidium parvum; it was postulated that since let-7i negatively regulates TLR4, its repression helps sustain the innate immune response (Chen et al, 2007; Hu et al, 2009).
Here, we identify the cytokines IL-6 and IL-10 as new immune-related targets of let-7 microRNAs (while our paper was in preparation, others identified regulation of IL-6 mRNA by let-7 during cell transformation; Iliopoulos et al, 2009). These two cytokines, along with IL-1 and TNFα, have important pro- and anti-inflammatory roles in Salmonella infections (Klimpel et al, 1995). LPS stimulation of macrophages highly induces cytokine TNFα which then promotes NF-κB translocation to the nucleus and the inflammatory response (Bouwmeester et al, 2004), but also antagonizes NF-κB effects by inducing apoptosis (Micheau and Tschopp, 2003). In concert with IL-1 and IL-6, TNFα also causes systemic inflammatory symptoms such as fever or septic shock (Carswell et al, 1975; Sundgren-Andersson et al, 1998).
IL-10 is important to counteract the above pro-inflammatory effects, and functions to moderate TNFα and IL-1α production, preventing autocrine-induced cell death or endotoxin shock in Salmonella-infected macrophages or mice, respectively (Arai et al, 1995a, 1995b). In light of these established functions of IL-10, we interpret the derepression of IL-10 production by downregulation of let-7 to prevent excessive immune-response outcomes such as septic shock and apoptosis during Salmonella infection. Intriguingly, lower levels of let-7 will also ensure the concomitant upregulation of pro-inflammatory IL-6, balancing elevated IL-10 production and limiting polarization of the immune response towards an anti-inflammatory outcome. Two additional microRNAs, miR-106a and miR-466l, were recently reported to inhibit the IL-10 mRNA by 3′ UTR interactions different from those by let-7 (Sharma et al, 2009; Ma et al, 2010). However, neither of them was differentially regulated in our data sets, suggesting that post-transcriptional regulation of IL-10 during Salmonella infection is primarily by the let-7 family. Nonetheless, whether IL-6 and IL-10 are always concomitantly derepressed upon LPS-triggered let-7 reduction remains to be determined.
We are yet to identify the factors that repress let-7. The RNA-binding protein Lin28 that inhibits let-7 at the precursor level (Heo et al, 2008, 2009) showed unaltered mRNA levels within 24 h of infection (data not shown), suggesting that downregulation of let-7 by Salmonella does not involve a block of microRNA maturation.
Network functions of infection-induced microRNAs
Based on studies of plant immunity and defence mechanisms, it has been suggested that eukaryotes restrict bacterial pathogens by a balanced induction of microRNAs that repress negative defence regulators and repression of microRNAs that repress positive effectors of defence (Ruiz-Ferrer and Voinnet, 2009). In part, this is also visible for Salmonella and mammalian microRNAs, for example, repression of let-7 that represses the positive effector of defence, IL-6. By contrast, the derepression of IL-10 constitutes a novel type of regulation, that is, the repression of a microRNA that represses a negative effector of defence.
Arguably, none of these regulations should be viewed in isolation since almost all Salmonella-regulated microRNAs engage in feedback loops of the inflammatory response. Examples include the complex signalling downstream of TLR4-mediated LPS sensing where upregulated miR-146 represses IL-1 receptor-associated kinase (IRAK-1) and TNF receptor-associated factor 6 (TRAF6), two key adaptor molecules downstream of TLR and cytokine receptors (Taganov et al, 2006). Likewise, miR-155 targets an adaptor in the TLR/IL-1 signalling cascade (TAB2) and the suppressor of cytokine signalling 1 (SOCS1) (Androulidaki et al, 2009; Ceppi et al, 2009; Lu et al, 2009), and let-7i feedback regulates TLR4 (Chen et al, 2007). Our study provides important reference data of regulated microRNAs to foster a better understanding of these complex regulations and their network motifs during natural infections.
Materials and methods
Cell culture and bacterial strains
RAW 264.7, HeLa and MEF-1 cells were grown in RPMI 1640 (Gibco) supplemented with 10% fetal calf serum (Biochrom), 2 mM L-glutamine (Gibco), 1 mM sodium pyruvate (Gibco) and 0.5% β-mercapto-ethanol (Gibco) in a 5% CO2, humidified atmosphere at 37°C.
Primary bone marrow-derived cells were differentiated in DMEM (4.5 g glucose/ml) supplemented with 10% fetal calf serum, 10% L929 differentiation supernatant, 2 mM L-glutamine and 1 mM sodium pyruvate.
Salmonella enterica serovar Typhimurium strain SL1344 is referred to as wild type throughout this study. The ΔSPI-1 (JVS-0405) or ΔSPI-2 (JVS-1103) strains with deletions of Salmonella Pathogenicity Islands 1 or 2 were provided by Paetzold et al (2007) or Karsten Tedin, respectively (Hansen-Wester et al, 2004). The ΔSPI-1/ΔSPI-2 strain (JVS-3614) was constructed by phage P22 transduction of strain JVS-0405 with a lysate of strain JVS-1103. Wild-type Salmonella constitutively expressing GFP from a chromosomal locus (strain JVS-3858) was recently described (Papenfort et al, 2009). E. coli strains E2348/69 (EPEC) and DH5a (K12) were kindly provided by JP Böttcher (MPI-IB Berlin).
Bacterial infection, antigen stimulation and endotoxin tolerance
Two days before infection, 2.5 × 105 host cells per well were seeded into 6-well plates. Overnight cultures of bacteria were diluted 1:100 in fresh L-broth medium and grown aerobically until an OD of 2. Bacteria were harvested by centrifugation and resuspended in complete RPMI medium. RAW 264.7 cells were infected at an MOI of 1 and HeLa cells at an MOI of 10. After addition of bacteria, the cells were centrifuged at 250 g for 10 min at room temperature followed by 20 min incubation in 5% CO2, humidified atmosphere, at 37°C. Medium was then replaced for complete RPMI containing gentamicin (50 μg/ml) to kill extracellular bacteria. Following 30 min incubation, cells were supplied with fresh complete RPMI containing 10 μg/ml gentamicin for the remainder of the experiment.
To quantify bacterial invasion and intracellular replication, infected host-cell cultures were solubilized with PBS (Gibco) containing 0.1% Triton X-100 at the respective time points. Samples were serially diluted in PBS and plated on LB plates. The number of colonies formed from the recovered bacteria was compared with that obtained from the input bacteria used for infection.
For stimulation with purified S. Typhimurium LPS (Sigma, #L6143) or FliC (Alexis; #ALX-522-028) cells were seeded at 2.5 × 105 per well of a 6-well plate 2 days before exposure. LPS and FliC were directly added to the cell-culture medium at the indicated final concentrations.
Endotoxin tolerance was induced by pre-exposing RAW 264.7 macrophages to heat-killed S. typhimurium (HKS) at an MOI of 10 for 20 h. Cells were then washed four times with PBS and cultured in fresh RPMI for 3 additional days. For subsequent experiments, cells were seeded into 6-well plates 2 days before the onset of the infection (assuring a total gap of 5 days between removal of the HKS stimulus and re-stimulation with bacteria).
Sequencing of small RNAs
Infected and mock-treated cells were stabilized with RNAlater (Qiagen) reagent and send to Vertis Biotech AG (Munich, Germany) for RNA isolation, size fractionation (10–29 nt fraction, recovered from denaturating gels), and cDNA library generation. The cDNA libraries were sequenced using a Roche FLX 454 platform. In all, 454 reads were aligned against the respective set of mature microRNAs using the R package mirMap454. Briefly, mirMap454 attempts to identify common 5′ and 3′ adaptor sequences in the reads and removes these before further analysis. Each read is aligned against all mature microRNAs in the database semi-globally using dynamic programming. The simple scoring scheme employed allows terminal gaps in the mature microRNA sequence at no costs, while indels in general are scored with +3, mismatches with +2 and mismatches to N with +1. Motivated by the error distribution of 454 reads, up to three terminal mismatches are ignored in scoring the alignment if the remainder of the sequences aligns without indels or mismatches. For each read, the alignment to a mature microRNA with lowest score is identified. If the score of this alignment is 5 or less, the read is considered to be mapped to this microRNA. If the read maps to n different microRNAs with equal scores, 1/n reads are counted as mapped to each of these microRNAs. The microRNA reads were normalized to the total read number of a given library. Data sets were filtered for microRNAs that covered at least 0.1% of all reads in one of the control data sets or in the respective infection data set.
RNA isolation, northern blots and qRT–PCR
Total RNA was isolated with the TRIzol Reagent (Invitrogen), according to the manufacturer's instructions. For northern blot detection of microRNAs, 25 μg of total RNA per lane was separated on a 15% PAA, 8 M urea gel at 250 V in TAE buffer. In all, 2 μl of 5′ γATP-labelled Decade Marker (Ambion, labelled according to the manufacturer's instructions) was used as a size marker. Wetblot transfer onto a Hybond XL membrane (Amersham) was carried out in 0.5 × TAE buffer for 50 min at 50 V. Transferred RNA was crosslinked at 120 mJ/cm2 (UV Crosslinker Biolink B, PeqLab). The following pre-designed LNA probes were used (Exiqon, Denmark): mmu-let-7a (#39510-00), mmu-miR-155 (#39471-00), mmu-miR-146a (#39466-00) and mmu-miR-21 (#39103-00). snoRNA-202 was detected using a DNA-oligo JVO-5873 (5′-AAGTACTTTCATCAAGTCAGTACAGC-3′). Probes were labelled at the 5′ end with γ-dATP [32P]. Hybridization was carried out overnight at 62°C (LNA) or at 42°C for 30 min (DNA) in Rapid-hyb hybridization buffer (GE Healthcare). Hybridization signals were detected on a Phosphoimager system (FLA 3000, Fuji).
To independently quantify microRNA expression changes, we used TaqMan miRNA assay kits (Applied Biosystems) according to the manufacturer's instructions. The following kits were used: hsa-miR-98, assay-ID 000577; hsa-let-7a, assay-ID 000377; hsa-let-7d, assay-ID 002283; mmu-miR-155, assay-ID 001806; hsa-miR-146a, assay-ID 000468; snoRNA202, assay-ID 001232. Fold changes were determined using the 2(−ΔΔCT)-method (Livak and Schmittgen, 2001). Expression of candidate miRNAs was normalized to expression levels of snoRNA202 (Wong et al, 2007).
Microarray experiments
Transcriptomics of RAW 264.7 macrophages were performed as dual-colour hybridizations on mouse whole genome G4122F 4 × 44k multipack microarrays (Agilent Technologies). To compensate for dye-specific effects, a dye-reversal colour-swap was applied (Churchill, 2002). Total RNA labelling was performed with the Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies). In brief, mRNA was reverse transcribed and amplified using an oligo-dT-T7-promotor primer and resulting cRNA was labelled either with Cyanine 3-CTP or Cyanine 5-CTP. After precipitation, purification and quantification 1.25 μg of each labelled cRNA was fragmented and hybridized to mouse whole genome microarrays according to the supplier's protocol (Agilent Technologies). Scanning of microarrays was performed with 5 μm resolution using a G2565CA microarray laser scanner (Agilent Technologies). Raw microarray image data were analysed with the Image Analysis/Feature Extraction software G2567AA (Version A.9.5, Agilent Technologies). The extracted MAGE-ML files were transferred to and analysed with the Rosetta Resolver Biosoftware, Build 7.1 (Rosetta Biosoftware). Ratio profiles comprising single hybridizations were combined in an error-weighted manner to create ratio experiments. A 1.5-fold change expression cutoff for ratio experiments was applied together with anti-correlation of ratio profiles rendering the microarray analysis highly significant (P-value>0.01), robust and reproducible. Genes identified as differentially expressed were processed further by calculating the respective fold changes based on the relative intensity values obtained from infection-specific and mock-treated samples in the same hybridization. Generation of a Heatmap diagram representing infection-specific mRNA expression changes was done by log2 transformation of fold changes using the Software ‘Cluster’ followed by visualization of the result table using the software ‘Treeview’ (Eisen et al, 1998). The data presented here are deposited in NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and accessible through GEO Series accession number (GSE27703).
Cell sorting
Cells infected with GFP expressing Salmonella were washed twice with PBS and harvested by centrifugation at 250 g for 10 min at room temperature. The cell pellet was resuspended in PBS, filtered through a MACS pre-separation filter (Miltenyi Biotec), and sorted using a FACS Aria II system (BD Biosciences). First gating was performed by selecting the fraction of intact cells based on granularity (side-scatter) and cell diameter (forward-scatter). Thereon, the GFP-positive and negative fractions were defined based on fluorescence signals as detected in the FITC channel (GFP-signal) and the PE channel (autofluorescence). Cells transfected with fluorescein-labelled LNA oligos (Exiqon) were sorted using a FACS Aria III system (BD Biosciences) using the same strategy as described above, detecting the fluorescein signal in the FITC channel.
Apoptosis and necrosis evaluation tests
LDH release from cells infected as described above was quantified using the Cytotox96 kit (Promega) according to the manufacturer's instructions. For quantification of apoptosis induction by Annexin V/PI staining, cells were washed twice with PBS and resuspended in binding buffer (BD Pharmingen) at 106 cells/ml. In all, 100 μl of cell suspension was mixed with 5 μl APC-labelled Annexin V (BD Pharmingen) and 1 μl of 500 μg/ml PI (lyophilized stock from Sigma). Upon incubation at room temperature for 15 min (light-protected) cells were subjected to flow cytometry using a BD LSR II flow-cytometer. Upon gating on the fraction of intact cells based on granularity (side-scatter) and cell diameter (forward-scatter) annexin-positive and PI-positive cell populations were determined using the appropriate single-stained controls and the Annexin V and PerCP channels versus forward-scatter. Subsequent gating was performed on the fraction of cells staining positive for Annexin V, but negative for PI to determine changes in the fractions of apoptotic but not necrotic cells.
Luciferase assays
The 3′ UTRs of murine IL-6 and IL-10 were amplified by PCR from an MEF-1 cDNA library and cloned downstream of the Renilla luciferase coding sequence, between the XhoI and NotI sites of the psicheck2 luciferase reporter vector (Promega). Primers used in PCR reactions were (sequences in 5′ to 3′ direction) for IL-6: JVO-4410 (ATCGGACTCGAGTGCGTTATGCCTAAGCATATC) and JVO-4411 (TCCGATGCGGCCGCAAATATAATATAATTTATTTGTTTGAAGACAGTCT); for IL-10: JVO-4414 (ATCGGACTCGAGAACACCTGCAGTGTGTATTGAG) and JVO-4415 (TCCGATGCGGCCGCCGAATAAGATCCATTTATTCAAAAT). Mutants of IL-6 and IL-10 3′ UTRs were generated by site-directed mutagenesis using the plasmid psicheck2-IL-6 and psicheck2-IL-10 as template, respectively, using the QuickChange mutagenesis kit from Stratagene. Primers for the IL-6 reporter let-7 binding site mutant: JVO-4540 (AGTTGCAAGACATGAATTGCTAATTTAAATATGTT); for IL-10 reporter deletion of let-7 binding sites: JVO-4686 (CTTGGAAAACCTCGTTTGTCTCCGAAATATTTATTAC), JVO-4804 (GTACCTCTCTCCGAAATATTTATGTTCCCATTCTATTTATTCACTG), JVO-4641 (CGAAATATTTATTACCTCTGAGTTCCCATTCTATTTATTCAC); for the IL-10 reporter mutant of let-7 binding sites: JVO-7688 (GGAAAACCTCGTTTGCCAACATCTCCGAAATATTTATC), JVO-7690 (CTCCGAAATATTTATCCAACATGACCAACAAGTTCCCATTCTATTTATTC) and the respective complementary oligonucleotides. To generate the IL-10 3′ UTR triple mutant devoid of all three let-7 binding sites, psicheck2-IL10-Δlet7(1) (template), primer JVO-4860 (CTCGTTTGTCTCCGAAATATTTATGTTCCCATTCTATTTATTCACTG) and its complement were used.
For the transfection experiments 5.0 × 104 MEF-1 cells were seeded into 24-well plates 24 h before transfection. Reporter vectors (1 μg) were transfected together with 50 nM pre-miR miRNA precursor molecules (Ambion; negative control ID#AM17110; hsa-let-7a ID#PM10050; hsa-let-7d ID#PM11778; mmu-miR-155 ID#PM13058) or with 50 nM of miRCURY LNA microRNA-specific knockdown probes (Exiqon; scrambled-miR cat#199002-00; hsa-let-7a cat#118000-00; mmu-miR-155 cat#139471-00) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were collected 48 h after transfection. Transfection of RAW 264.7 cells with the pre-miR precursor molecules or fluorescein-labelled LNA knockdown probes (Exiqon; scrambled-miR cat#199004-08; hsa-let-7a cat#410017-08; hsa-let-7d cat#410020-08; mmu-miR-155 cat#411222-08) for the comparison of IL-6 and IL-10 reporter activity and cytokine production was performed using Lipofectamine 2000. Salmonella infections were carried out 24 h later (pre-miR transfections) or 48 h later (LNA transfections; cell sorting step 24 h post transfection). For analysis of luciferase activity in response to endogenous let-7 regulation transfection of RAW 264.7 cells with the reporter vectors was performed using the Amaxa Cell Line Nucleofector® Kit V (Lonza), according to the manufacturer's instructions. Salmonella infection was performed 24 h after transfection/electroporation, and cells were collected 24 h later. Firefly and Renilla luciferase activities were measured on a Wallac 1420 Victor3 multilabel counter (Perkin-Elmer) using beetle-juice and Renilla-juice (PJK GmbH).
IL-6 and IL-10 quantification
The amount of IL-6 and IL-10 present in culture supernatants of RAW 264.7 cells was determined by ELISA using the OptEIA Mouse IL-6 immunoassay (BD Pharmingen) or the Quantikine Mouse IL-10 immunoassay (R&D Systems) according to the manufacturer's instructions.
Statistical analysis
Statistical tests were performed using a two-tailed Student's t-test with equal variances. P-values ⩽0.05 were considered as a measure for statistical significance. Results of statistical analysis are mentioned in the respective figure legends.
Supplementary Material
Acknowledgments
We thank Jörg Hackermüller for data analysis; and Avinash Sonawane and Kathrin Fröhlich for technical contributions in the early phase of the project. Work in the Vogel lab was supported by funds from SIROCCO (EU FP7) and the German Ministry of Education and Research (grants NGFN 01GS0806/JV-BMBF-01 and 0315836).
Footnotes
The authors declare that they have no conflict of interest.
References
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C (2009) The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31: 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hiromatsu K, Nishimura H, Kimura Y, Kobayashi N, Ishida H, Nimura Y, Yoshikai Y (1995a) Effects of in vivo administration of anti-IL-10 monoclonal antibody on the host defence mechanism against murine Salmonella infection. Immunology 85: 381–388 [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hiromatsu K, Nishimura H, Kimura Y, Kobayashi N, Ishida H, Nimura Y, Yoshikai Y (1995b) Endogenous interleukin 10 prevents apoptosis in macrophages during Salmonella infection. Biochem Biophys Res Commun 213: 600–607 [DOI] [PubMed] [Google Scholar]
- Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB (2008) MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol 45: 1995–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD (2008) MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9: 839–845 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Lopez-Collazo E (2009) Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol 30: 475–487 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Burgyan J (2008) Role of silencing suppressor proteins. Methods Mol Biol (Clifton, NJ) 451: 69–79 [DOI] [PubMed] [Google Scholar]
- Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B (1975) An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci USA 72: 3666–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P (2009) MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA 106: 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, Splinter PL, O’Hara SP, LaRusso NF (2007) A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem 282: 28929–28938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA (2002) Fundamentals of experimental design for cDNA microarrays. Nat Genet 32(Suppl): 490–495 [DOI] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science (New York, NY) 304: 242–248 [DOI] [PubMed] [Google Scholar]
- de Vries W, Berkhout B (2008) RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol 40: 2007–2012 [DOI] [PubMed] [Google Scholar]
- Diacovich L, Gorvel JP (2010) Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol 8: 117–128 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T, Churin Y, Gerhard M, Meyer TF (2010) Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS ONE 5: e9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Galan JE (2009) Common themes in the design and function of bacterial effectors. Cell Host Microbe 5: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR (2007) A viral microRNA functions as an orthologue of cellular miR-155. Nature 450: 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36 (Database issue): D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen-Wester I, Chakravortty D, Hensel M (2004) Functional transfer of Salmonella pathogenicity island 2 to Salmonella bongori and Escherichia coli. Infect Immun 72: 2879–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN (2008) Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A (1999) The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA 96: 2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Zhou R, Liu J, Gong AY, Eischeid AN, Dittman JW, Chen XM (2009) MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J Immunol 183: 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K (2009) An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimpel GR, Asuncion M, Haithcoat J, Niesel DW (1995) Cholera toxin and Salmonella typhimurium induce different cytokine profiles in the gastrointestinal tract. Infect Immun 63: 1134–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O (2005) A cellular microRNA mediates antiviral defense in human cells. Science (New York, NY) 308: 557–560 [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lindsay MA (2008) microRNAs and the immune response. Trends Immunol 29: 343–351 [DOI] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A (2009) Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol 10: 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E (2009) miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci USA 106: 15819–15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, CA) 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY (2009) Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Liu X, Li D, Wang P, Li N, Lu L, Cao X (2010) MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J Immunol 184: 6053–6059 [DOI] [PubMed] [Google Scholar]
- Mastroeni P, Grant A, Restif O, Maskell D (2009) A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol 7: 73–80 [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V (2009) Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12: 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114: 181–190 [DOI] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM (2009) Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34: 696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science (New York, NY) 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science (New York, NY) 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA 104: 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzold S, Lourido S, Raupach B, Zychlinsky A (2007) Shigella flexneri phagosomal escape is independent of invasion. Infect Immun 75: 4826–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158 [DOI] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449: 919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T (2004) Identification of virus-encoded microRNAs. Science (New York, NY) 304: 734–736 [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA (New York, NY) 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A (2007) Requirement of bic/microRNA-155 for normal immune function. Science (New York, NY) 316: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ (2008) The let-7 family of microRNAs. Trends Cell Biol 18: 505–516 [DOI] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485–510 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B (2009) Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci USA 106: 5761–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Sundgren-Andersson AK, Ostlund P, Bartfai T (1998) IL-6 is essential in TNF-alpha-induced fever. Am J Physiol 275 (6 Part 2): R2028–R2034 [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D (2006) NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179: 5082–5089 [DOI] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR (2009) The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23: 1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez Y, Ferreira RB, Finlay BB (2009) Molecular mechanisms of Salmonella virulence and host resistance. Curr Top Microbiol Immunol 337: 93–127 [DOI] [PubMed] [Google Scholar]
- Wong L, Lee K, Russell I, Chen C (2007) Endogenous controls for real-time quantification of miRNA using TaqMan MicroRNA assays. Appl Biosyst, Application Note 127AP11-01: 1–8 [Google Scholar]
- Wyllie DH, Kiss-Toth E, Visintin A, Smith SC, Boussouf S, Segal DM, Duff GW, Dower SK (2000) Evidence for an accessory protein function for Toll-like receptor 1 in anti-bacterial responses. J Immunol 165: 7125–7132 [DOI] [PubMed] [Google Scholar]
- Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM (2009) Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis 200: 916–925 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H (2008) miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest 88: 1358–1366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.