Abstract
One of the key consequences of exposure of human cells to genotoxic agents is the activation of DNA damage responses (DDR). While the mechanisms underpinning DDR in fully differentiated somatic human cells have been studied extensively, molecular signaling events and pathways involved in DDR in pluripotent human embryonic stem cells (hESC) remain largely unexplored. We studied changes in the human genome-wide transcriptome of H9 hESC line following exposures to 1 Gy of gamma-radiation at 2 hrs and 16 hrs post-irradiation. Quantitative real-time PCR was performed to verify the expression data for a subset of genes. In parallel, the cell growth, DDR kinetics, and expression of pluripotency markers in irradiated hESC were monitored. The changes in gene expression in hESC after exposure to ionizing radiation (IR) are substantially different from those observed in somatic human cell lines. Gene expression patterns at 2 hr post-IR showed almost an exclusively p53-dependent, predominantly pro-apoptotic, signature with a total of only 30 up-regulated genes. In contrast, the gene expression patterns at 16 hr post-IR showed 354 differentially expressed genes, mostly involved in pro-survival pathways, such as increased expression of metallothioneins, ubiquitin cycle, and general metabolism signaling. Cell growth data paralleled trends in gene expression changes. DDR in hESC followed the kinetics reported for human somatic differentiated cells. The expression of pluripotency markers characteristic of undifferentiated hESC was not affected by exposure to IR during the time course of our analysis. Our data on dynamics of transcriptome response of irradiated hESCs may provide a valuable tool to screen for markers of IR exposure of human cells in their most naive state; thus unmasking the key elements of DDR; at the same time, avoiding the complexity of interpreting distinct cell type-dependent genotoxic stress responses of terminally differentiated cells.
Keywords: Human embryonic stem cell, Ionizing radiation, Gene expression, DNA microarray
1. Introduction
The genetic material of every living being is constantly challenged by environmental agents such as background IR and endogenous threats arising as a by-product of normal metabolism. For example, every cell in the human organism on average receives tens of thousands of DNA lesions per day [1]. To counter these deleterious effects produced by genotoxic agents, several cellular mechanisms have evolved to detect the various types of DNA lesions, to signal their presence, and to mediate their repair and removal; all collectively known as DDR. The biological significance of these mechanisms can hardly be overestimated since the accumulation of DNA damage has been thought to play a critical role in the cancerogenesis and the aging processes [2]. Therefore, a great deal of effort has been invested in elucidating the details of molecular signaling and cellular events associated with DDR.
Early work based on using conventional biochemical and genetics techniques identified many components of DDR in human cells [3, 4]. One of the key aspects of DDR is the widespread changes in the level of expression of many genes involved in various pathways of cellular metabolism. With the development of newer, high-throughput technology it became possible to interrogate the expression of thousands of genes simultaneously following specific treatment conditions [5], including genotoxic agent exposures [6]. These functional genomics experiments vastly extended our knowledge how human cells of different origins respond to IR [7–11]. DNA microarray profiling enabled identification of novel radiation-responsive signaling pathways, further advancing our understanding of IR effects on humans [12]. However, practically all these experiments were performed on terminally differentiated cells either grown in tissue culture or derived from tissue biopsy specimens. Little is known about how genotoxic stress, particularly IR exposure, affects the global signaling events in pluripotent hESC. The available data primarily focus only on specific genes and distinct pathways involved in IR response of hESC [13–15]. It is known that the human fetus is very susceptible to genotoxic insults [16, 17]; but the underlying molecular mechanisms are not fully understood. In addition, hESC are currently under intense research in a context of toxicological studies in which hESC are being employed as a model for therapeutic drug screening [18]. Therefore, identification of novel molecular targets in genotoxic agent screening in pluripotent human cells could significantly contribute to their future use not only in regenerative-based cell replacement clinical strategies, but also for establishing optimized treatment schemes in a personalized medicine [19]. Only one report [20] aimed to define the effects of IR on global changes in gene expression in hESC. However, this study interrogated hESC response only at a single, late timepoint after IR exposures. In the present study, we set out to characterize the dynamics of the transcriptional response of cultured irradiated hESC, to determine the key components of DDR in hESC, and to identify signaling pathways possibly responsible for the sensitivity of early human developmental stage cells to genotoxic IR exposures. To this end, we used a whole human genome-wide functional genomics approach. We show that following 1 Gy of gamma-radiation exposure of H9 hESC “early” DDR is almost exclusively p53-dependent. In contrast, the “late” gene expression signature of irradiated hESC is characterized by robust involvement of a wide range of signaling pathways, many of which seem to be responsible for the survival of hESC.
2. Materials and methods
2.1. Cell Culture
Cultured hESCs (H9 cell line, WiCell, Madison, WI, passage 35 – 40) were routinely grown in mTeSR-1 medium (Stemcell Technologies, Vancouver, Canada) on a BD Matrigel hESC-qualified matrix (BD Biosciences, San Jose, CA) at 37°C and 5% CO2. Cell cultures were maintained and expanded following supplier’s protocol. Cells were passaged every 5–7 days using collagenase IV (Invitrogen, Carlsbad, CA). The medium was changed every day.
Cell cultures were divided into two groups, and were either exposed to 1 Gy of X-ray irradiation using X-RAD 320 Biological Irradiator unit (Precision X-Ray, Inc., North Branford, CT; dose rate about 1 Gy/min; 320 kV, 12.5 mA); or, alternatively, were mock-irradiated. Cells then were allowed to recover in CO2 incubator and collected at 2 h and 16 h post-irradiation for analysis.
2.2. Cell proliferation assay
The cells were seeded on Matrigel in equal aliquots before the start of experiment. After IR exposures (0.2 Gy or 1 Gy) and time of incubation post-IR (17 hrs, 41 hrs and 65 hrs), cell cultures were rinsed with phosphate buffer saline (PBS; Invitrogen) to remove detached cells. These floating cells were collected by gentle centrifugation for subsequent cell counting. The attached cells, that are presumably alive, were collected by treatment with Trypsin-EDTA (Invitrogen) for 3 min at 37°C, and washed with PBS buffer supplemented with 0.5% BSA. Cell count was performed using Z1 Coulter Cell Counter system (Beckman Coulter, Inc., Brea, CA) at least in triplicate for each experimental data point.
2.3. Immunocytochemistry
The cell cultures were grown on glass-bottom LabTek Chamber Slide (BD Biosciences) as described above. Cells were fixed with 4% paraformaldehyde for 10 minutes, and then permeabilized with 0.1% Triton-X-100 for 5 minutes. Primary antibodies dissolved in blocking solution containing 3% bovine serum albumin (BSA; Sigma, St. Louis, MO) were added for 1 hr, and then appropriately coupled Alexa Fluor secondary antibodies (Invitrogen) were used for indirect immunofluorescent detection of primary antigens, as desribed in [21]. All secondary antibodies were tested for nonspecific immunoreactivity. The following primary antibodies were chosen: Oct-4, SSEA4, TRA-1-81 and 53BP1 (Santa Cruz Biotechnology, Santa Cruz, CA). DAPI stain was used to identify the nuclei. The antifade media (VectaShield, Vector Laboratories, Inc., Burlingame, CA) was used to mount the coverslips; the samples were examined by Axioplan Zeiss epifluorescent microscope (Carl Zeiss, Thornwood, NY). The microscope and CCD camera image acquisition settings were the same for all corresponding samples. 53BP1 foci assay was performed as described in [22], and at least 200 cells were examined per each datapoint. Foci scoring by eye were typically performed in a blinded manner.
2.4. Cell cycle analysis
Cell cycle analysis was performed using propidium iodide (PI) / Triton-X-100 staining solution (0.1 mg/ml PI; 0.1 mg/ml RNAse A; 0.1 % (v/v) Triton-X-100, Sigma). Human ESCs were harvested by collagenase IV treatment and counted with hemocytometer. In total, 500,000 cells were fixed after incubation with 70% ethanol at 4°C for 2 h and stained in PI / Triton-X-100 staining solution for 30 min at room temperature in dark. The samples were analyzed by flow cytometry (FACS Calibur, BD Biosciences) measuring FL2 area versus total counts and with ModFit 3.0 (Verity Software House) to generate percentages of cells in G1, S and G2/M phases
2.5. RNA sample preparation, probe labeling and DNA microarray procedure
The extraction of total RNA was performed using Trizol (Invitrogen), and then RNA preparations were purified with RNeasy kit (Qiagen, Valencia, CA) and TURBO DNA-free kit (Ambion, Inc., Austin, TX) per manufacturers’ instructions. The amount and quality of RNA samples were assessed on the Agilent 2100 Bioanalyzer with RNA 6000 Nano Reagents and Supplies (Agilent, Santa Clara, CA) [23]. Agilent RNA Spike-In Mix was added to the RNA samples prior to the labeling reactions following the RNA Spike-In Kit protocol. Subsequently, cRNA targets were synthesized from 1 μg of total RNA in each reaction and fluorescently labeled with Cy5-CTP (PerkinElmer, Waltham, MA) in separate labeling reactions using the Agilent Quick-Amp Labeling kit. The Universal Reference RNA (Stratagene, La Jolla, CA) was used to synthesize Cy-3-CTP (PerkinElmer, Waltham, MA) labeled cRNA sample to include as a common reference target throughout all experiments. The dual-labeled cRNA targets corresponding both to experimental and reference samples were combined and hybridized to 4x44k Agilent Human Whole Genome oligo microarrays using Agilent SureHyb hybridization chambers. Protocols for microarray hybridization and washing were as suggested by manufacturer. Hybridized DNA microarrays were scanned with a resolution of 5 μm on an Agilent DNA microarray scanner enabled by SureScan High-Resolution Technology (Agilent), and TIFF images were subsequently processed by Feature Extraction 10.5 software (Agilent). All samples had four independent biological replicates, and each replicate was run on a separate array.
2.6. Data analysis
Raw data derived from processing TIFF image files were analyzed using BRB-Array Tools Version 3.8.0 software developed by Dr. Richard Simon and Amy Peng Lam (Biometric Research Branch, National Cancer Institute, NIH). Time-matched irradiated versus mock-irradiated samples were used for analysis to determine the radiation-responsive genes from quadruplicate independent experiments per each data point. Differentially expressed genes were identified using a Significance Analysis of Microarrays (SAM) method [24]. We identified genes that were differentially expressed among the two classes (irradiated hESC versus mock-irradiated hESC cultures) by using a multivariate permutation test [25]. We used the multivariate permutation test to provide a median false discovery rate (FDR) of 10%. The FDR is the proportion of the list of genes claimed to be differentially expressed that are false positives. The test statistics used are random variance t-statistics for each gene [26]. The sets of differentially expressed genes were tested for functional significance using the DAVID Bioinformatics Resources 2008 [27]. This tool obtains the Gene Ontology (GO) annotations from a database and generates a statistical analysis of the functional annotations that are overrepresented in the selected set of genes. A Bonferroni correction for multiple comparisons was included in the analysis [28]. GO biological processes with EASE scores less than 0.05 were considered to be statistically significant [29]. Minimum Information About a Microarray Experiment (MIAME)-compliant raw data for our experiments have been uploaded onto the ArrayExpress database maintained by the European Bioinformatics Institute (accession no. E-MEXP-2596).
2.7. Quantitative real-time PCR
The quantitative RT-PCR was done on RNA samples from three independent cell culture experiments. The complementary DNA was synthesized from total RNA using One-Step RNA-to-Ct kit (Applied Biosystems) according to the manufacturer’s protocol. For each gene, PCR reactions were run in triplicate on one sample. RT-PCR was performed on iCycler iQ (Bio-Rad, Inc.) in 20-μl reactions by using TaqMan Assay-on-Demand primers/probe sets (Applied Biosystems) for the following genes: CDKN1A, GADD45A, DHFR, CCDC88A and MT1F. Quantitative RT-PCR data were analyzed as in [30].
3. Results and discussion
3.1. Assessment of growth of cultured hESC population in response to IR exposure
To determine how growth of hESC in culture is affected by IR exposures, we studied the kinetics of changes in cell numbers following IR exposures (Additional Fig. 1). We found that after 0.2 Gy exposures cell count reaches a plateau between 17 – 41 hrs post-IR, presumably due to ongoing cell cycle arrest and/or death of a subpopulation of cells. Some cell death at 17 hrs after 0.2 Gy irradiation of hESC is substantiated by the increase in the fraction of floating, presumably dead, cells. By 65 hr post 0.2 Gy exposures, the cell numbers are comparable to those in sham-irradiated hESC cultures (Additional Fig. 1). In contrast, irradiation of hESC with 1 Gy dose leads to a dramatic decrease in cell numbers following this exposure. This drop in cell population was apparent already at 17 hrs post-IR, peaked at 41 hrs, and then, by 65 hrs, gradually returned to values observed after 17 hrs post-IR exposures. However, even at 65 hrs post 1 Gy irradiation the cell numbers were twice as low as those in sham-irradiated hESC and cells post 0.2 Gy exposures. At 17 hrs post 1 Gy irradiation, about one third of the cell population is apparently dead. Presumably, the drop in cell number of hESC population after 1 Gy irradiation is also dependent on cell cycle arrest, since the level of cell death is decreased by 41 hrs post-IR compared to 17 hrs; however, the cell count was still declining at 41 hrs compared to 17 hrs (Additional Fig. 1). The cell survival data are in general agreement with our previous data on hESC apoptosis after exposure to IR [31]. There we found that after 6 hrs post 1 Gy IR exposure approximately 30% of hESC stained positively for cleaved caspase 3, an apoptotic marker.
3.2. DNA damage response, cell cycle profile and maintenance of pluripotency in irradiated hESC
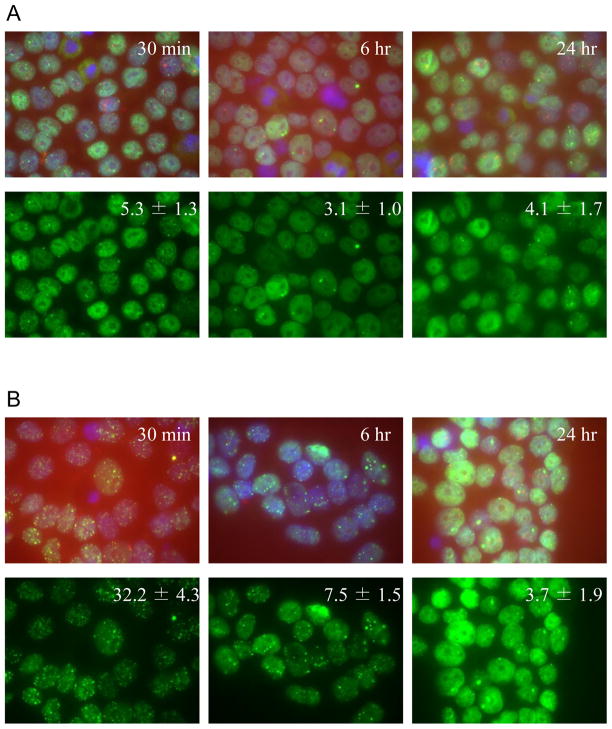
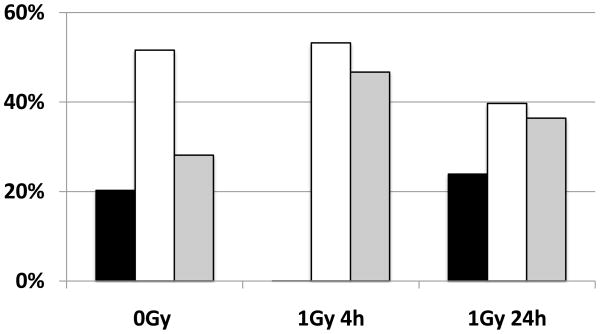
One of the key aspects of cellular DDR is activation of DNA double strand breaks (DSB) repair mechanisms [32]. 53BP1 is known to be one of the key components of DDR, is crucially involved in DNA DSB repair, and in the maintenance of genomic stability [33]. We investigated the DNA DSB repair in hESC by studying the kinetics of so-called ionizing radiation-induced foci (IRIF), specifically, the timing of appearance and turnover of 53BP1 nuclear foci [34]. As can be seen from Fig. 1, the constitutive level of 53BP1-foci in sham-irradiated hESC (referring to “background” foci level between 30 min till 24 hrs post-sham exposures) varied within the range of 3 – 5 foci per cell, on average. However, at 30 min after 1 Gy IR exposure, the number of foci per cell increased dramatically (32.2 ± 4.3), reflecting the formation of DNA double strand breaks (DSB). At 6 hrs post 1 Gy exposures, the level of IRIF decreased significantly (7.5 ± 1.5) reflecting intensive DNA repair, but was still considerably higher than in the sham-irradiated hESC cultures (3.1 ± 1.0). However, at 24 hrs following irradiation, the level of DNA DSB returned to the background level (Fig. 1) reflecting the completion of repair of DNA DSBs. Therefore, the results of our study suggest that DNA DSB repair in hESC exposed to IR generally follows the same kinetics as in human differentiated cells [35]. Cell cycle profile of non-irradiated hESC is characteristic for this type of cells and shows that the majority of the cells are in S phase (Fig. 2). Four hours after 1 Gy of IR all cells that were in G1 phase entered S-phase, while none of the cells underwent cell division and entered G1 phase. However, after 24 hours post IR cells cycle profile returned to almost normal. These data demonstrate that hESC were arrested at G2/M phase following 1 Gy of IR exposures, but they recovered by 24 hrs post-IR (Fig. 2). At the same time there were no G1/S phase arrest in hESC, consistent with report published by others [14]. However, a major difference between fully differentiated cells and hESC is the ability of the latter to give rise to virtually all cells of a human body (pluripotency), whereas the former are restricted to their specific lineage. We examined if hESC in our experiments maintain the pluripotency following IR exposure. To this end, we performed immunocytochemical analysis of cultured hESC at selected time points after 1 Gy irradiation (6 hrs, 24 hrs and 48 hrs) as shown on Additional Fig.2. As a negative control, we included human BJ fibroblasts, human adult bone-marrow mesenchymal stromal cells (hMSC), and immunostaining of hESC with secondary fluorescent label-coupled antibodies (Additional Fig.3). Based on results of in situ immunostaining of hESC with the established markers of pluripotency such as Oct-4, SSEA4 and TRA-1-81, we found no observable difference between irradiated and sham-irradiated hESC cultures (Additional Fig. 2). These data are in accordance with our previous observations on continuous expression of markers of pluripotency in irradiated hESC [31], and on ability of hESC exposed to IR to form teratoma in mice [20].
Figure 1. Immunocytochemical analysis of DNA damage response in cultured H9 cells after 1 Gy irradiation.
The experimental procedure was performed as indicated in the Materials and methods section. 53BP1 immunostaining is shown in green; cell nuclei were stained with DAPI (blue). Upper rows: merged images, lower rows – 53 BP1 imaging only. Results on 53BP1 foci counts are presented as means and the standard error of the means. Panel A: shown is 53BP1 foci kinetics in sham-irradiated cells (0 Gy), panel B: irradiated cells (1 Gy).
Figure 2. Cell cycle distribution of cultured H9 cells after 1 Gy irradiation.
Percentages of cells in G1 (black bars), S (white bars) and G2/M phases (gray bars) are shown for non-irradiated cells (0Gy) and for cells 4 and 24 hrs after 1Gy irradiation.
3.3. The dynamics of hESC response to IR exposures at the level of global transcriptome
In order to gain insight into the changes in gene expression elicited by exposure of hESC to IR, the whole-genome wide DNA microarray technique was used. We studied changes in the level of messenger RNA across virtually all known genes/transcripts in human genome (more than 40,000). We analyzed transcriptome of hESC at two time points after exposure to ionizing radiation; 2 hrs to assess an immediate or early response and 16 hrs to assess the later response.
The results of our transcriptome screening showed that at 2 hr post 1 Gy exposure of cultured H9 cells there were only 30 statistically significant differentially expressed genes (Table 1). Interestingly, all of these genes were up-regulated by more than two-fold compared to sham-irradiated control cell cultures handled in parallel with the irradiated ones. Many of induced genes have been already shown to participate in DDR in somatic adult differentiated cells, such as fibroblasts and peripheral blood cells [9, 36, 37]. Indeed, CDKN1A, BTG2, SESN1, IER5 and GADD45A are among the best studied and extensively characterized markers of IR exposure of human cells, and their induction is usually associated with temporary cell cycle arrest. CDKN1A is known to act as the cyclin-dependent kinase inhibitor [38]; BTG2 exerts its anti-proliferative functions mainly via degradation of messenger RNA [39]. SESN1 provides a mechanistic link between genotoxic p53-mediated stress signaling and metabolic mTOR checkpoint [40], also being one of five genes constituting gene expression signature of IR exposure [37]. IER5 affects radiosensitivity via disturbing radiation-induced cell cycle checkpoints [41]. GADD45A has been shown to be implicated in G2/M arrest of the cell cycle. This finding is in agreement with our results on the accumulation of hESC in G2/M phase at the early timepoint after IR exposure (Fig. 2). The activation of G2/M checkpoint in irradiated hESC was also observed by others [14], although the specific involvement of GADD45A was not elucidated. At the same time, several genes induced by IR exposure in hESC are known to dampen cell cycle checkpoints. For example, PPM1D serves to reverse the p53 and Chk1-induced cell cycle arrest and return the cell to a homeostatic state following completion of DNA repair [42]. In addition, we found up-regulation of some other key genes involved in G2/M cell cycle transition (PLK2, PLK3). It was shown that p53-dependent activation of PLK2 prevents mitotic catastrophe following spindle damage [43]. Over-expression of PLK2 is thought to increase cell survival upon stress conditions [44]. In contrast, PLK3 has been demonstrated to be a multifunctional gene rapidly activating by reactive oxygen species (ROS) and functioning in proapoptotic pathways [45, 46].
Table 1.
Comprehensive list of differentially expressed genes in irradiated H9 cells (1 Gy, 2 hr)
| Gene symbol | GeneBank accession | Gene description | Fold-change |
|---|---|---|---|
| SPAG9 | NM_001130528 | Homo sapiens sperm associated antigen 9 (SPAG9), transcript variant 1, mRNA | 26.40 |
| DHFR | NM_000791 | Homo sapiens dihydrofolate reductase (DHFR), mRNA | 15.89 |
| ZNF79 | NM_007135 | Homo sapiens zinc finger protein 79 (ZNF79), mRNA | 6.91 |
| BTG2 | NM_006763 | Homo sapiens BTG family, member 2 (BTG2), mRNA | 6.58 |
| CDKN1A | NM_000389 | Homo sapiens cyclin-dependent kinase inhibitor 1A (p21, Cip1) (CDKN1A), transcript variant 1, mRNA | 5.81 |
| PSMF1 | NM_006814 | Homo sapiens proteasome (prosome, macropain) inhibitor subunit 1 (PI31) (PSMF1), transcript variant 1, mRNA | 5.30 |
| GDF15 | NM_004864 | Homo sapiens growth differentiation factor 15 (GDF15), mRNA | 5.17 |
| MGC5370 | BC006795 | Homo sapiens hypothetical protein MGC5370, mRNA (cDNA clone IMAGE:3049213), partial cds. | 4.61 |
| ADRBK1 | NM_001619 | Homo sapiens adrenergic, beta, receptor kinase 1 (ADRBK1), mRNA | 4.45 |
| PLK3 | NM_004073 | Homo sapiens polo-like kinase 3 (Drosophila) (PLK3), mRNA | 4.33 |
| PLK2 | NM_006622 | Homo sapiens polo-like kinase 2 (Drosophila) (PLK2), mRNA | 4.28 |
| SESN1 | NM_014454 | Homo sapiens sestrin 1 (SESN1), mRNA | 3.54 |
| BBC3 | NM_014417 | Homo sapiens BCL2 binding component 3 (BBC3), transcript variant 4, mRNA | 3.49 |
| MTFMT | NM_139242 | Homo sapiens mitochondrial methionyl-tRNA formyltransferase (MTFMT), nuclear gene encoding mitochondrial protein, mRNA | 3.48 |
| HTATIP2 | NM_001098523 | Homo sapiens HIV-1 Tat interactive protein 2, 30kDa (HTATIP2), transcript variant 5, mRNA | 3.45 |
| LIF | NM_002309 | Homo sapiens leukemia inhibitory factor (cholinergic differentiation factor) (LIF), mRNA | 3.37 |
| PAPD5 | NM_001040285 | Homo sapiens PAP associated domain containing 5 (PAPD5), transcript variant 2, mRNA | 3.26 |
| PPM1D | NM_003620 | Homo sapiens protein phosphatase 1D magnesium-dependent, delta isoform (PPM1D), mRNA | 3.24 |
| THC2502237 | THC2502237 | Q69QZ1_ORYSA (Q69QZ1) F-box protein-like, partial | 3.16 |
| ZNF761 | NM_001008401 | Homo sapiens zinc finger protein 761 (ZNF761), mRNA | 3.03 |
| THC2545454 | THC2545454 | Unknown | 3.02 |
| IER5 | NM_016545 | Homo sapiens immediate early response 5 (IER5), mRNA | 2.96 |
| PINX1 | ENST00000314787 | Pin2-interacting protein X1 (TRF1-interacting protein 1)(Liver- related putative tumor suppressor)(Protein 67-11-3) [Source:UniProtKB/Swiss-Prot;Acc:Q96BK5] | 2.93 |
| CARD8 | NM_014959 | Homo sapiens caspase recruitment domain family, member 8 (CARD8), mRNA | 2.92 |
| FAS | NM_000043 | Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA | 2.80 |
| GADD45A | NM_001924 | Homo sapiens growth arrest and DNA-damage-inducible, alpha (GADD45A), mRNA | 2.76 |
| TP53INP1 | NM_033285 | Homo sapiens tumor protein p53 inducible nuclear protein 1 (TP53INP1), transcript variant 1, mRNA | 2.73 |
| ZSCAN20 | NM_145238 | Homo sapiens zinc finger and SCAN domain containing 20 (ZSCAN20), mRNA | 2.50 |
| ZNF135 | NM_003436 | Homo sapiens zinc finger protein 135 (ZNF135), mRNA | 2.50 |
| ANKRA2 | NM_023039 | Homo sapiens ankyrin repeat, family A (RFXANK-like), 2 (ANKRA2), mRNA | 2.32 |
In concert with our previously obtained data [31] that showed presence of ca. 30% apoptotic cells in hESC population after 1 Gy IR exposure, several of early-induced differentially expressed genes in irradiated hESC are known to participate in apoptosis (GDF15, CARD8, HTATIP2, TP53INP1, others). Some of them are among the most characterized cell death-promoting factors, such as FAS and BBC3 also known as PUMA. Specifically, HTATIP2 was shown to be involved in cellular oxidative stress surveillance and a potent inductor of apoptosis through stabilization of p53 mRNA [47]. BBC3 is a crucial mediator of p53-dependent as well as p53-independent apoptosis induced by a wide variety of stimuli. BBC3 directly binds and antagonizes all known pro-survival Bcl-2 family members to induce mitochondrial dysfunction and downstream caspase activation [48]. FAS has been shown to play a central role in the physiological regulation of programmed cell death as well [49]. It is noteworthy that several induced IRresponsive genes are known to participate in some aspects peculiar to stem-cell-unique homeostasis. Specifically, PINX1 positively regulates telomerase activity [50] which is apparently abundant in hESC. Probably, stressed hESC temporarily modulate the activity of telomerase shortly after IR exposures in a way to preserve the genome stability. Another interesting target of IR exposure in hESC is LIF, which is a pleiotropic cytokine best known for its key role in maintenance of pluripotency and self-renewal of mouse embryonic stem cells [51]; although the importance of LIF expression in irradiated hESC is not obvious. A few up-regulated genes in hESC at 2 hr post-IR are presumably acting as transcription factors (ZNF79, ZSCAN20, ZNF135, and ZNF761), the other few genes are still uncharacterized.
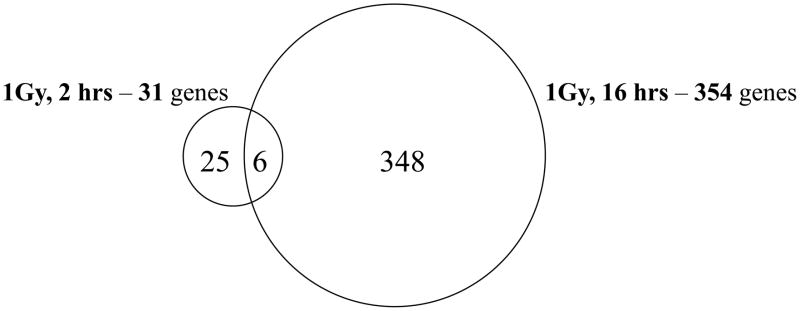
In order to examine the transcriptome of hESC underpinning the “late” response of these cells to IR exposure, we undertook genomic profiling of irradiated H9 cells at 16 hr post IR exposure. At this time point most apoptotic cells were cleared from hESC colonies (data not shown). About 354 differentially expressed genes were identified; all of them showed up-regulation, varying in range from about 1.5-fold till 25-fold over time-matched sham-irradiated cell cultures (Additional Table 1). The list of genes most highly overexpressed (more than 3-fold) are presented in Table 2. Interestingly, we found that irradiated hESC robustly overexpress many genes belonging to metallothionein gene superfamily (MT1M, MT1L, MT1H, MT1G), which is observed also in many types of somatic human cells after IR as well [9, 36]. Metallothionein expression was shown to be induced by cytokines, growth factors, and oxidative stress conditions [52]. One of the primary functions of metallothioneins is thought to involve the protection of stressed cell populations from the deleterious effects of ROS, including ROS detoxification. However, the most highly overexpressed characterized gene at 16 hr post-1 Gy exposure was HIST1H4I. The regulation of histone H4 subfamily gene expression in hESC under normal cell culture conditions was interrogated before [53]. HIST1H4I was found to be moderately expressed compared to other genes in H4 subfamily under nonstressed conditions, but the relevance of more than 22-fold overexpression of this specific histone species in irradiated H9 cells is presently unknown. Another histone gene HIST1H4E belonging to the same gene cluster was also identified as being IR-induced genes, albeit to a much lesser degree compared to HIST1H4I. We observed up-regulation of many transcription factors at 16 hr post 1 Gy exposure, such as ZNF302, ZNF33A, ZNF697, SP5, ZFYVE16 and others. Our DNA microarray analysis of irradiated H9 cells revealed that the gene expression signatures characterizing “early” (2 hr) and “late” (16 hr) response to 1 Gy are profoundly distinct (Fig. 3). Only 6 genes were differentially expressed at both timepoints examined; they were: CDKN1A, BTG2, GDF15, SESN1, PLK3 and ANKRA2. Sustained expression of these genes in irradiated hESC over prolonged time periods may constitute the specific “gene expression signature” which can serve as a marker of IR exposure of hESC, although further experiments are needed for a proper validation.
Table 2.
IR-induced genes showing more than 3-fold changes in irradiated H9 cells (1 Gy, 16 hr)
| Gene symbol | GeneBank accession | Gene description | Fold-change |
|---|---|---|---|
| A_24_P755169 | A_24_P755169 | Unknown | 25.86 |
| HIST1H4I | NM_003495 | Homo sapiens histone cluster 1, H4i (HIST1H4I), mRNA | 22.87 |
| CCDC88A | NM_018084 | Homo sapiens coiled-coil domain containing 88A (CCDC88A), transcript variant 2, mRNA | 13.80 |
| CCDC144A | NM_014695 | Homo sapiens coiled-coil domain containing 144A (CCDC144A), mRNA | 10.03 |
| KIAA0564 | NM_001009814 | Homo sapiens KIAA0564 (KIAA0564), transcript variant 2, mRNA | 9.70 |
| ZNF302 | NM_018443 | Homo sapiens zinc finger protein 302 (ZNF302), transcript variant 1, mRNA | 6.56 |
| MT1M | NM_176870 | Homo sapiens metallothionein 1M (MT1M), mRNA | 5.07 |
| BGLAP | NM_199173 | Homo sapiens bone gamma-carboxyglutamate (gla) protein (BGLAP), mRNA | 4.95 |
| SP5 | NM_001003845 | Homo sapiens Sp5 transcription factor (SP5), mRNA | 4.95 |
| TMCO3 | NM_017905 | Homo sapiens transmembrane and coiled-coil domains 3 (TMCO3), mRNA | 4.42 |
| PIGQ | NM_004204 | Homo sapiens phosphatidylinositol glycan anchor biosynthesis, class Q (PIGQ), transcript variant 2, mRNA | 4.37 |
| FAM71F1 | NM_032599 | Homo sapiens family with sequence similarity 71, member F1 (FAM71F1), mRNA | 4.09 |
| SAPS3 | NM_018312 | Homo sapiens SAPS domain family, member 3 (SAPS3), mRNA | 3.99 |
| DDX46 | NM_014829 | Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 (DDX46), mRNA | 3.84 |
| ZNF33A | NM_006954 | Homo sapiens zinc finger protein 33A (ZNF33A), transcript variant 1, mRNA | 3.82 |
| HNRNPU | NM_004501 | Homo sapiens heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (HNRNPU), transcript variant 2, mRNA | 3.58 |
| LOC391777 | CR609307 | Full-length cDNA clone CS0DH003YG13 of T cells (Jurkat cell line) of Homo sapiens (human). | 3.55 |
| WDR90 | NM_145294 | Homo sapiens WD repeat domain 90 (WDR90), mRNA | 3.54 |
| FAM36A | NM_198076 | Homo sapiens family with sequence similarity 36, member A (FAM36A), mRNA | 3.43 |
| ZNF697 | NM_001080470 | Homo sapiens zinc finger protein 697 (ZNF697), mRNA | 3.41 |
| MDP-1 | NM_138476 | Homo sapiens magnesium-dependent phosphatase 1 (MDP-1), mRNA | 3.34 |
| THC2648397 | THC2648397 | ALU1_HUMAN (P39188) Alu subfamily J sequence contamination warning entry, partial (8%) | 3.29 |
| IL27RA | NM_004843 | Homo sapiens interleukin 27 receptor, alpha (IL27RA), mRNA | 3.25 |
| SKP1 | NM_170679 | Homo sapiens S-phase kinase-associated protein 1 (SKP1), transcript variant 2, mRNA | 3.10 |
| MT1L | NR_001447 | Homo sapiens metallothionein 1L (gene/pseudogene) (MT1L), non-coding RNA | 3.10 |
| PPIL5 | NM_152329 | Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 5 (PPIL5), transcript variant 1, mRNA | 3.07 |
| BX101252 | BX101252 | BX101252 NCI_CGAP_Lu24 Homo sapiens cDNA clone IMAGp998I115625, mRNA sequence | 3.04 |
| ZFYVE16 | NM_001105251 | Homo sapiens zinc finger, FYVE domain containing 16 (ZFYVE16), transcript variant 2, mRNA | 3.03 |
Figure 3. Venn diagram presenting the number of differentially expressed genes in H9 cells after 1 Gy of IR exposures.
The transcriptional profiling assays were conducted as indicated in Materials and methods section. The SAM analysis was done with BRB ArrayTools (FDR less than 10%). The number of genes in an intersection corresponds to transcripts upregulated both at 2 hr and 16 hr timepoints following IR exposures.
3.4. Classification of differentially expressed genes in functional groups
To comprehensively identify the biological pathways involved in IR response of H9 cells, the Gene Ontology analysis was done (Table 3). P53 signaling pathway critically predominates in “early” 2 hr post-IR exposure response. Out of total 30 genes that were differentially expressed at this timepoint about 24 genes are known or predicted to be direct transcriptional targets of p53 [54]. Although many of these genes are responsible for apoptosis induction/execution, some of induced genes are thought to act in pro-survival pathways. Therefore, it is conceivable to suggest that the subpopulation of hESCs committed to cell death within the bulk cell population overexpressed the positive regulation of apoptosis genes, whereas the surviving cell subpopulation induced the negative regulators of cell death; however, further studies need to be done to address this issue in more detail. Negative regulation of progression through the cell cycle also scored very high in pathway analysis at the 2 hr timepoint following IR exposure of hESC. Gene Ontology analysis revealed that at 16 hr post-IR exposure among the mostly affected pathways were metabolic processes, such as the ubiquitin cycle involved in protein turnover, intracellular transport, transferase and carbohydrate metabolic pathways, and cell division processes, especially associated with G2/M phase events. Interestingly, ubiquitin-dependent signaling recently sparked a lot of attention owing to its crucial involvement in DNA damage signaling [55]. Negative regulation of transcription was also identified as one of the top events characterizing “late” response of hESC to IR exposures.
Table 3.
Gene Ontology analysis of overrepresented biological processes/pathways in H9 cells after IR exposures
| Exposures | Overrepresented categories | EASE score |
|---|---|---|
| 1Gy, 2 hrs | P53 signaling pathway | 5.1E-8 |
| Regulation of progression through cell cycle | 0.00012 | |
| Apoptosis | 0.00014 | |
| Negative regulation of progression through cell cycle | 0.00029 | |
| Positive regulation of apoptosis | 0.0006 | |
| Response to DNA damage stimulus | 0.0015 | |
| Negative regulation of apoptosis | 0.0053 | |
| Response to stress | 0.024 | |
|
| ||
| 1Gy, 16 hrs | Ubiquitin cycle | 3.1E-5 |
| Transferase activity, transferring glycosyl groups | 0.0006 | |
| Metallothionein superfamily | 0.00077 | |
| Negative regulation of transcription, DNA-dependent | 0.0017 | |
| Intracellular transport | 0.0019 | |
| Carbohydrate metabolic process | 0.004 | |
| Cell division and chromosome partitioning | 0.01 | |
The predominant p53-elicited transcriptional response of hESC to IR exposures at early timepoint such as 2 hr resembles that observed in adult differentiated human somatic cells [11]. However, induction of p53 in pluripotent cells was shown to have some stem cell-specific consequences. For example, p53 was reported to downregulate Nanog in mouse embryonic stem cells resulting in differentiation of these cells [56]. In hESC, p53 was shown to be required for apoptosis induction both under suboptimal cell culture conditions and after genotoxic stress [15].
3.5. Transcriptional response to IR in hESC is different from somatic cells
In comparison with the whole-genome-wide gene expression changes observed after 1 Gy exposure of human IMR-90 fibroblasts, H9 hESC show much more robust and focused response in terms of the magnitude of the induction of a small subset of IRresponsive genes. IR-induced transcriptome alterations in IMR-90 cells, on the other hand, involve a relatively modest modulation of more than a thousand of genes belonging to a wide range of metabolic pathways [57]. We found only 2 genes that were induced by IR exposures both in hESC and IMR-90 fibroblasts, namely, CDKN1A and BTG2. The lack of similarity in responses to IR exposures between pluripotent and fully differentiated human cells may reflect the distinct stress-defense mechanisms operating within human cells possessing so different epigenetic landscapes [58]. It was shown that the genetic material of embryonic stem cells is composed primarily of euchromatin, in contrast to a more compacted chromatin state of differentiated cells [59].
Cellular response to IR is known to be multifaceted involving, but not limited to, cell-cycle arrest, induction of DDR, and the changes in global gene expression. Today, still relatively little is known about even the key aspects of hESC biology, including cell cycle governing mechanisms, although it has been shown that the relevant processes might be distinct from that observed in fully differentiated somatic human cells [53, 60]. One of the reasons that no coherent picture emerged yet is due to a lack of high-throughput studies addressing the radioresponse of hESC at the systems biology level. One of the earliest observations made by researchers studying hESC biology was that p21 (CDKN1A) is rapidly upregulated following genotoxic stress exposures (up to 200-fold after 5 Gy IR) [53]. However, in our studies CDKN1A was overexpressed less than 6-fold at 2 hr post IR exposure, and less than 2-fold compared to sham-irradiated cell cultures at a later timepoint (16 hr). These discrepancies could be attributed to a much lower dose of IR exposure we used in our studies (1 Gy versus 5 Gy), the differences in cell culture protocol (feeder-free versus mouse cell feeder-based cultures), and other variables. It was shown recently that messenger RNA levels of several DNA repair genes are elevated in human embryonic stem cells compared with their differentiated forms; although no studies were undertaken to assess the expression of DNA repair genes in hESC after genotoxic stress exposure such as IR treatment [61]. In our experiments we have observed a lack of substantial changes in expression of DNA repair genes in H9 cells following 1 Gy of gamma-ray exposures. This is in concert with what was reported for irradiated human adult somatic cells as well [57] and with the fact that DDR mostly manifests on post-translational level [32]. Therefore, hESC and human differentiated somatic cells, quite interestingly, in many ways behave similarly upon exposure to IR.
3.6. Chromosomal distribution of IR-modulated genes in hESC
Previously, we reported that there is a non-random chromosomal distribution of IRresponsive genes in human primary somatic cells following DNA-targeted 125I-based irradiation [30]. Here, we show that such non-randomness is characteristic of hESC exposed to external irradiation as well (Additional Fig. 4). IR-induced genes (1 Gy, 2 hr) are enriched on chromosomes 1, 5, 6 and 19, whereas “late” response genes are on chromosomes 5, 7 and 13. It is worth mentioning that in human primary differentiated somatic cells IR-responsive genes were also disproportionately over-represented on chromosomes 5, 7 and 19 [30]. The mechanisms underlying this phenomenon are currently unknown and need to be examined in the future.
3.7. Real-time PCR validation of DNA microarray results
To verify the gene expression changes observed with DNA microarray technique with an independent method, we analyzed the relative abundance of transcripts of five genes showing different patterns of expression in our studies (Table 4). In general, a good concordance between DNA microarray results and RT-PCR data was observed; the trends of expression alterations were preserved in all cases, however, for some datapoints the magnitude of gene expression changes varied to some extent. This is in accordance with reports published elsewhere [30, 57, 62]. Therefore, we partially verified our functional genomics studies with an assay complementary to DNA microarray technique to strengthen the validity of our main conclusions.
Table 4. The comparison between DNA microarray and quantitative RT-PCR datasets.
Data are presented as mean ± SEM
| CDKN1A | GADD45A | DHFR | CCDC88A | MT1F | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 hr | 16 hr | 2 hr | 16 hr | 2 hr | 16 hr | 2 hr | 16 hr | 2 hr | 16 hr | |
| DNA microarray | 2.54±0.19 | 0.93±0.09 | 1.46±0.30 | 0.59±0.05 | 2.70±1.12 | −0.60±0.40 | 0.41±0.36 | 1.00±0.46 | −0.24±0.10 | 0.52±0.19 |
| Real-time PCR | 5.03±0.68 | 1.03±0.47 | 2.87±0.64 | 0.37±0.45 | 0.93±0.25 | −0.53±0.04 | 0.35±0.04 | 0.50±0.41 | 0.43±0.41 | 0.85±0.20 |
4. Conclusions
In this study, we report the first comprehensive data on the dynamics of the whole-genome-wide transcriptional changes in hESC subjected to IR exposures. We have shown that “early” (2 hr) gene expression alterations of hESC following 1 Gy of IR exposure feature almost exclusively a p53-dependent, apparently pro-apoptotic, pattern of response. The “late” (16 hr) gene expression signature of irradiated hESC is characterized by predominant expression of pro-survival pathways. None of the statistically significant, IR responsive, differentially expressed genes in hESC are down-regulated. We found a unique subset of 6 genes that were consistently up-regulated after IR at both timepoints examined. This gene expression signature may serve as a marker of IR exposure of hESC. Additionally, the importance of our work lies in a fact that we identified novel markers of IR exposure of human cells in their most pluripotent state; thus, unmasking the key genes of DDR, which may not be otherwise revealed in case of studying DDR in settings of fully established epigenetic landscape of terminally differentiated cells.
Supplementary Material
Acknowledgments
We thank W. DeGraff for his invaluable help with cell culture irradiation. This research was supported by the Intramural Research Program of the NIH, Clinical Center.
Role of the funding source
The funding source had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Abbreviations
- hESC
human embryonic stem cell
- IR
ionizing radiation
- DDR
DNA damage response
- PBS
phosphate buffer saline
- BSA
bovine serum albumin
- PI
propidium iodide
- SAM
Significance Analysis of Microarrays
- FDR
false discovery rate
- GO
Gene Ontology
- DNA DSB
DNA double strand break
- IRIF
ionizing radiation–induced foci
- qRT-PCR
quantitative real-time polymerase chain reaction
- SEM
standard error of the mean
Footnotes
Authors’ contributions
MVS participated in the design of the study, conducted experimental procedures including cell culture and irradiation, did immunocytochemical and array experiments, data mining and statistical analysis, validated array data with qRT-PCR, and drafted the manuscript. IVP conducted cell cycle studies and drafted the manuscript. IGP participated in the design of the study and drafted the manuscript. RDN conceived of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mykyta V. Sokolov, Email: sokolovm@mail.nih.gov.
Irina V. Panyutin, Email: ipanyutinv@mail.nih.gov.
Igor G. Panyutin, Email: ipanyuting@mail.nih.gov.
Ronald D. Neumann, Email: rneumann@mail.nih.gov.
References
- 1.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 2.Bassi P, Sacco E. Cancer and aging: the molecular pathways. Urol Oncol. 2009;27:620–627. doi: 10.1016/j.urolonc.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 4.Zhan Q, Bae I, Kastan MB, Fornace AJ., Jr The p53-dependent gamma-ray response of GADD45. Cancer Res. 1994;54:2755–2760. [PubMed] [Google Scholar]
- 5.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci U S A. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ., Jr Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 7.Berglund SR, Rocke DM, Dai J, Schwietert CW, Santana A, Stern RL, Lehmann J, Hartmann Siantar CL, Goldberg Z. Transient genome-wide transcriptional response to low-dose ionizing radiation in vivo in humans. Int J Radiat Oncol Biol Phys. 2008;70:229–234. doi: 10.1016/j.ijrobp.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Ghilardi-Netto T, Donadi EA, Passos GA, Sakamoto-Hojo ET. Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. J Radiat Res (Tokyo) 2009;50:61–71. doi: 10.1269/jrr.08034. [DOI] [PubMed] [Google Scholar]
- 9.Rodningen OK, Overgaard J, Alsner J, Hastie T, Borresen-Dale AL. Microarray analysis of the transcriptional response to single or multiple doses of ionizing radiation in human subcutaneous fibroblasts. Radiother Oncol. 2005;77:231–240. doi: 10.1016/j.radonc.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Sanzari JK, Nuth M, Kennedy AR. Induction of cytokine gene expression in human thyroid epithelial cells irradiated with HZE particles (iron ions) Radiat Res. 2009;172:437–443. doi: 10.1667/RR1363.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warters RL, Packard AT, Kramer GF, Gaffney DK, Moos PJ. Differential gene expression in primary human skin keratinocytes and fibroblasts in response to ionizing radiation. Radiat Res. 2009;172:82–95. doi: 10.1667/RR1677.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amundson SA. Functional genomics in radiation biology: a gateway to cellular systems-level studies. Radiat Environ Biophys. 2008;47:25–31. doi: 10.1007/s00411-007-0140-1. [DOI] [PubMed] [Google Scholar]
- 13.Filion TM, Qiao M, Ghule PN, Mandeville M, van Wijnen AJ, Stein JL, Lian JB, Altieri DC, Stein GS. Survival responses of human embryonic stem cells to DNA damage. J Cell Physiol. 2009;220:586–592. doi: 10.1002/jcp.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momcilovic O, Choi S, Varum S, Bakkenist C, Schatten G, Navara C. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells. 2009;27:1822–1835. doi: 10.1002/stem.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- 16.De Santis M, Cesari E, Nobili E, Straface G, Cavaliere AF, Caruso A. Radiation effects on development. Birth Defects Res C Embryo Today. 2007;81:177–182. doi: 10.1002/bdrc.20099. [DOI] [PubMed] [Google Scholar]
- 17.Romanova LK, Zhorova ES. The radiation effects of low doses of irradiation on human embryos and fetuses. Ontogenez. 1994;25:55–65. [PubMed] [Google Scholar]
- 18.Jensen J, Hyllner J, Bjorquist P. Human embryonic stem cell technologies and drug discovery. J Cell Physiol. 2009;219:513–519. doi: 10.1002/jcp.21732. [DOI] [PubMed] [Google Scholar]
- 19.Davila JC, Cezar GG, Thiede M, Strom S, Miki T, Trosko J. Use and application of stem cells in toxicology. Toxicol Sci. 2004;79:214–223. doi: 10.1093/toxsci/kfh100. [DOI] [PubMed] [Google Scholar]
- 20.Wilson KD, Sun N, Huang M, Zhang WY, Lee AS, Li Z, Wang SX, Wu JC. Effects of ionizing radiation on self-renewal and pluripotency of human embryonic stem cells. Cancer Res. 2010;70:5539–5548. doi: 10.1158/0008-5472.CAN-09-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokolov MV, Smilenov LB, Hall EJ, Panyutin IG, Bonner WM, Sedelnikova OA. Ionizing radiation induces DNA double-strand breaks in bystander primary human fibroblasts. Oncogene. 2005;24:7257–7265. doi: 10.1038/sj.onc.1208886. [DOI] [PubMed] [Google Scholar]
- 22.Sokolov MV, Neumann RD. Radiation-induced bystander effects in cultured human stem cells. PLoS One. 2010;5:e14195. doi: 10.1371/journal.pone.0014195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokolov MV, Dickey JS, Bonner WM, Sedelnikova OA. gamma-H2AX in bystander cells: not just a radiation-triggered event, a cellular response to stress mediated by intercellular communication. Cell Cycle. 2007;6:2210–2212. doi: 10.4161/cc.6.18.4682. [DOI] [PubMed] [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon RM, Dobbin K. Experimental design of DNA microarray experiments. Biotechniques. 2003;(Suppl):16–21. [PubMed] [Google Scholar]
- 26.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 29.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Sokolov MV, Neumann RD, Panyutin IG. Effects of DNA-targeted ionizing radiation produced by 5-[125I]iodo-2′-deoxyuridine on global gene expression in primary human cells. BMC Genomics. 2007;8:192. doi: 10.1186/1471-2164-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokolov MV, Panyutin IV, Onyshchenko MI, Panyutin IG, Neumann RD. Expression of pluripotency-associated genes in the surviving fraction of cultured human embryonic stem cells is not significantly affected by ionizing radiation. Gene. 2010;455:8–15. doi: 10.1016/j.gene.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 34.Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene. 2006;25:3258–3266. doi: 10.1038/sj.onc.1209352. [DOI] [PubMed] [Google Scholar]
- 35.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokolov M, Panyutin IG, Neumann R. Genome-wide gene expression changes in normal human fibroblasts in response to low-LET gamma-radiation and high-LET-like 125IUdR exposures. Radiat Prot Dosimetry. 2006;122:195–201. doi: 10.1093/rpd/ncl423. [DOI] [PubMed] [Google Scholar]
- 37.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 39.Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding KK, Shang ZF, Hao C, Xu QZ, Shen JJ, Yang CJ, Xie YH, Qiao C, Wang Y, Xu LL, Zhou PK. Induced expression of the IER5 gene by gamma-ray irradiation and its involvement in cell cycle checkpoint control and survival. Radiat Environ Biophys. 2009;48:205–213. doi: 10.1007/s00411-009-0213-4. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23:5556–5571. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto T, Wang PY, Ma W, Sung HJ, Matoba S, Hwang PM. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2009;106:14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie S, Wu H, Wang Q, Kunicki J, Thomas RO, Hollingsworth RE, Cogswell J, Dai W. Genotoxic stress-induced activation of Plk3 is partly mediated by Chk2. Cell Cycle. 2002;1:424–429. doi: 10.4161/cc.1.6.271. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Niu J, Uwagawa T, Peng B, Chiao PJ. Function of polo-like kinase 3 in NF-kappaB-mediated proapoptotic response. J Biol Chem. 2005;280:16843–16850. doi: 10.1074/jbc.M410119200. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J, Chen J, Lu B, Dong L, Wang H, Bi C, Wu G, Guo H, Wu M, Guo Y. TIP30 induces apoptosis under oxidative stress through stabilization of p53 messenger RNA in human hepatocellular carcinoma. Cancer Res. 2008;68:4133–4141. doi: 10.1158/0008-5472.CAN-08-0432. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7:1001–1012. doi: 10.1038/nrd2637. [DOI] [PubMed] [Google Scholar]
- 50.Zhang B, Bai YX, Ma HH, Feng F, Jin R, Wang ZL, Lin J, Sun SP, Yang P, Wang XX, Huang PT, Huang CF, Peng Y, Chen YC, Kung HF, Huang JJ. Silencing PinX1 compromises telomere length maintenance as well as tumorigenicity in telomerase-positive human cancer cells. Cancer Res. 2009;69:75–83. doi: 10.1158/0008-5472.CAN-08-1393. [DOI] [PubMed] [Google Scholar]
- 51.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laukens D, Waeytens A, De Bleser P, Cuvelier C, De Vos M. Human metallothionein expression under normal and pathological conditions: mechanisms of gene regulation based on in silico promoter analysis. Crit Rev Eukaryot Gene Expr. 2009;19:301–317. doi: 10.1615/critreveukargeneexpr.v19.i4.40. [DOI] [PubMed] [Google Scholar]
- 53.Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- 54.Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D’Erchia AM, Navarro B, Tullo A, Saccone C, Gisel A. p53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinformatics. 2007;8(Suppl 1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selvarajah J, Moumen A. Role of ubiquitination in the DNA damage response: proteomic analysis to identify new DNA-damage-induced ubiquitinated proteins. Biochem Soc Trans. 38:87–91. doi: 10.1042/BST0380087. [DOI] [PubMed] [Google Scholar]
- 56.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 57.Sokolov MV, Smirnova NA, Camerini-Otero RD, Neumann RD, Panyutin IG. Microarray analysis of differentially expressed genes after exposure of normal human fibroblasts to ionizing radiation from an external source and from DNA-incorporated iodine-125 radionuclide. Gene. 2006;382:47–56. doi: 10.1016/j.gene.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 58.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasmussen TP. Developmentally-poised chromatin of embryonic stem cells. Front Biosci. 2008;13:1568–1577. doi: 10.2741/2781. [DOI] [PubMed] [Google Scholar]
- 60.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 61.Maynard S, Swistowska AM, Lee JW, Liu Y, Liu ST, Da Cruz AB, Rao M, de Souza-Pinto NC, Zeng X, Bohr VA. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells. 2008;26:2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassi C, Mello SS, Cardoso RS, Godoy PD, Fachin AL, Junta CM, Sandrin-Garcia P, Carlotti CG, Falcao RP, Donadi EA, Passos GA, Sakamoto-Hojo ET. Transcriptional changes in U343 MG-a glioblastoma cell line exposed to ionizing radiation. Hum Exp Toxicol. 2008;27:919–929. doi: 10.1177/0960327108102045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.