Abstract
Even though nicotine has been shown to modulate mRNA expression of a variety of genes, a comprehensive high-throughput study of the effects of nicotine on the tissue-specific gene expression profiles has been lacking in the literature. In this study, cDNA microarrays containing 1117 genes and ESTs were used to assess the transcriptional response to chronic nicotine treatment in rat, based on four brain regions, i.e. prefrontal cortex (PFC), nucleus accumbens (NAs), ventral tegmental area (VTA), and amygdala (AMYG). On the basis of a non-parametric resampling method, an index (called jackknifed reliability index, JRI) was proposed, and employed to determine the inherent measurement error across multiple arrays used in this study. Upon removal of the outliers, the mean correlation coefficient between duplicate measurements increased to 0.978±0.0035 from 0.941 ±0.045. Results from principal component analysis and pairwise correlations suggested that brain regions studied were highly similar in terms of their absolute expression levels, but exhibited divergent transcriptional responses to chronic nicotine administration. For example, PFC and NAs were significantly more similar to each other (r=0.7; P<10−14) than to either VTA or AMYG. Furthermore, we confirmed our microarray results for two representative genes, i.e. the weak inward rectifier K+ channel (TWIK-1), and phosphate and tensin homolog (PTEN) by using real-time quantitative RT-PCR technique. Finally, a number of genes, involved in MAPK, phosphatidylinositol, and EGFR signaling pathways, were identified and proposed as possible targets in response to nicotine administration. © 2001 Elsevier Science B.V. All rights reserved.
Keywords: Microarray, Normalization, mRNA expression, Brain, Nicotine, Pathway
1. Introduction
Nicotine is believed to be the primary component in tobacco smoke, which rewards habitual smoking. Animal studies have indicated that nicotine stimulates dopamine secretion in the outer shell of the nucleus accumbens (NAs) in a manner similar to that of cocaine, amphetamine, and morphine [38]. Moreover, nicotine was shown to increase the extracellular levels of excitatory amino acids, glutamate and aspartate, in the ventral tegmental area (VTA) upon stimulation of nicotinic acetylcholine receptors (nAChRs) [46]. Involvement of nicotine in both dopaminergic and glutamergic neurotransmission may underlie its addictive potential and association with neuropsychiatric disorders, such as Alzheimer’s disease (AD), Parkinsonism, and Schizophrenia [7,33].
Previous studies have demonstrated that nicotine administration modulates expression level of a variety of genes, including those involved in the catecholamine and neuropeptide synthesis, and transcriptional activation. For example, acute single injection of nicotine was reported to increase mRNA levels of tyrosine hydroxylase, a rate-limiting step in catecholamine synthesis, in NAs and VTA [6,49]. Chronic exposure to nicotine induced long-term increases in the mRNA expression levels of genes involved in the regulation of food intake and energy expenditure, such as neuropeptide Y (NPY), orexins, and their receptors [27,22]. Similar to other addictive substances, such as cocaine and alcohol, nicotine also was shown to induce immediate early gene expression, e.g. c-fos, jun-B, in various brain regions [18,53,34,37]. Upon nicotine administration, Fos immunoreactivity was induced particularly in the accessory optic system [36,35,39] and the mesocorticolimbic system [43,44]. Nevertheless, a comprehensive mRNA expression profiling in regards to nicotine treatment is yet to be performed. Clearly, the characterization of region-specific response to nicotine using high-throughput mRNA expression data may further enhance our knowledge of the mechanisms involved in nicotine addiction/withdrawal as well as the pharmacological links between nicotine and neural degeneration.
DNA chip technology makes possible to obtain gene expression data on thousands of genes simultaneously. Consequently, cDNA microarrays have been used to analyze differential expression of genes in various contexts and organisms, e.g. differentiation and development [50], disease diagnosis [15,1], and drug discovery [20,55]. Majority of the microarray data analysis in the literature was performed using clustering techniques that allow grouping of genes with similar expression profiles [15,10]. These analyses allow for the identification of regulatory mechanisms that are common to genes with similar expression patterns as well as the allocation of unknown genes to known functional gene groups. Recent studies also revealed that expression profiling in response to a pharmacological agent is crucial for identifying candidate genes as possible drug targets [20].
In this study, we used a neural-focused cDNA microarray, which contains 1117 unique genes and ESTs, to characterize (a) the extent of regional differentiation in brain gene expression of saline-control rats, and (b) transcriptional response to chronic nicotine in rat brain. We also analyzed the reliability of our microarray data, and developed an index to normalize the error associated with printing and hybridization across multiple arrays.
2. Materials and methods
2.1. Production of microarrays
A set of 1117 known genes and ESTs were selected from 15K human cDNA set (Research Genetics, AL) based on their expression patterns in brain and other tissues, biological functions and/or similarity with known brain-expressed genes. The clones were cultured in Luria-Bertani medium; purified using 96-well alkaline lysis miniprep kit (Qiagen, Inc., CA) and amplified by a primer pair (forward: 5′-CTGCAAGGCGATTAAGTTGGGTAAC; and reverse: 5′-GTGAGCGGATAACAATTTCACACAGGAAACAGC). Two duplicates of 1117 cDNA clones plus 35 PCR products containing no inserts were printed on each array (600 μm apart) on an S&S Nitran SuperCharge nylon membrane (Schelicher & Schuell, NH) using GMS 417 Arrayer (Affymetrix Inc., CA) with solid steel pins (pin size: 300 μm).
2.2. Animals and nicotine self-administration
Male Holtzman rats (225–250 g; Harlan Sprague–Dawley, Madison, WI) were used for all experiments. The self-administration procedure was essentially that described previously [54,26]. Briefly, rats were anesthetized, implanted with jugular catheters and tethers, and placed into operant chambers for the duration of experiment. During recovery, daily injections of gentamycin (4 mg/kg, i.v.) and hourly injection of saline (50 μl containing 200 U/ml heparin, i.v.) were administered. On the 3rd day of recovery, the light over each lever was turned on and i.v. nicotine (0.03 mg/kg/injection, pH 7.0; expressed as the free base of nicotine sulfate; Sigma) was made available contingent upon one press of one of the two levers (the rats do not receive any prior training). A 7-s time-out, during which the light over the active lever was off, followed each injection. Nicotine was always available except for a 1–2-h period at the end of the light portion of the light cycle, when the nicotine solutions were changed and the animals were cared for; the cue lights over the levers were not on during this period. At the end of 21 days of self-administration of nicotine or saline, brains were removed immediately after decapitation (following a lethal overdose of sodium pentobarbital, 125 mg/kg, i.p.). Two millimeter slices were made with a Stoelting tissue slicer; each slice was placed on an ice-cold dish and punches were made as described [48] to obtain specific brain regions.
2.3. Total RNA isolation
Total RNA was isolated from the corresponding pooled regions of three rats per group using RNeasy kits (Qiagen Inc., CA). Four brain regions were examined in this study: prefrontal cortex (PFC), nucleus accumbens (NAs), ventral tegmental area (VTA), and amygdala (AMYG), which have been shown to be important sites involved in drug addiction. Before use, RNA samples were treated with RNAse-free DNAse I at 37°C for 30 min. RNA concentrations were determined and assessments were made with respect to the integrity of the samples.
2.4. Radioactive cDNA probe preparation and microarray hybridization
cDNA probes were prepared as previously described [51]. Briefly, 3–10 μg total RNA were reverse-transcribed in a reaction mixture containing 8 μl of 5× first strand RT buffer, 1 μl of 1 μg/μl 24-mer poly(dT) primer, 4 μl of 20 mM dNTPs (-dCTP), 4 μl of 0.1 M DTT, 40 U of RNasin, 6 μl of 3000 Ci/mmol α-33P-dCTP and Dep-water to a final volume of 40 μl. The RT mixture was first heated at 70°C for 5 min, followed by incubation on ice for 2 min. Two μl of Superscript II reverse transcriptase (Life Technologies Inc., CA) was then added followed by incubation at 42°C for 60 min. At the end of incubation, 5 μl of 0.5 M EDTA was added to chelate divalent cations. After addition of 10 μl of 1.0 M NaOH, the samples were incubated at 65°C for 30 min to hydrolyze the remaining RNA. Following the addition of 25 μl of 1 M Tris (pH 8.0), the samples were purified using Bio-Rad 6 purification columns (Hercules, CA). cDNA microarrays were pre-hybridized in a 4-ml hybridization buffer containing 3.2 ml Microhyb (Research Genetics, AL) and 0.8 ml 50% dextran sulfate, 100 μl of 10 mg/ml denatured human Cot 1 DNA (Life Technologies Inc., CA) and 100 μl of 8 mg/ml denatured poly(dA) (Pharmacia Inc., NJ). After atleast 4 h of pre-hybridization at 65°C, approximately 106 cpm/ml of heat-denatured cDNA probes were added followed by 17 h of incubation at 65°C. Hybridized arrays were washed in 2× SSC and 1% SDS twice at room temperature followed by 1–2 times of washing in 2× SSC and 0.1% SDS at 55°C for 15 min each [51]. The microarrays were exposed to phosphorimager screens for 1–3 days. The screens were then scanned in a Molecular Dynamics STORM PhosphorImager (Sunnyvale, CA) at 50 μm resolution.
2.5. Image analysis, array normalization, and measurement of duplicate reliability
The software ImageQuant (Molecular Dynamics Inc., CA) was used to convert the hybridization signals on the image into intensity values, and the clone ID numbers and expression data were stored in the MS Excel format. In order to adjust for the hybridization differences among multiple arrays used in this study, we normalized the intensity of each spot to its pin-specific mean hybridization intensity value (pin-wise normalization). Furthermore, the average background intensity was calculated based on the hybridization intensities of the spots without inserts.
Differences (absolute) between the normalized intensities of two duplicates, Dijk, were calculated, where i refers to the clone ID number (N = 1117; ESTs and unique genes), j to type of the treatment (saline or nicotine), and k to the brain region (PFC, NAs, VTA, AMYG). The inherent error associated with the microarray data was estimated by analyzing the sample distribution of the absolute differences between duplicates, pooled from all arrays under investigation (i.e. error at the population level). Consequently, we developed an index, called jackknifed reliability index (JRI), which estimates the influence of each duplicate difference on a sample statistics, i.e. kurtosis, using a resampling routine written in Matlab [31]. Kurtosis, k, fourth central moment divided by the standard deviation, estimates the height of the sample distribution:
where E(x) is the expected value of sample vector x (Mathworks, Inc., MA). The jackknifed kurtosis value for each duplicate difference (JRIijk) corresponds to the sample (N = 9216) kurtosis value without that particular observation. A similar jackknifing approach has been applied for the diagnosis of outliers in regression analysis, e.g. Cook’s distance [11]. Based on the distribution of JRI values obtained for the absolute duplicate differences, one can set a cut-off point (Cpool) that consistently estimates the inherent error level above which an expression reading is considered as unreliable. Theoretically (i.e. error-free), no difference between duplicate measurements should be expected. Expected JRI value, JRIexp, is then the magnitude of JRI when Dijk equals to zero. Consequently, we obtained the minimum duplicate difference, Dmin, which is greater than zero and with JRI value equaling to JRIexp, as the cut-off value (Cpool). Even though each array has its own sampling error, Cpool represents the reasonable error value permitted across microarrays.
2.6. Statistical analysis
Pairwise correlations between brain regions with respect to (a) the magnitude of expression in saline-control experiments and (b) the transcriptional response to nicotine were assessed using Pearson’s correlation coefficient (Matlab, Inc., MA). Principal components analysis (PCA) was performed using average expression levels calculated from four brain regions (variables) for saline and nicotine treatments, separately (S-PLUS Inc., WA). Principle components were extracted as the linear combinations of the variables, where a, b, c, and d are loadings, i is index for a component, j is either the saline or nicotine treatment, and E refers to average normalized expression level:
2.7. Confirmation by reverse transcription and real time polymerase chain reaction (real time quantitative RT-PCR)
We confirmed our microarray results from two representative genes, i.e. the weak inward rectifier K+ channel 1 (TWIK-1), and the phosphatase and tensin homolog (PTEN), by using the real-time quantitative RT-PCR technique. Total RNA was extracted from brain punches (VTA and AMYG) obtained from an independent time-course experiment of nicotine self-administration described in a previous study [26]. Optimal RT-PCR conditions for PTEN and TWIK-1 reported here were determined by the strategy described previously [25]. Some modifications were made to optimize conditions for real time RT-PCR protocol. Briefly, total RNA concentration of each sample was measured using a UV spectrophotometer at 260 nm, whereby all samples were diluted to 0.2 μg/μl. Immediately prior to reverse transcription, sample total RNA concentration was measured in duplicate using RIBOGreen™ kit (Molecular Probes, Inc., Eugene, OR), and later was used to normalize the expression data obtained from real time RT-PCR as suggested by the manufacturer [3]. Then, 0.5 μg of total RNA was reverse-transcribed in a final volume of 20 μl containing 4 μl of 5× reverse transcriptase buffer (0.1 M Tris–HCl, pH 8.8; 0.5 M KCl, 1% Triton X-100), 5 mM MgCl2, 10 mM dithiothreitol, 0.625 mM of each dNTP, 20 U RNasin, 1 μl of 50 μM random hexamers and 200 U SuperScript II RNase H− reverse transcriptase (Gibco BRL Life Technologies, Grand Island, NY). The RT mixtures were incubated at 42°C for 1 h and then heated at 95°C for 5 min to inactivate the reverse transcriptase. Amplification of 4 μl RT mixture (equivalent to 0.1 μg total RNA) was carried out using core reagents from the SYBR® Green Kit (PE Biosystems, Foster City, CA), 5 μl 10× SYBR® Green PCR buffer, 4.0 μl 25 mM MgCl2, 1.0 μl 12.5 mM dNTP mix with dUTP, 1 μl of sense or antisense primers (0.1 μg/μl) and 2.5 U AmpliTaq DNA polymerase in a total volume of 50 μl. The RT-PCR reactions were initially denatured at 94°C for 3 min, and then were subjected to cycles of denaturation (94°C, 30 s), annealing (60°C, 30 s) and extension (72°C, 45 s). The Bio-Rad iCycler thermal cycler (Bio-Rad, Hercules, CA) was used to carry out the real time PCR. The number of amplification cycles was set to 40 for both genes. After the last cycle, the extension was continued for another 7 min at 72°C. A 15-μl sample was resolved on a 1% agarose gel containing 25 μl ethidium bromide (500 μg/ml) to check for product purity. Expression level of each gene was determined after specifying an amplification cycle that corresponds to the exponential growth phase of the PCR reaction for each gene (23 cycles for TWIK-1; 28 cycles for PTEN; data not shown). Intensity values for control and treatment samples were measured at these specified amplification cycles. Intensity values were based on the incorporation of fluorescent SYBR® green dye (excitation/emission, 497/520 nm; Molecular Probes, Inc., Eugene, OA) into the double strand PCR products and were measured by the Bio-Rad iCycler thermal cycler. The primer sequences used for PCR amplification were: (1) PTEN (264 bp): sense 5′-TCTACTCCTCCAACTCAGGAC-3′ and antisense 5′-CATTATCCGCACGCTCTATAC-3′, and (2) TWIK (382 bp): sense 5′-TGTCCTGCTTCTTCTTCATCCC-3′ and antisense 5′-TCGTTTTGCTTCTGCTCCTCC-3′.
3. Results
3.1. Microarray normalization and reliability
The jackknifed reliability index, JRIijk, tagged each clone per treatment per region with a reliability value that estimates its influence on the sample distribution (see Section 2.5 for details). Cut-off value for unreliable duplicate measurements (Cpool) was estimated as 0.14 at the log 10 scale (i.e. 1.38- or 0.72-fold difference). Based on this Cpool value, we discarded approximately 13.5% of the total number of observations (Ntotal = 9216). However, among the multiple microarray membranes, the percentage of reliable measurements ranged from 72 to 93% (see Fig. 1 for an example) with an average reliability of 86.6% per array (±7.0% S.D.). Upon filtering out the outliers from each array, the mean correlation coefficient between duplicate measurements equaled to 0.978 (S.D.: ±0.0035; Narray = 8).
Fig. 1.
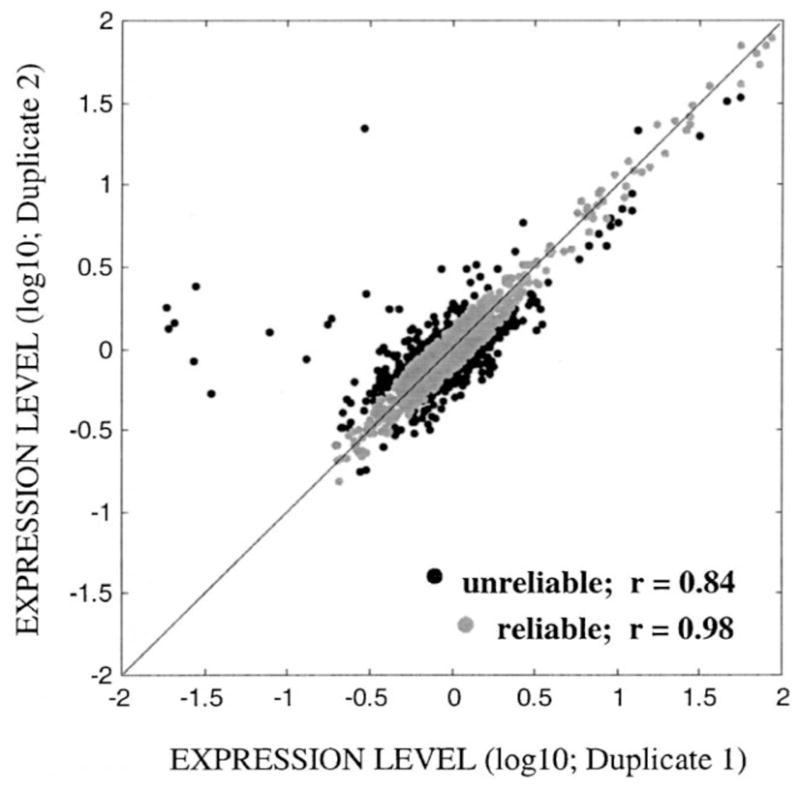
A representative plot of the duplicate normalized expression levels of a microarray (i.e. PFC of saline control rats) before (dark shade) and after (light shade) removing the outliers based on the Cpool value (0.14 at log 10 scale). Straight line is the theoretical 1:1 correspondence between duplicates. Upon outlier removal (Noutlier=324), Pearson’s correlation coefficient increased from 0.84 to 0.98.
3.2. Variability in gene expression level within and among brain regions
In order to compare the expression levels of genes among multiple brain regions or time points, one has to use only those clones reliably measured across all arrays under investigation. In our data set, 391 clones whose expression levels were reliably measured among four brain regions under investigation have met this criterion. Based on the saline-control experiments, pairwise correlation coefficients between regions in terms of the magnitude of gene expression, ranged between 0.96 and 0.98, all of which were highly significant (P<10−5; Fig. 2). Furthermore, the first principle component of microarray data from saline-control rats, PC1saline, accounted for 97.6% of the variation within the expression data, suggesting that most of the variability in the microarray data was due to differences within a brain region.
Fig. 2.
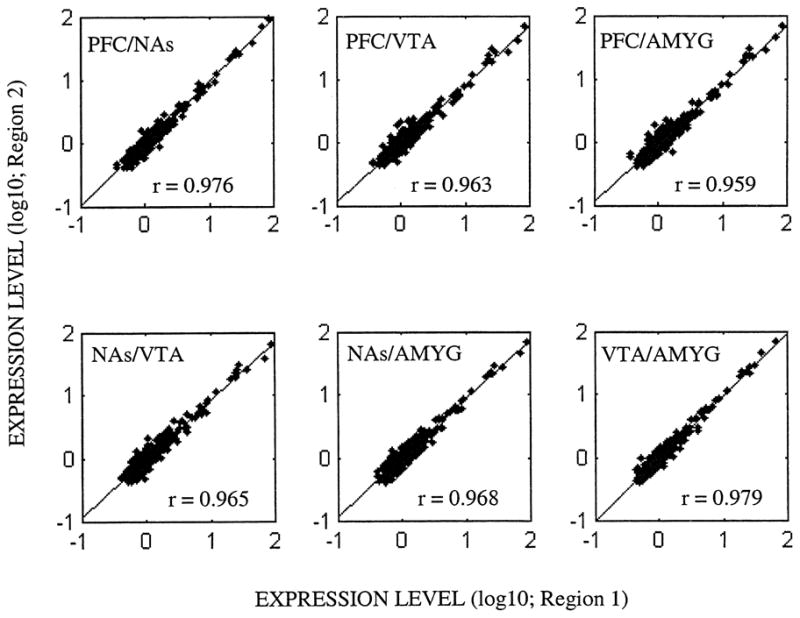
Pairwise correlations between brain regions in terms of the magnitude of mRNA expression (Ngenes=391; reliable measurements across all arrays) or saline control rats. All pairwise correlations were significant at P<10−5. PFC: prefrontal cortex; NAs: nucleus accumbens; VTA: ventral tegmental area; AMYG: amygdala.
3.3. Region-specific expression profiles in response to chronic nicotine treatment
Unlike glass slides hybridized with fluorescent probes, no universal reference exists for the membrane-based (radioactive) microarrays. However, our experiments were designed in such a way that for each brain region, a saline/nicotine treatment pair existed. On the basis Cpool value (1.38- or 0.72-fold difference between nicotine and control experiments; see Section 3.1), we found that only 3.6–12.4% of genes, depending on the brain region, showed any alteration in their gene expression levels (Table 1). Accordingly, PFC and NAs were found to be the most responsive brain regions to chronic nicotine treatment, followed by VTA and AMYG regions (Table 1).
Table 1.
Overall transcriptional response to nicotine in different brain regions
| Brain regions |
||||
|---|---|---|---|---|
| PFC | NAs | VTA | AMY | |
| Number of clones | 669 | 844 | 774 | 878 |
| Over-expressed (%) | 40 (6.0) | 59 (7.0) | 25 (3.2) | 20 (2.3) |
| Under-expressed (%) | 43 (6.4) | 43 (5.1) | 29 (3.7) | 11 (1.3) |
| Total (%) | 83 (12.4) | 102 (12.1) | 54 (6.9) | 31 (3.6) |
The number of genes/ESTs with a reduction or induction in gene expression above the inherent replicate error (Cpool=0.14, log 10) was reported. PFC: prefrontal cortex; NAs: nucleus accumbens; VTA: ventral tegmental area; AMYG: amygdala.
Based on the Pearson’s correlation coefficients obtained between regions with respect to the transcriptional response to nicotine, we found that PFC and NAs shared a significantly greater degree of similarity to each other (rPFC,NAs=0.71; P=1.3×10−15) than they did to either VTA or AMYG (P>0.05). On the other hand, VTA and AMYG were significantly more similar to each other (rVTA,AMYG=0.29, P=0.004). To visualize these similarities among brain regions with respect to the expression response to nicotine administration, we used a multidimensional scaling map, which summarized the multivariate gene expression data in a bivariate manner (Fig. 3; Matlab Inc., MA). As seen in Fig. 3, PFC and NAs were tightly clustered together indicating that the gene expression profiles were more similar to each other. Similarly, VTA and AMYG were more closely associated with each other when compared with their distance to either PFC or NAs.
Fig. 3.
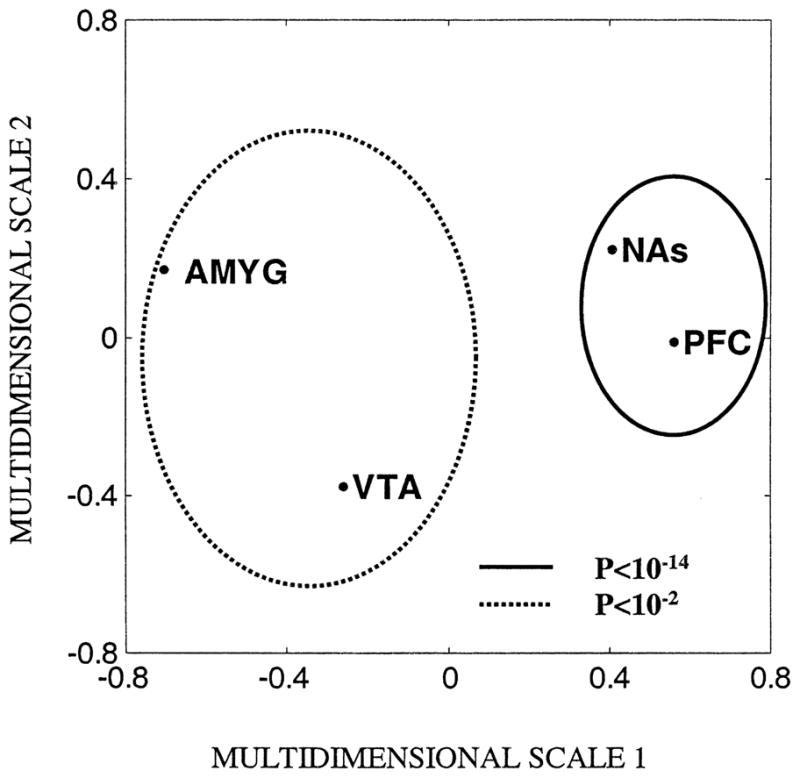
Multidimensional scaling map of the regional transcriptional response to nicotine in rat (Ngenes=95). Gene expression profiles from brain regions (1) PFC and NAs (clustered with a solid line), and (2) VTA and AMYG (clustered with a broken line) were significantly correlated. PFC: prefrontal cortex; NAs: nucleus accumbens; VTA: ventral tegmental area; AMYG: amygdala.
Even though microarray technology allows us to gain information about hundreds of genes simultaneously, only a small percentage of these genes may be informative with respect to the treatment in consideration [12]. Accordingly, 95 of the 391 clones with reliable measurements across all arrays, exhibited alterations above the Cpool level within at least one brain region. Herein, we report on the region-specific transcriptional response of a set of representative genes whose nicotine to control gene expression ratio is greater than 1.75-fold or less than 0.6-fold (Ngenes=8; Table 2). We identified the signaling pathways, such as mitogen-activated protein kinase (MAPK), epidermal growth factor receptor (EGRF), phosphatidylinositol, to which these genes belong. The relevance of these pathways to nicotine administration was also assessed based on the available literature [52,16,32].
Table 2.
Transcriptional response (nicotine to saline ratio of expression levels) of a set of representative genes upon chronic nicotine treatment
| Accession no. | Gene name | Symbol | Pathway | PFC | NAs | VTA | AMYGG |
|---|---|---|---|---|---|---|---|
| R35665 | Epidermal growth factor receptor | EGFR | EGFR, MAPK | 1.12 | 1.00 | 0.81 | 0.57 |
| N26062 | WD40 protein Ciao1 | CIAO1 | EGFR | 0.83 | 1.23 | 1.00 | 0.57 |
| W37864 | Phosphatase and tensin homolog | PTEN | EGFR, PI, MAPK | 0.89 | 0.88 | 0.80 | 1.83 |
| AA284234 | Ribosomal protein S6 kinase, 70 kDa, 2 | RPS6KB2 | PI, MAPK | 0.85 | 0.86 | 0.53 | 1.07 |
| R94153 | Inositol 1,4,5-trisphosphate 3-kinase B | ITPKB | PI | 1.08 | 2.46 | 1.22 | 1.84* |
| AA521431 | Profilin 1 | PFN1 | PI | 0.52 | 0.56 | 1.03 | 1.11 |
| AA453498 | Rap1 guanine-nucleotide exchange factor | EPAC | MAPK | 1.94 | 1.41 | 0.92 | 0.95 |
| H70047 | Regulator of G-protein signaling 13 | RGS13 | MAPK | 0.57 | 0.83 | 0.83 | 0.96 |
Accession number, gene symbol, and assignment of a signaling pathway(s) for each gene were obtained from the website at http://www.ncbi.nlm.nih.gov. Results in bold indicate significant upregulation or downregulation upon nicotine treatment (>1.75- or <0.6-fold, respectively).
Unreliable measurement.
PFC: prefrontal cortex; NAs: nucleus accumbens; VTA: ventral tegmental area; AMYG: amygdala; MAPK: mitogen-activated protein kinase; PI: phosphatidylinositol; EGFR: epidermal growth factor receptor.
3.4. Confirmation of microarray results by real-time quantitative RT-PCR
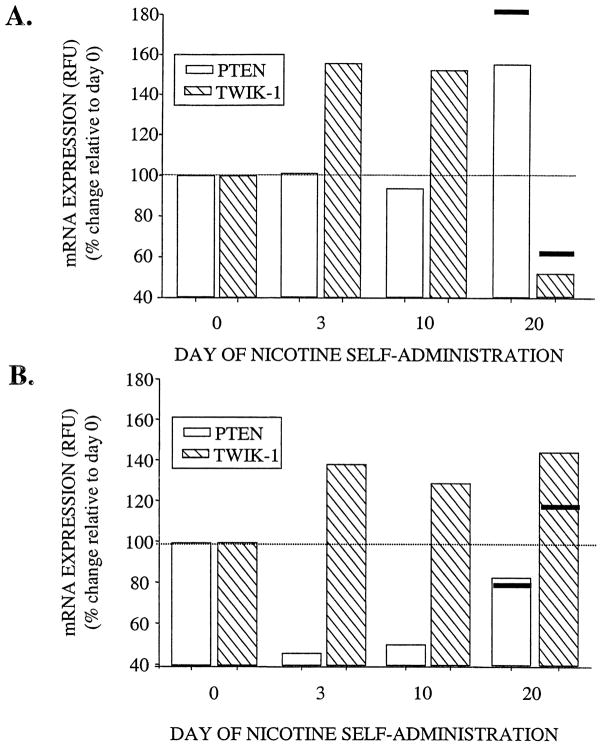
TWIK-1, which belongs to the two-pore-domain K+ channels is believed to play an important role in the regulation of resting membrane potential [24]. In the present study, we found that TWIK-1 expression in AMYG was drastically downregulated (0.62-fold) upon chronic nicotine administration as measured by the cDNA microarrays. Results from the real-time quantitative RT-PCR confirmed this finding, i.e. 0.5-fold decrease in TWIK-1 expression was observed in AMYG at day 20 of nicotine administration (Fig. 4). PTEN, exhibiting a dual-specificity phosphatase activity, also dephosphorylates the 3rd position of phosphatidylinositol (PtdIns) phosphates [30,5]. Our findings from microarray experiments indicated that PTEN was specifically over-expressed in the AMYG upon chronic administration of nicotine (1.83-fold of control rats). Similarly, results of the real-time quantitative RT-PCR confirmed that PTEN expression of nicotine administrating individuals at day 20 is at least 1.6-fold greater than that of the control rats in AMYG (Fig. 4).
Fig. 4.
Effects of nicotine on PTEN and TWIK-1 mRNA expression levels in the amygdala (A) and ventral tegmental area (B). Holtzman rats self-administered 0.03 mg/kg nicotine for 0, 3, 10, and 20 days. Total RNA was isolated from each region as described in Section 2.2. Gene expression levels were measured as fluorescent intensities using semi-quantitative real time PCR, and were normalized to the initial RNA concentration as quantified by the RIBOGreen™ kit (see Section 2.7). Data were presented as percent change relative to day 0 (dotted line), and the solid lines at day 20 represent the respective percent change obtained from cDNA microarray experiments.
4. Discussion
4.1. Reliability of microarray data
Even though multivariate data analysis is crucial in summarizing and visualization of gene expression data sets, outcomes of these analyses are greatly influenced by the algorithms used, reliability of microarray data, and the degree of variability in the magnitude of treatment effects [13]. In fact, it has been reported that up to 30% of the microarray data maybe discarded based on errors associated with printing, hybridization and/or measurements [19]. Nevertheless, we are not aware of any study that reports on a technique(s) for the normalization of the measurement error across multiple arrays, despite its importance for the comparative analysis of microarray data. In this study, we used the jacknifed reliability index (JRI) to determine a standardized cut-off value for filtering out unreliable measurements across multiple arrays. We suggest that a cut-off value based on the pooled duplicate differences is advantageous, because (a) as the number of duplicate expression differences in the pool increases, true error value (population parameter) associated with the microarray procedure is better estimated, and (b) a pooled cut-off value normalizes the variability with respect to the measurement error among different experiments (or arrays). Accordingly, we have estimated that 13.5% of all observations had relatively large influences on the sample kurtosis value, corresponding to an absolute duplicate difference greater than 0.14 at log 10 scale (1.38 or 0.72-fold of control expression level).
4.2. Regional differences in the magnitude of mRNA expression
Principal component analysis (PCA) enables one to extract components, which describe different aspects of the variability associated with the data set (e.g. magnitude and shape of expression data). Furthermore, PCA can be a powerful tool to detect the trends in the expression data obtained from multiple individuals. Nevertheless, most studies in the microchip field concentrated on the comparisons of expression patterns using various clustering algorithms with less attention given to the comparison of the magnitude of gene expression. Herein, we used PCA to summarize the range of expression level within and across different brain regions from the saline/control rats. A large portion of the observed variability (ca. 98%) in gene expression levels was attributable to the differences in the magnitude of expression levels within brain regions. On the other hand, differences among brain regions accounted for only 2% of the total variability, indicating that brain regions reported in this study are highly similar in their magnitude of gene expression. Our results were in accord with those from a recent microarray study assessing region- and strain-specific differences in brain gene expression levels of mouse [45], which indicated that structures of medial temporal lobe (hippocampus, amygdala, and entorhinal cortex) were highly similar in their gene expression levels.
4.3. Region-specific transcriptional response to chronic nicotine: involvement of MAPK, phosphatidylinositol, EGFR signaling pathways
Microarray data analysis is a powerful tool for identifying functional clusters among genes under investigation. Among the genes /ESTs included on the arrays, we found that genes whose expression levels seem to be affected by nicotine administration mainly fell into three distinct functional groups: cell signaling/communication, cell structure/motility, and gene/protein expression. In this report, we limit our focus on a set of representative genes that are involved in MAPK, phosphatidylinositol, and EGFR signaling pathways, all of which were previously implicated with nicotine administration. Nicotine was previously shown to induce mitogen-activated protein kinase (MAPK) signaling pathway in pheochromocytoma (PC12) cells via activation of protein kinase C/Raf/RSKII/CREB [52]. In the present study, we found that, in PFC, nicotine self-administration increased the expression of Rap1 guanine-nucleotide-exchange factor activated by cAMP (EPAC; 1.93-fold increase), which mediates the activation of B-raf in neurons [56,9]. The involvement of Rap family of ras-related GTPases in response to nicotine administration was further accentuated by the changes observed in two additional genes involved in the Rap signaling pathway, namely Rap2A (1.66-fold increase in PFC) and MR-GEF (0.71-fold decrease in PFC and NAs). On the other hand, mRNA expression of RGS13, a member of the regulator of G-protein signaling (RGS) family was reduced by 0.57-fold in response to nicotine. RGS proteins are known to impair the activation of MAP kinases by inhibiting G-protein signaling [8]. For example, a recent study has shown that RGS4, upon inhibiting the coupling of 5-HT(1B) receptors to cellular signals, attenuated the activation of extracellular signal-regulated kinase, ERK [28]. Overexpression of EPAC and downregulation of RGS13 were observed in PFC and to a lesser degree (1.41- and 0.82-fold, respectively) in NAs of the nicotine treated rats. These findings suggest that nicotine may activate the MAPK pathway via EPAC/B-raf in a region-specific manner.
Nicotine also has been shown to elevate intracellular calcium levels upon activation of the store-operated calcium channels possibly by an increase in the D-myo-inositol 1,4,5-trisphosphate (Ins(1,4,5)P3) production [16].In this study we identified a number of genes, such as profilin 1, Ins(1,4,5)P3 3-kinase B, and PTEN, involved in Ins(1,4,5)P3 turnover as well as intracellular calcium homeostasis, as potential modulators/targets of nicotine in rat brain. Profilin negatively regulates the phophoinositide signaling pathway by binding with phosphatidylinositol 4,5-bisphosphate (PIP2) and preventing the formation of Ins(1,4,5)P and diacylglycerol via inhibition of the phospholipase C gamma 1 hydrolysis [14]. We observed that the expression level of profilin was reduced by 0.52- and 0.56-fold, in PFC and NAs of nicotine treated rats, respectively, suggesting an increase in the concentration of Ins(1,4,5)P3 in these brain regions. Ins(1,4,5)P3 3-kinase catalyzes the production of D-myo-inositol 1,3,4,5-tetra-kisphosphate (Ins(1,3,4,5)P4) from Ins(1,4,5)P3 Interestingly, Ins(1,4,5)P3 3-kinase expression was specifically increased in NAs of rats treated with nicotine (2.45-fold). These results suggested that activation of phosphatidylinositol signaling pathway in PFC and NAs upon chronic nicotine administration maybe due to a potential increase in the production of Ins(1,4,5)P3 in PFC and NAs, and Ins(1,3,4,5)P4 only in NAs.
On the other hand, PTEN exhibits 3-phosphatase activity towards Ins(1,3,4,5)P4 and thus reduces the availability of Ins(1,3,4,5)P4 [30]. Interestingly, PTEN expression was increased only in AMYG upon chronic nicotine administration, implicating the presence of a negative feedback on the phosphatidylinositol pathway in this region. Genes whose expression levels were altered by nicotine in AMYG also included epidermal growth factor receptor (EGFR) and Ciao1. Previous studies have shown that a tight relationship exists between EGFR and PTEN, such that an increase in PTEN leads to down regulation of EGFR expression [41,23]. EGFR also is transcriptionally regulated by another gene, namely Wilms tumor (WT1), which physically interacts with Ciao1 [29,21]. Recently, PTEN has gained enormous attention from the scientific community since it was found to be a tumor suppressor gene whose inactivation constitutively activates PI3 /AKT pathway [17,4,40]. Involvement of PI3 /AKT pathway in response to nicotine was also supported by our observation that the expression of p70S6, a downstream target of AKT gene [42], was downregulated in VTA (Table 2). The observed effects of nicotine on the mRNA expression of PTEN and AKT/p70S6 warrant further investigation, which may shed light on the associations between nicotine and cancer [32,47,2].
In summary, we demonstrate that the JRI proposed in this study can be used to normalize the duplicate measurement error across multiple arrays, as it allows for the identification of a cut-off value for filtering out duplicate differences with relatively large influence on the sample statistics (i.e. kurtosis). Upon application of principal component analysis and multidimensional scaling on the filtered microarray data, we found that brain regions under investigation were highly similar in regards to their absolute expression levels, but exhibited highly region-specific transcriptional response to nicotine. We also confirmed the microarray results obtained for the TWIK-1 and PTEN gene using real-time quantitative RT-PCR on Bio-Rad iCycler. Finally, a number of genes that are involved in MAPK, phosphatidylinositol, and EGFR signaling pathways have been identified as possible targets/modulators in response to nicotine administration.
Acknowledgments
This project was in part supported by National Institute of Health grant R01-DA12844 and R01-DA13783 to MDL.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Baron JA. Beneficial effects of nicotine and cigarette smoking: the real, the possible and the spurious. Br Med Bull. 1996;52:58–3. doi: 10.1093/oxfordjournals.bmb.a011533. [DOI] [PubMed] [Google Scholar]
- 3.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 4.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, Jen J, Isaacs WB, Bova GS, Sidransky D. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 5.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase /AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr LA, Rowell PP, Pierce WM., Jr Effects of subchronic nicotine administration on central dopaminergic mechanisms in the rat. Neurochem Res. 1989;14:511–515. doi: 10.1007/BF00964911. [DOI] [PubMed] [Google Scholar]
- 7.Dalack GW, Healy DJ, Meador-Woodruff JH. Nicotine dependence in schizophrenia: clinical phenomena and laboratory findings. Am J Psychiatry. 1998;155:1490–1501. doi: 10.1176/ajp.155.11.1490. [DOI] [PubMed] [Google Scholar]
- 8.Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 9.Dugan LL, Kim JS, Zhang Y, Bart RD, Sun Y, Holtzman DM, Gutmann DH. Differential effects of cAMP in neurons and astrocytes. Role of B-raf. J Biol Chem. 1999;274:25842–25848. doi: 10.1074/jbc.274.36.25842. [DOI] [PubMed] [Google Scholar]
- 10.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J. Regression Diagnostics. Sage Publications; Newbury Park, CA: 1991. [Google Scholar]
- 12.Fuhrman S, Cunningham MJ, Wen X, Zweiger G, Seilhamer JJ, Somogyi R. The application of shannon entropy in the identification of putative drug targets. Biosystems. 2000;55:5–14. doi: 10.1016/s0303-2647(99)00077-5. [DOI] [PubMed] [Google Scholar]
- 13.Getz G, Levine E, Domany E. Coupled two-way clustering analysis of gene microarray data. Proc Natl Acad Sci USA. 2000;97:12079–12084. doi: 10.1073/pnas.210134797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- 15.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 16.Gueorguiev VD, Zeman RJ, Hiremagalur B, Menezes A, Sabban EL. Differing temporal roles of Ca2+ and cAMP in nicotineelicited elevation of tyrosine hydroxylase mRNA. Am J Physiol. 1999;276:C54–C65. doi: 10.1152/ajpcell.1999.276.1.C54. [DOI] [PubMed] [Google Scholar]
- 17.Guldberg P, Thor SP, Birck A, Ahrenkiel V, Kirkin AF, Zeuthen J. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 18.Harlan RE, Garcia MM. Drugs of abuse and immediate-early genes in the forebrain. Mol Neurobiol. 1998;16:221–267. doi: 10.1007/BF02741385. [DOI] [PubMed] [Google Scholar]
- 19.Herwig R, Poustka AJ, Muller C, Bull C, Lehrach H, O’Brien J. Large-scale clustering of cDNA-fingerprinting data. Genome Res. 1999;9:1093–1105. doi: 10.1101/gr.9.11.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone RW, Wang J, Tommerup N, Vissing H, Roberts T, Shi Y. Ciao 1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J Biol Chem. 1998;273:10880–10887. doi: 10.1074/jbc.273.18.10880. [DOI] [PubMed] [Google Scholar]
- 22.Kane JK, Parker SL, Matta SG, Fu Y, Sharp BM, Li MD. Nicotine up-regulates expression of orexin and its receptors in rat brain. Endocrinology. 2000;141:3623–3629. doi: 10.1210/endo.141.10.7707. [DOI] [PubMed] [Google Scholar]
- 23.Kleihues P, Ohgaki H. Phenotype vs. genotype in the evolution of astrocytic brain tumors. Toxicol Pathol. 2000;28:164–170. doi: 10.1177/019262330002800121. [DOI] [PubMed] [Google Scholar]
- 24.Lesage F, Lazdunski M. Molecular and functional properties of two-pore-domain potassium channels. Am J Physiol. 2000;279:F793–F801. doi: 10.1152/ajprenal.2000.279.5.F793. [DOI] [PubMed] [Google Scholar]
- 25.Li MD, MacDonald GJ, Ford JJ. Breed differences in expression of inhibin/activin subunits in porcine anterior pituitary glands. Endocrinology. 1997;138:712–718. doi: 10.1210/endo.138.2.4949. [DOI] [PubMed] [Google Scholar]
- 26.Li MD, Kane JK, Matta SG, Blaner WS, Sharp BM. Nicotine enhances the biosynthesis and secretion of transthyretin from the choroid plexus in rats: implications for beta-amyloid formation. J Neurosci. 2000;20:1318–1323. doi: 10.1523/JNEUROSCI.20-04-01318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li MD, Kane JK, Parker SL, McAllen K, Matta SG, Sharp BM. Nicotine administration enhances NPY expression in the rat hypothalamus. Brain Res. 2000;867:157–164. doi: 10.1016/s0006-8993(00)02283-6. [DOI] [PubMed] [Google Scholar]
- 28.Lione AM, Errico M, Lin SL, Cowen DS. Activation of extracellular signal-regulated kinase (ERK) and Akt by human serotonin 5-HT(1B) receptors in transfected BE(2)-C neuroblastoma cells is inhibited by RGS4. J Neurochem. 2000;75:934–938. doi: 10.1046/j.1471-4159.2000.0750934.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu XW, Gong LJ, Guo LY, Katagiri Y, Jiang H, Wang ZY, Johnson AC, Guroff G. The Wilms’ tumor gene product WT1 mediates the down-regulation of the rat epidermal growth factor receptor by nerve growth factor in PC12 cells. J Biol Chem. 2000;275:1. doi: 10.1074/jbc.M008776200. [DOI] [PubMed] [Google Scholar]
- 30.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 31.Manly BFJ. Multivariate Statistical Methods: A Primer. Chapman & Hall; London: 1994. [Google Scholar]
- 32.Mathur RS, Mathur SP, Young RC. Up-regulation of epidermal growth factor-receptors (EGF-R) by nicotine in cervical cancer cell lines: this effect may be mediated by EGF. Am J Reprod Immunol. 2000;44:114–120. doi: 10.1111/j.8755-8920.2000.440207.x. [DOI] [PubMed] [Google Scholar]
- 33.Mihailescu S, Drucker-Colin R. Nicotine, brain nicotinic receptors, and neuropsychiatric disorders. Arch Med Res. 2000;31:131–144. doi: 10.1016/s0188-4409(99)00087-9. [DOI] [PubMed] [Google Scholar]
- 34.Nisell M, Nomikos GG, Chergui K, Grillner P, Svensson TH. Chronic nicotine enhances basal and nicotine-induced Fos immuno- reactivity preferentially in the medial prefrontal cortex of the rat. Neuropsychopharmacology. 1997;17:151–161. doi: 10.1016/S0893-133X(97)00040-7. [DOI] [PubMed] [Google Scholar]
- 35.Pagliusi SR, Tessari M, DeVevey S, Chiamulera C, Pich EM. The reinforcing properties of nicotine are associated with a specific patterning of c-fos expression in the rat brain. Eur J Neurosci. 1996;8:2247–2256. doi: 10.1111/j.1460-9568.1996.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 36.Pang Y, Kiba H, Jayaraman A. Acute nicotine injections induce c-fos mostly in non-dopaminergic neurons of the midbrain of the rat. Mol Brain Res. 1993;20:162–170. doi: 10.1016/0169-328x(93)90122-6. [DOI] [PubMed] [Google Scholar]
- 37.Pich EM, Pagliusi SR, Tessari M, Talabot-Ayer D, van Hooft H, Chiamulera C. Common neural substrates for the addictive properties of nicotine and cocaine. Science. 1997;275:83–86. doi: 10.1126/science.275.5296.83. [DOI] [PubMed] [Google Scholar]
- 38.Pontieri FE, Tanda G, Orzi F, Di Chiara G. Effects of nicotine on the nucleus accumbens and similarity to those of addictive drugs. Nature. 1996;382:255–257. doi: 10.1038/382255a0. [DOI] [PubMed] [Google Scholar]
- 39.Ren T, Sagar SM. Induction of c-fos immunostaining in the rat brain after the systemic administration of nicotine. Brain Res Bull. 1992;29:589–597. doi: 10.1016/0361-9230(92)90127-j. [DOI] [PubMed] [Google Scholar]
- 40.Risinger JI, Hayes AK, Berchuck A, Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 41.Rodrigues GA, Falasca M, Zhang Z, Ong SH, Schlessinger J. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol Cell Biol. 2000;20:1448–1459. doi: 10.1128/mcb.20.4.1448-1459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roques M, Vidal H. A phosphatidylinositol 3-kinase/p70 ribosomal S6 protein kinase pathway is required for the regulation by insulin of the p85alpha regulatory subunit of phosphatidylinositol 3-kinase gene expression in human muscle cells. J Biol Chem. 1999;274:34005–34010. doi: 10.1074/jbc.274.48.34005. [DOI] [PubMed] [Google Scholar]
- 43.Salminen O, Seppa T, Gaddnas H, Ahtee L. The effects of acute nicotine on the metabolism of dopamine and the expression of Fos protein in striatal and limbic brain areas of rats during chronic nicotine infusion and its withdrawal. J Neurosci. 1999;19:8145–8151. doi: 10.1523/JNEUROSCI.19-18-08145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salminen O, Seppa T, Gaddnas H, Ahtee L. Effect of acute nicotine on Fos protein expression in rat brain during chronic nicotine and its withdrawal. Pharmacol Biochem Behav. 2000;66:87–93. doi: 10.1016/s0091-3057(00)00203-3. [DOI] [PubMed] [Google Scholar]
- 45.Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, Mayford M, Lockhart DJ, Barlow C. From the cover: regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schilstrom B, Fagerquist MV, Zhang X, Hertel P, Panagis G, Nomikos GG, Svensson TH. Putative role of presynaptic alpha7* nicotinic receptors in nicotine stimulated increases of extracellular levels of glutamate and aspartate in the ventral tegmental area. Synapse. 2000;38:375–383. doi: 10.1002/1098-2396(20001215)38:4<375::AID-SYN2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Schuller HM. Nitrosamine-induced lung carcinogenesis and Ca2+/calmodulin antagonists. Cancer Res. 1992;52:2723s–2726s. [PubMed] [Google Scholar]
- 48.Sharp BM, Matta SG. Detection by in vivo microdialysis of nicotine-induced norepinephrine secretion from the hypothalamic paraventricular nucleus of freely moving rats: dose-dependency and desensitization. Endocrinology. 1993;133:11–19. doi: 10.1210/endo.133.1.8391419. [DOI] [PubMed] [Google Scholar]
- 49.Shim IS, Won JS, Lee JK, Song DK, Kim SE, Huh SO, Kim YH, Suh HW. Modulatory effect of ginseng total saponin on dopamine release and tyrosine hydroxylase gene expression induced by nicotine in the rat. J Ethnopharmacol. 2000;70:161–169. doi: 10.1016/s0378-8741(99)00166-x. [DOI] [PubMed] [Google Scholar]
- 50.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, III, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang K, Wu H, Mahata SK, O’Connor DT. A crucial role for the mitogen-activated protein kinase pathway in nicotinic cholinergic signaling to secretory protein transcription in pheochromocytoma cells. Mol Pharmacol. 1998;54:59–69. doi: 10.1124/mol.54.1.59. [DOI] [PubMed] [Google Scholar]
- 53.Torres G, Horowitz JM. Drugs of abuse and brain gene expression. Psychosom Med. 1999;61:630–650. doi: 10.1097/00006842-199909000-00007. [DOI] [PubMed] [Google Scholar]
- 54.Valentine JD, Hokanson JS, Matta SG, Sharp BM. Self-administration in rats allowed unlimited access to nicotine. Psychopharmacology (Berl) 1997;133:300–304. doi: 10.1007/s002130050405. [DOI] [PubMed] [Google Scholar]
- 55.Young RA. Biomedical discovery with DNA arrays. Cell. 2000;102:9–15. doi: 10.1016/s0092-8674(00)00005-2. [DOI] [PubMed] [Google Scholar]
- 56.Zwartkruis FJ, Bos JL. Ras and Rap1: two highly related small GTPases with distinct function. Exp Cell Res. 1999;253:157–165. doi: 10.1006/excr.1999.4695. [DOI] [PubMed] [Google Scholar]