Abstract
Electrotransfer of molecules is a well established technique which finds extensive use for gene transfer and holds great promise for anticancer treatment. Despite its widespread application, the mechanisms governing the entry of DNA into the cell and its intracellular trafficking are not yet known. The aim of this study is to unravel the role of the actin cytoskeleton during gene electrotransfer in cells. We performed single-cell level approaches to observe the organization of the actin cytoskeleton in Chinese hamster ovary (CHO) cells. In addition, we performed experiments at the multiple-cell level to evaluate the efficiency of DNA transfer after alteration of the actin cytoskeleton using the drug latrunculin B. Actin patches colocalizing with the DNA at the plasma membrane were observed with additional characteristics similar to those of the DNA aggregates in terms of time, number, and size. The disruption of the microfilaments reduces the DNA accumulation at the plasma membrane and the gene expression. This is the first direct experimental evidence of the participation of the actin cytoskeleton in DNA electrotransfer.
Introduction
The cell plasma membrane acts as a barrier that hinders the transfer of molecules such as nucleic acids. Therefore, a variety of techniques have been developed to transfer DNA to the cytoplasm. These include the use of viruses,1 chemical methods such as cell-penetrating peptides,2 or liposome-mediated transfer,3 as well as the physical approaches of sonoporation4 and electroporation.5,6,7
Electroporation or electropermeabilization relies on the application of pulsed electric fields to cells. Thereby, plasma membranes become permeable to a large variety of nonpermeant molecules.8,9,10 Electroporation has proven to be a versatile technique. It has the advantages of being simple, safe, and highly efficient. Successful clinical applications of this method such as cancer treatment and gene therapy have been developed. A local antitumor drug delivery treatment in patients, called electrochemotherapy, has emerged.11 Electrotransfer has also been used to transfer DNA in vivo into the skin, liver, tumors, skeletal muscle cells, and a large variety of other tissues (for a review see ref. 12). Electrochemotherapy is now being accepted in a number of countries as a palliative treatment, and clinical trials of gene electrotransfer are now under investigation.13,14
Many models have been proposed to explain the basic mechanism of electropermeabilization and gene transfer as well as the difference in the efficiency of penetration between small sized molecules (such as anticancer drugs) and macromolecules (such as plasmid DNA).15,16,17,18 It is especially important to understand why only a few percent of cells express the electrotransfered gene while nearly 100% of a cell population can be electropermeabilized. Experiments show that membrane permeabilization takes place facing the electrodes and that it is asymmetric, the membrane area facing the anode being more permeabilized than the one facing the cathode.19 It allows the free and rapid diffusion of small (below 4kDa) external compounds into the cytoplasm. Gene transfer and expression appear as a multistep and complex process, which requires (i) membrane permeabilization, (ii) electrophoretic migration of the DNA, (iii) DNA/membrane interaction, (iv) translocation and migration of the plasmid in the cytoplasm, and (v) its passage through the nuclear envelope.6,19,20 However, little is known about the details of these steps, in particular, the behavior and future of the DNA in steps (iv) and (v) while the electric field is turned off. This complex process involves an anchoring step connecting plasmid DNA to the permeabilized membrane facing the negative electrode during the electric pulses, followed by a postpulse transfer into the cytoplasm.19,21,22 Recent results reveal the existence of two classes of plasmid DNA/membrane complexes: (i) complexes of low stability from which plasmid DNA can leave and return to the pulsation buffer, and (ii) complexes of high stability, where plasmid DNA cannot be removed even by applying electric pulses of reversed polarity. Only plasmid DNA belonging to the second class leads to effective gene expression.22 After being internalized, DNA must cross the cytoplasm to reach the nucleus. DNA molecules larger than 2,000 base pairs are unable to diffuse freely in the cytoplasm.23 However, electrotransfer leads to gene expression. Therefore, plasmids must be able to cross the cytoplasm by other means than free diffusion. Characterizing how plasmids move through the cytoplasm is imperative to understanding and enhancing transfection efficiencies. One possibility is that plasmids use the cell's own machinery to be transported through the cytoplasm. While the active transport of electrotransfered plasmids along microtubules has been previously observed,24,25 an active participation of the actin network has not yet been reported. In this study we present results which clearly indicate that the actin cytoskeleton also has an active role during the electric field mediated gene transfer in mammalian cells.
Results
For our investigations, single-cell level approaches were preferred. We performed two complementary strategies of visualization of the actin organization. The first was to use phalloidin-rhodamine 123 as a fluorescent label specific for filamentous actin. It gives a good spatial resolution but requires fixation of the samples.26 The second strategy was to transiently express actin fused with enhanced green fluorescence protein (EGFP) allowing for dynamical observations. In addition, we performed gene expression experiments at the multiple-cell level. This approach gives access to the efficiency of the complete transfer process.
Effect of the electric field on the actin cytoskeleton in the presence of DNA
The first objective was to determine to what extent the application of electric field pulses in the presence of DNA could affect the organization of the actin cytoskeleton. To this end, we performed single-cell level observations of the filamentous actin labeled with the phalloidin-rhodamine 123 dye. We observed the formation of actin patches which corresponded in location, time, number, and size to the DNA aggregate occurrence (Figures 1 and 3 and Supplementary Video S1). Indeed, these patches were localized only at the plasma membrane facing the negative electrode which is the area where the DNA interacts with the membrane. They were observable 10 minutes after the electric field application. This value corresponds to the average persistence time of the DNA aggregates. The average number of actin patches counted was 8.4 ± 3.9 per cells. The number of DNA aggregates was 7.2 ± 3.8. Their size was comparable to the one of the DNA aggregates. These actin structures were never observed in the absence of DNA during the electric field application. These observations suggest that actin could be involved in DNA/membrane interaction and/or DNA crossing of the membrane.
Figure 1.
Effect of the electric field applied in the presence of plasmid DNA on the actin cytoskeleton organization in CHO cells. The cells were pulsed in the presence of the pEGFP-C1 plasmid DNA with the following parameters: 10 pulses of 5 ms at 0.4 kV/cm and 1 Hz. (a–h) Cells were fixed 10 minutes after the application of the electric field and stained with phalloidin-rhodamine 123 dye (32 cells with actin patches were observed). The observations were performed using confocal microscopy. Panel b is a projection of a z-stack. The black arrow on the right side indicates the direction of the electric field. The white arrows on the pictures point to the actin patches formed at the plasma membrane facing the negative electrode. Bar = 20 µm. CHO, Chinese hamster ovary; EGFP, Enhanced green fluorescence protein.
Figure 3.
Visualization of the DNA/membrane interaction and the actin cytoskeleton in CHO cells after application of the electric field. CHO cells transiently expressing the EGFP-actin protein were pulsed in the presence of POPO-3 labeled plasmid DNA (pEGFP-C1) with the following parameters: 10 pulses of 5 ms at 0.4 kV/cm and 1 Hz. The observations were performed using wide field microscopy between 5 and 30 minutes after application of the electric field. (a,d,g,j) POPO-3 labeled DNA, (b,e,h,k) EGFP-actin protein expression, (c,f,i,l) merge of the two channels (18 cells clearly showed colocalization between DNA and actin). The black arrow on the right side indicates the direction of the electric field. Bar = 5 µm. CHO, Chinese hamster ovary; EGFP, enhanced green fluorescence protein.
To complete and confirm these results, we performed similar experiments with another strategy. Instead of using phalloidin-conjugated label, we observed the actin cytoskeleton of live Chinese hamster ovary (CHO) cells transiently expressing EGFP-actin. Once again, the formation of actin patches at the plasma membrane facing the negative electrode was observed (Figure 2 and Supplementary Video S2), with an average number of 8.5 ± 3.9 patches per cell. Time series acquisitions gave information about the lifetimes of these structures. They began to be visible around 5 minutes after the electric field application, i.e., ~5 minutes after the formation of the DNA aggregates (Figure 2b,g). The disappearance of some of the actin patches started after ~15–20 minutes (Figure 2c,d), while other patches persisted at the plasma membrane for up to 45 minutes (Figure 2e,h–j). In conclusion, actin polymerized in patches a few minutes after the DNA/membrane interaction was established and could stay in this stage for 45 minutes.
Figure 2.
Effect of the electric field applied in the presence of plasmid DNA on the actin cytoskeleton organization in CHO EGFP-actin expressing cells. CHO cells transiently expressing the EGFP-actin protein were pulsed in the presence of pEGFP-C1 plasmid DNA with the following parameters: 10 pulses of 5 ms at 0.4 kV/cm and 1 Hz. The observations were performed using wide field microscopy. (a–e) Time series of a cell, (f–g) another cell observed before the electric field application and 6 minutes after, (h–j) different cells at different times (25 cells with actin patches were observed). The black arrow on the right side indicates the direction of the electric field. The white arrows on the pictures point to the actin patches formed at the plasma membrane facing the negative electrode. Bar = 10 µm. CHO, Chinese hamster ovary; EGFP, enhanced green fluorescence protein.
In order to confirm that the actin patches were a direct consequence of the presence of DNA and coincided with the location of the DNA aggregates, we performed two color colocalization studies on living cells. We electrotransfered fluorescent DNA labeled with POPO-3 into CHO cells which transiently expressed EGFP-actin. Our observations showed that DNA aggregates and actin patches colocalized well (Figure 3). Actin polymerized exactly at the positions where the DNA interacted with the plasma membrane. These observations strongly suggest that actin is actively involved in the uptake of DNA by the CHO cells.
Effect of actin disruption on DNA/membrane interaction
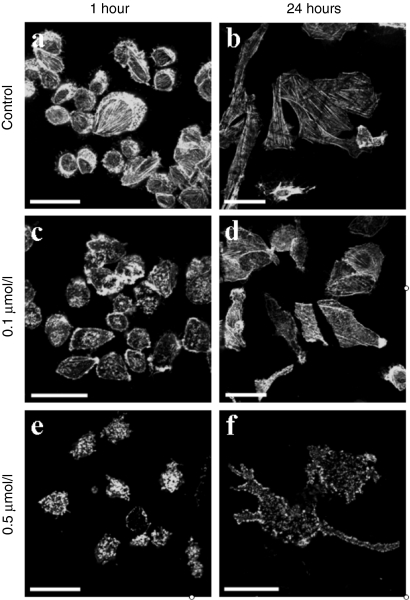
From the observations described above, it seems plausible that actin has an effect on the first steps of electrotransfection, i.e., on the DNA/membrane interaction. To investigate this further, we disrupted the actin cytoskeleton with latrunculin B and observed the effect on DNA behavior at the plasma membrane. Latrunculin B binds to a monomer of actin causing the depolymerization of the actin filaments. We used concentrations which were high enough to destroy filaments but did not affect the cell viability and morphology. The optimum conditions of latrunculin B incubation were 0.1 µmol/l for 1 hour at 37 °C (Figure 4c). Under these conditions the density of actin filaments was reduced, but cell viability and morphology were unaffected (Figure 4c). Higher concentrations, however, led to changes in cell morphology and viability (Figure 4e). CHO cells were pulsed in the presence of fluorescently labeled DNA (TOTO-1). In nontreated cells, the DNA/membrane interaction was characterized by aggregate formation at the plasma membrane facing the negative electrode (Figure 5a,c,e,g) as previously described.19 In the treated cells, the DNA/membrane interaction was clearly less visible but still present (Figure 5b,d,f,h). We counted the number of aggregates present as a function of fluorescence intensity (Figure 5i). For the control cells, the mean fluorescence intensity of the aggregates was 12,949 ± 476 arbitrary units while for the treated cells the mean fluorescence intensity was 6,480 ± 514 arbitrary units (Figure 5i inset). The difference was highly significant (P = 5 × 10−10). This hints at the actin cytoskeleton being involved in the accumulation of the DNA inserted into the plasma membrane. It is noteworthy that even after disruption of the actin cytoskeleton some DNA was still located in patches on the membrane. Thus it appears that actin can enhance the amount of DNA in the patches on the membrane, but is not needed for patch formation.
Figure 4.
Effect of latrunculin B on the actin cytoskeleton organization in CHO cells. Filamentous actin was stained with phalloidin-rhodamine 123 dye after fixation of the sample. The observations were performed using confocal microscopy. (a,b) nontreated cells, (c,d,e,f) latrunculin B treated cells at 0.1 µmol/l (c,d) or 0.5 µmol/l (e,f), 37 °C for 1 hour (c,e) or 24 hours (d,f) (for each condition, a minimum of 50 cells were observed). Bar = 20 µm. CHO, Chinese hamster ovary.
Figure 5.
Visualization of the DNA/membrane interaction and fluorescence intensity distribution of the DNA aggregates in CHO cells with and without latrunculin B drug treatment. The cells were pulsed in the presence of TOTO-1 labeled plasmid DNA (pEGFP-C1) with the following parameters: 10 pulses of 5 ms at 0.4 kV/cm and 1 Hz. (a,c,e,g) control cells, (b,d,f,h) latrunculin B treated cells (0.1 µmol/l, 37 °C, 1 hour before the electric field application) (in both conditions, 34 cells were visualized), (i) number of aggregates counted as a function of the fluorescence intensity in control (gray) and treated cells (black) (number of aggregates counted n = 330 for the control cells and n = 154 for the treated cells), (i inset) mean fluorescence intensity (mean + SEM). The black arrow on the right side indicates the direction of the electric field. Bar = 20 µm. CHO, Chinese hamster ovary; EGFP, enhanced green fluorescence protein.
Effect of actin disruption on gene expression
Subsequent to internalization into the cell, the DNA has to move through the cytoplasm to reach the nucleus for gene expression to occur. In order to investigate whether the actin cytoskeleton had an effect on the DNA intracellular movement, we measured EGFP reporter gene expression on cells which were treated with latrunculin B. We evaluated both the percentage of fluorescent cells (i.e., percentage of EGFP transfected cells) and the mean fluorescence intensity (i.e., the transfection efficiency). Latrunculin B incubations were performed 1 hour before application of the electric field in the presence of plasmid DNA (L1hT on Figure 6). The objective was to disturb the DNA/membrane interaction step as in the previous experiment (Figure 5b,d,f,h). Treatments were also performed 1 hour and 24 hours after the electric field application (TL1h and TL24h respectively on Figure 6) with the aim to potentially disturb the translocation and the transport of the DNA through the cytoplasm. The concentration of latrunculin B was chosen to be 0.1 µmol/l (Figure 4c,d). Three different control experiments without any drug treatment were additionally performed. The first control corresponded to cells not exposed to an electric field (NT on Figure 6), the second to cells exposed to an electric field (E on Figure 6) and the third one corresponded to cells exposed to electric pulses in the presence of plasmid DNA (T on Figure 6). This last control was used as a reference, all the data being normalized in relation to it.
Figure 6.
Gene expression in CHO cells treated with latrunculin B before or after electric field application. The cells were transfected with pEGFP-C1 plasmid DNA with the following parameters: 10 pulses of 5 ms at 0.4 kV/cm and 1 Hz and analyzed by flow cytometry. (a) Percentage of transfected cells, (b) transfection efficiency both normalized in relation to T. Experiments were performed in triplicate and repeated seven times (mean + SEM). NT, nontreated and nontransfected cells; E, cells exposed to electric field but without DNA; T, nontreated but transfected cells (our reference); L1hT, latrunculin B incubation performed for 1 hour before the transfection; TL1h/TL24h, latrunculin B incubation performed for 1 hour /24 hours after the transfection; CHO, Chinese hamster ovary; EGFP, enhanced green fluorescence protein.
In the case of pretreatment with latrunculin B (L1hT), both the transfection level and efficiency decreased. We measured a decrease of 30% and 40%, respectively, compared to the reference cells (T) (Figure 6), indicating that the actin network alteration before the electric field application had an effect on gene expression. For the latrunculin B treatments performed after the DNA electrotransfer (TL1h and TL24h), the percentage of transfected cells was the same as for the reference cells (Figure 6a). At the moment of application of the electric pulses, the control cells and the post-treated cells were exactly in the same state. However, the transfection efficiency decreased by about 20% for the 1 hour post-treated cells (TL1h) and increased by about 40% for the 24 hours post-treated (TL24h) (Figure 6b). These results show that the actin cytoskeleton has an influence on the gene expression. In the first step(s) of DNA transfer, the actin cytoskeleton seems to be necessary for optimal DNA expression, whereas later on, interactions with the actin network appear to be a limiting factor in the plasmid DNA transfer process.
Discussion
A primary limitation of gene therapy, specifically nonviral gene therapy, is the difficulty in achieving high levels of gene expression. The plasma membrane and nuclear envelope as well as the dense cytoplasmic meshwork are all obstacles to DNA transfer which must be overcome by the DNA in order to reach the nucleus and to be transcribed. A number of studies have been devoted to understanding the mechanisms by which the DNA overcomes the barriers presented by the plasma membrane and nuclear envelope, while processes occurring in the cytoplasm are only recently being investigated. Our study has focused on the actin cytoskeleton and its implication in the mechanism of gene delivery mediated by electric field.
Our results show that when DNA was present during the electric field application, and only in that case, the formation of actin patches at the plasma membrane occured. Their characteristics were similar to the ones of the DNA aggregates.19 They had the same location (plasma membrane facing the negative electrode), the same persistence (10 minutes after application of pulses), the same number (7–8 per cell), and comparable size. These observations were made using two different labeling strategies (phalloidin-conjugated dye and EGFP-actin transient expression). The actin patches colocalized with the DNA aggregates at least at the plasma membrane level facing the negative electrode. The observation of the EGFP-actin CHO cells gave information about the lifetime of these patches and their future in cells. They became detectable at the earliest 3 minutes after the application of the electric field and stayed localized at the plasma membrane until 45 minutes. The observed colocalization and the similarities in the behavior of actin patches and DNA aggregates lead us to suggest that actin is actively involved in the process of DNA entry and transport in the cytoplasm. It is the first time in the case of gene transfer mediated by electric fields that a correlation between changes in the actin cytoskeleton and DNA electrotransfer is seen.
Disruption of the actin network led to a significant decrease of the DNA accumulation at the plasma membrane. It did not change the way the DNA interacts with the membrane i.e., the formation of aggregates. Despite the fact that actin does not appear to play a role in the initial formation of the DNA/membrane complexes, it could play an important role in the quantity of DNA interacting with the plasma membrane. The decrease in accumulation of DNA at the membrane level when the actin network is altered could mean that the actin cytoskeleton participates in the transition from the low to the high stability complexes. It has been suggested that the DNA is trapped in the membrane during the application of the electric field19,22 but it remains accessible until 5 minutes after the last electric pulse.27 Indeed, the DNA is sensitive to DNAse action in the few minutes following the electropermeabilization indicating access to external molecules. The DNA is protected later on, at a time corresponding to the end of the permeabilized state.28 This time coincides with the appearance of the actin patches. In addition, it was shown that the lifetime of the permeable state is dependent on the cytoskeleton.29 It seems that there is a correlation between the permeable state of the membrane, the enclosing of the DNA, and the actin polymerization at the membrane. It is possible that the DNA trapping and thus DNA accumulation are actin-dependent even if in this time range no actin polymerization was detectable. The actin polymerization in patches colocalizing with the DNA was observable after 3 minutes. These observations suggest that the DNA may be enclosed in structures where actin polymerization occurs. During the time in which these structures are formed, the membrane is permeable and the DNA remains accessible.
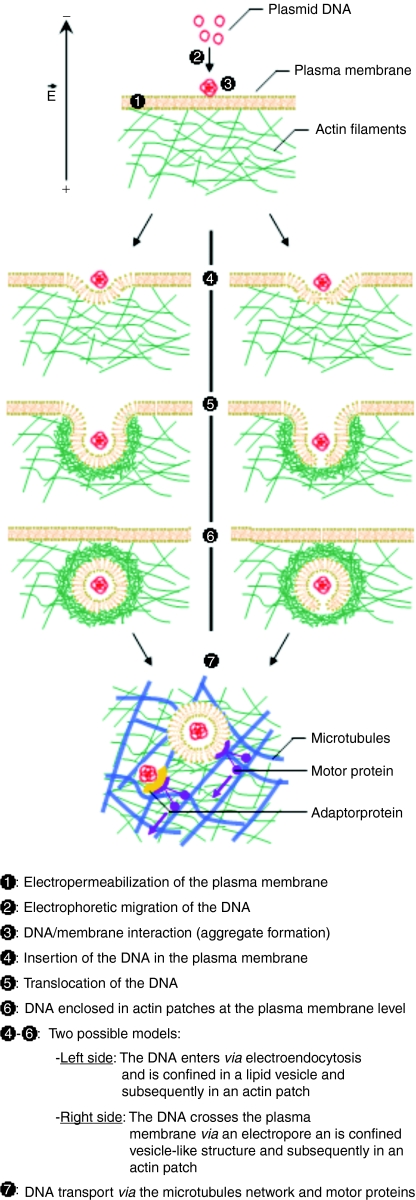
The way the DNA crosses the plasma membrane is still unknown. It is possible an endocytotic process could be occurring (Figure 7, left side). Actin has been shown to be important in a number of modes of exogenous molecular uptake (pinocytosis, phagocytosis, and macropinocytosis) and is also expected to play a role in clathrin or caveolin independent endocytosis.30 The budding structure from the plasma membrane is a prerequisite for any endocytotic pathway.31 All types of endocytosis require the involvement of actin for both the budding step and the early stage of the endosomal transport. The time needed for the internalization step is in agreement with what we have observed.32,33 Electroendocytotic processes have already been proposed by other research groups.16,34,35,36 Membrane actin polymerization is known to occur when a high amount of phosphatidylinositol 4,5-bisphosphate, which is highly negatively charged, is present in the membrane.37 Phosphatidylinositol 4,5-bisphosphate recruits dynamin proteins which polymerize at areas with high membrane curvature.38 Dynamin subsequently initiates the actin polymerization. It is possible that the high local density of negative charge in the DNA aggregate could trigger a similar response from the actin network. The electrophoretically driven insertion of the DNA into the membrane could pull the plasma membrane and initiate a lipid vesicle (Figure 7, left side, steps 4, 5). As it is the case for extracellular pathogens, membrane invaginations could also form due to the DNA aggregates, without any assistance from the cell machinery.39 Thus, binding to the membrane and a subsequent connection to the actin network could be a very general means for particle engulfment and transport which could be exploited by both pathogens (bacteria, viruses) and nonviral vectors (Figure 7, step 6). The second model suggested to explain molecular uptake mediated by the electric field is internalization through electropores, resulting in direct access to the cytoplasm (Figure 7, right side).5,7,18 In addition to the fact that pores were never observable, one of the limits of this model is that molecules above 4 kDa do not show any diffusion motion through the plasma membrane.7 The DNA interacts during tens of minutes with the membrane and moreover forms aggregates whose size was evaluated to be between 100 and 500 nm.19 This is much higher than the pore size estimation of 1–10 nm.7 However, it remains possible that the DNA accumulates where pores are formed and its electrophoretically driven insertion in the membrane pulls the pore (Figure 7, right side steps 4, 5).16 The vesicle-like structure formed could be recognized as an endocytotic vesicle and induce a similar response from the cell as for an endocytotic process, with the recruitment of dynamin at the neck of the vesicle-like structure and as a consequence polymerization of actin in a patch around it (Figure 7, step 6).
Figure 7.
DNA electrotransfer possible mechanisms. It is a multistep process. During application of the electric pulses, the plasma membrane is permeabilized (step 1), the DNA, because of its negative charge, electrophoretically migrates toward the positive electrode (step 2). It interacts with the electropermeabilized membrane facing the negative electrode and forms aggregates (step 3). The electrophoretically driven insertion (step 4) and the translocation (step 5) of the DNA through the plasma membrane are not yet known but two models can be proposed. The DNA could pull the membrane during its insertion and initiate the formation of a vesicle; we can in this case speak about electroendocytosis (left side), or the DNA could be inserted where an electropore is formed, pull it, and initiate the formation of a vesicle-like structure (right side). The plasma membrane is in interaction with the actin network thanks to connecting proteins. The presence of vesicle or vesicle-like structure may induce the recruitment actin associated proteins and initiate its polymerization where the lipid–DNA complex is located (step 5 and 6). Other data indicate that DNA migration in the cytoplasm might occur via motor proteins interacting with the microtubule network (step 7).24,25 The DNA could either be enclosed in the lipid vesicle or naked, and interacting with protein(s) providing the connection between it and the microtubule network. Subsequent steps are the crossing of the nuclear envelope and the expression in protein of the DNA sequence (not represented in the illustration here).
Disruption of the actin network leads to a lower gene expression when 1 hour treatments were performed before or after electric field application. The lower expression level for the 1 hour pretreatment can be explained by the lower accumulation of DNA observed at the membrane. Indeed, if less DNA interacts with the plasma membrane, less DNA enters/migrates within the cell to the nucleus and less DNA is available for expression. Ondrej et al.40 did similar experiments on fibroblasts transfected via lipoplex formation. After transfection, they performed latrunculin B treatment using higher concentrations and counted much lower numbers of plasmid copies in the nucleus. The lower expression for the 1 hour post-treatment suggested that the internalization and early transport of the DNA is actin-dependant. This could arise from an eventual disturbance of actin patch formation by the presence of latrunculin B. Surprisingly, the expression level was higher when the treatment was performed for 24 hours. We propose that the dense actin network, while being useful at earlier stages of transfection (interaction with the membrane, internalization, early transport), could restrain the later stage of the DNA transport/diffusion. Verkman and colleagues have shown that DNA fragments longer than 2,000 base pairs showed no translational diffusion through the cytoplasm.23 More recent work from this group has shown that the actin cytoskeleton is the principal structure that limits this passive diffusion through the cytoplasm and that disruption of the actin cytoskeleton by drugs results in an increase in the diffusion of large macromolecules or linear DNA fragments.41 Our findings can thus be interpreted by an endocytosis-like phenomenon which explains the entrance and trafficking of electrotransfered plasmid DNA within the cell, with the actin cytoskeleton being involved in the membrane invagination and the early stage of the transport. Further long range transport takes place via the tubulin network.30,42,43 Vaughan et al. have previously shown that a stabilization of the microtubule network enhances electrotransfered plasmid expression.24,25 They also demonstrated the ability of plasmid DNA to interact with the microtubules via other proteins, which suggests an active movement along the microtubule network. Our results are in agreement with their findings, that a disruption of the actin network may make the movement of the DNA easier by reducing the crowding around it (Figure 7, step7). While it has been previously shown that electrotransfered plasmids move along the microtubule network and likely use dynein as the molecular motor that facilitates movement toward the nucleus,24 our work sheds light on the involvement of actin in this process.
In conclusion, we have shown that the actin cytoskeleton plays an important role in the gene transfer process mediated by the electric field in CHO cells. While it appears to be actively involved in the success of the first transfer step(s), it seems that it hinders DNA motion at later stages. If the actin cytoskeleton has a role in an active transport process, it should have it only at the early stage. We propose that the DNA, after being electrophoretically pushed to the membrane, accumulates in sites surrounded by actin anchored in the plasma membrane. The actin could polymerize around the DNA aggregates as a consequence of an endocytosis-like process. This involvement of actin in the crossing and/or transport of plasmid DNA is observed in gene delivery methods mediated by endocytosis such as liposome-mediated transfer.40,44 Certain bacterial pathogens and many viruses use the actin cytoskeleton and its associated motor proteins for intracellular movement.45,46,47,48,49 We will continue to investigate the role of the actin cytoskeleton and also the role of the microtubules in the intracellular trafficking of the DNA. In particular, the use of single particle tracking may help in characterizing the behavior of the DNA while in the cytoplasm, and confirm our hypothesis of the active involvement of the cytoskeleton in the migration of the DNA through the cytoplasm.
Materials and Methods
Cells. The wild-type Chinese hamster ovary clone (Toronto strain) was selected for its ability to grow in suspension or plated on petri dishes or on 22 × 22 mm microscope glass coverslips (Lab-Tek II; Nunc, Roskilde, Denmark). They were grown in Eagle's minimum essential medium (Eurobio, Les Ulis, France) supplemented with 8% heat inactivated fetal bovine serum (Gibco-Invitrogen, Carlsbad, CA), 0.584 g/l -glutamine (Gibco), 3.5 g/l -glucose (Sigma, St Louis, MO), 2.95 g/l tryptose-phosphate (Sigma), 100 U/ml penicillin (Gibco), 100 mg/l streptomycin (Gibco), and BME vitamins (Sigma).19
Electropulsation apparatus. Electropulsation was operated by using a CNRS cell electropulsator (Jouan, St Herblain, France), which delivered square-wave electric pulses. An oscilloscope (Enertec, St Etienne, France) monitored the pulse shape. Two stainless-steel parallel plate electrodes (length = 10 mm, interelectrode distance = 4 or 10 mm), connected to the voltage generator, gave a uniform electric field.
Electropulsation procedures. Cells in suspension were washed, resuspended in 100 µl plasmid-containing pulsation buffer (10 mmol/l K2HPO4/KH2PO4 pH 7.4, 1 mmol/l MgCl2, 250 mmol/l sucrose) and placed between the electrodes with a 4 mm gap. The electric field parameters were 10 pulses of 5 ms duration, 0.7 kV/cm intensity and 1 Hz frequency at room temperature. The electrotransfer of genes into CHO cells is optimal under these conditions.19 For plated cells, the culture medium was replaced by the same buffer described above. To allow for the immersion of the electrodes, the volume of the pulsation buffer was raised to 300 µl containing 1.25 µg plasmid DNA. Identical electric field conditions were used except for the intensity which was reduced to 0.4 kV/cm and the electrode gap which was 10 mm.19 In both cases, cells were maintained in the pulsation buffer for a few minutes after application of the electric pulses to allow for the return to the impermeable state.
DNA staining. A 4.7 kb plasmid (pEGFP-C1, Clontech, Palo Alto, CA) carrying the EGFP gene controlled by the cytomegalovirus promoter was stained stoichiometrically with DNA intercalating dyes TOTO-1 or POPO-3 (Molecular Probes-Invitrogen, Carlsbad, CA).19 The plasmid was stained with 7.6 × 10−5 mol/l dye at a DNA concentration of 1 µg/µl for 60 minutes on ice. This concentration yields an average base pair to dye ratio of 20. The fluorescence intensity detected is directly related to the amount of DNA. Plasmids were prepared from transformed Escherichia coli by using the Maxiprep DNA purification system according to manufacturer instructions (Qiagen kit, Qiagen, Chatsworth, CA).
Actin staining. The following protocol is provided by Molecular Probes. The cells, cultured in 24-well plates, were fixed with 4% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS) for 10 minutes at room temperature and permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 5 minutes. The cells were incubated with 1% BSA (Sigma) in PBS for 30 minutes. The phalloidin-rhodamine 123 (Molecular Probes) was added to the cells for 20 minutes (1U in 200 µl PBS per well) in the dark. The phalloidin-rhodamine 123 specifically stains filamentous actin. Between each step, the cells were washed two times with PBS containing Ca2+ and Mg2+. After drying, coverslips were mounted on a glass slide with mowiol and observed by confocal microscopy. The excitation and emission wavelengths of phalloidin-rhodamine 123 are 540 nm and 565 nm respectively.
EGFP-actin transient expression. The cells were suspended in the pulsation buffer (5 × 105 cells in 100 µl) containing 2 µg of pEGFP-actin (Clontech) and electrotransfected with parameters defined for the cell suspensions (see electropulsation procedures) 24–48 hours before use.
Latrunculin B treatment. The latrunculin B (Molecular Probes) incubations were made at 0.1 µmol/l at 37 °C for 1 hour before or 1 hour and 24 hours after the application of the electric pulses.50 The latrunculin B was diluted in dimethyl sulfoxide and added to the culture medium of cells plated 24 hours prior.
Gene expression procedure. The cells were cultured on plastic coverslips (Melinex; Agar scientific, Essex, UK) cut in relation to the size of the electrodes (10 × 10 mm) and deposed in 24-well plates. One 50 µl drop containing 25 × 103 cells in the culture medium was deposited in the centre of the coverslip and spread out with the pipette tip. After adhesion of the cells (around 20 minutes at 37 °C), 1 ml culture medium was added and the cells were cultured for 24 hours. The electrotransfection with pEGFP-C1 was performed in 200 µl pulsation buffer containing 2 µg plasmid. The parameters were those defined for plated cells (see electropulsation procedures). After 24 hours, the cells were washed, harvested with trypsin-EDTA (5 minutes, 37 °C) and suspended in 300 µl PBS. The samples were analyzed by flow cytometry (FACScan) via the channel FL1 (510 nm ≤ Λem ≤ 540 nm) to evaluate both the percentage of fluorescent cells (i.e., percentage of EGFP transfected cells) and the mean level of fluorescence associated (i.e., the efficiency of transfection). All the values were normalized in relation to the control cells which were transfected but not treated with latrunculin B.
Cell viability. Cell viability was determined by the ability of cells to grow and divide over a 24 hours period (corresponding to more than one generation). Viability was measured by monitoring cell growth trough a coloration method with crystal violet. The cells were incubated with 0.1% crystal violet in PBS for 20 minutes at room temperature and lysed with 10% acetic acid for 10 minutes at room temperature. 100 µl were diluted in 2 ml PBS and the optical density was measured at 595 nm. The optical density is proportional to the amount of cells and the measurements were normalized in relation to the control cells which were transfected but not treated with latrunculin B.21
Wide field microscopy. Cells were observed with a Leica ×100, 1.3 numerical aperture oil immersion objective mounted in a Leica DMIRB inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany). The wavelengths were selected by using the Leica L4 filter block (450 nm ≤ Λex ≤ 490 nm; dichroic mirror 500 nm, 500 nm ≤ Λem ≤ 550 nm) for the TOTO-1-labeled DNA and the EGFP-actin fusion protein and the Leica N2.1 filter block (515 nm ≤ Λex ≤ 560 nm; dichroic mirror cutoff 580 nm, 580 nm ≤ Λem) for the POPO-3-labeled DNA. Images were recorded and analyzed with the MetaVue Imaging System (Molecular Devices, MDS Analytical Technologies, Downingtown, PA) fitted with a cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ). This digitizing set-up allowed quantitative localized analysis of the fluorescence emission. This was performed along the cell membrane. Plot histograms detected local fluorescence intensity increase above the background level outside of the cells which is directly related to the number of fluorescent molecules locally present.
Confocal microscopy. Cells were observed with a Leica ×100, 1.4 numerical aperture oil immersion objective mounted in a Leica DMRE upright microscope equipped with the Leica TCS SP2 confocal system and monitored using the Leica Confocal Software. A 543 nm laser was used to excite the rhodamine 123 fluorophore and a suitable region of the emission spectrum was selected.
Statistical analysis. Errors bars represent the SE of the mean (SEM). The statistical significance of differences between fluorescence intensity of DNA aggregates in the control and the latrunculin B treated cells was evaluated using a paired Student's t-test. The statistical significance of differences in gene expression efficiency (percentage of transfected cells and their mean fluorescence level) between the control and the latrunculin B treated cells was evaluated by the nonparametric Mann–Whitney–Wilcoxon test. The degree of significance is given with these following labels: NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
SUPPLEMENTARY MATERIAL Video S1. z-stack projection of a CHO cell 10 minutes after the electric field exposure in the presence of plasmid DNA. Video S2. Time series of a CHO cell, expressing EGFP-actin, after application of the electric field in the presence of plasmid DNA.
Acknowledgments
We thank Favard Cyril, Elisabeth Bellard, Muriel Golzio, and Justin Teissié for useful discussions on this work. We acknowledge financial support from the Association Française contre les Myopathies (to M.-P. R.) and the Ministère des Affaires Etrangères et Européennes (programme PHC PROCOPE 2010–2011).
Supplementary Material
z-stack projection of a CHO cell 10 minutes after the electric field exposure in the presence of plasmid DNA.
Time series of a CHO cell, expressing EGFP-actin, after application of the electric field in the presence of plasmid DNA.
REFERENCES
- Young LS, Searle PF, Onion D., and, Mautner V. Viral gene therapy strategies: from basic science to clinical application. J Pathol. 2006;208:299–318. doi: 10.1002/path.1896. [DOI] [PubMed] [Google Scholar]
- Deshayes S, Morris MC, Divita G., and, Heitz F. Cell-penetrating peptides: tools for intracellular delivery of therapeutics. Cell Mol Life Sci. 2005;62:1839–1849. doi: 10.1007/s00018-005-5109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali PP., and, Chaudhuri A. Cationic liposomes as non-viral carriers of gene medicines: resolved issues, open questions, and future promises. Med Res Rev. 2007;27:696–722. doi: 10.1002/med.20090. [DOI] [PubMed] [Google Scholar]
- Newman CM., and, Bettinger T. Gene therapy progress and prospects: ultrasound for gene transfer. Gene Ther. 2007;14:465–475. doi: 10.1038/sj.gt.3302925. [DOI] [PubMed] [Google Scholar]
- Neumann E, Schaefer-Ridder M, Wang Y., and, Hofschneider PH. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982;1:841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio M, Rols MP., and, Teissié J. In vitro and in vivo electric field-mediated permeabilization, gene transfer, and expression. Methods. 2004;33:126–135. doi: 10.1016/j.ymeth.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Escoffre JM, Portet T, Wasungu L, Teissié J, Dean D., and, Rols MP. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol Biotechnol. 2009;41:286–295. doi: 10.1007/s12033-008-9121-0. [DOI] [PubMed] [Google Scholar]
- Neumann E., and, Rosenheck K. Permeability changes induced by electric impulses in vesicular membranes. J Membr Biol. 1972;10:279–290. doi: 10.1007/BF01867861. [DOI] [PubMed] [Google Scholar]
- Escoffre JM, Dean DS, Hubert M, Rols MP., and, Favard C. Membrane perturbation by an external electric field: a mechanism to permit molecular uptake. Eur Biophys J. 2007;36:973–983. doi: 10.1007/s00249-007-0194-7. [DOI] [PubMed] [Google Scholar]
- Gehl J. Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol Scand. 2003;177:437–447. doi: 10.1046/j.1365-201X.2003.01093.x. [DOI] [PubMed] [Google Scholar]
- Mir LM, Gehl J, Sersa G, Collins CG, Garbay JR, Billard V.et al. (2006Standard operating procedures of the electrochemotherapy: Instructions for the use of bleomycin or cisplatin administered either systemically or locally and electric pulses delivered by the Cliniporator (TM) by means of invasive or non-invasive electrodes Ejc Suppl 414–25. [Google Scholar]
- André F., and, Mir LM. DNA electrotransfer: its principles and an updated review of its therapeutic applications. Gene Ther. 2004;11 Suppl 1:S33–S42. doi: 10.1038/sj.gt.3302367. [DOI] [PubMed] [Google Scholar]
- Daud AI, DeConti RC, Andrews S, Urbas P, Riker AI, Sondak VK.et al. (2008Phase I trial of interleukin-12 plasmid electroporation in patients with metastatic melanoma J Clin Oncol 265896–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low L, Mander A, McCann K, Dearnaley D, Tjelle T, Mathiesen I.et al. (2009DNA vaccination with electroporation induces increased antibody responses in patients with prostate cancer Hum Gene Ther 201269–1278. [DOI] [PubMed] [Google Scholar]
- Hristova NI, Tsoneva I., and, Neumann E. Sphingosine-mediated electroporative DNA transfer through lipid bilayers. FEBS Lett. 1997;415:81–86. doi: 10.1016/s0014-5793(97)01097-1. [DOI] [PubMed] [Google Scholar]
- Klenchin VA, Sukharev SI, Serov SM, Chernomordik LV., and, Chizmadzhev YuA. Electrically induced DNA uptake by cells is a fast process involving DNA electrophoresis. Biophys J. 1991;60:804–811. doi: 10.1016/S0006-3495(91)82115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie TD., and, Tsong TY. Study of mechanisms of electric field-induced DNA transfection. V. Effects of DNA topology on surface binding, cell uptake, expression, and integration into host chromosomes of DNA in the mammalian cell. Biophys J. 1993;65:1684–1689. doi: 10.1016/S0006-3495(93)81208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gennes PG. Passive entry of a DNA molecule into a small pore. Proc Natl Acad Sci USA. 1999;96:7262–7264. doi: 10.1073/pnas.96.13.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio M, Teissie J., and, Rols MP. Direct visualization at the single-cell level of electrically mediated gene delivery. Proc Natl Acad Sci USA. 2002;99:1292–1297. doi: 10.1073/pnas.022646499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoffre JM, Portet T, Favard C, Teissié J, Dean DS., and, Rols MP.2010Electromediated formation of DNA complexes with cell membranes and its consequences for gene delivery Biochim Biophys Actaepub ahead of print). [DOI] [PubMed]
- Golzio M, Mora MP, Raynaud C, Delteil C, Teissié J., and, Rols MP. Control by osmotic pressure of voltage-induced permeabilization and gene transfer in mammalian cells. Biophys J. 1998;74:3015–3022. doi: 10.1016/S0006-3495(98)78009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurie C, Rebersek M, Golzio M, Kanduser M, Escoffre JM, Pavlin M.et al. (2010Electro-mediated gene transfer and expression are controlled by the life-time of DNA/membrane complex formation J Gene Med 12117–125. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N., and, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- Vaughan EE., and, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol Ther. 2006;13:422–428. doi: 10.1016/j.ymthe.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan EE, Geiger RC, Miller AM, Loh-Marley PL, Suzuki T, Miyata N.et al. (2008Microtubule acetylation through HDAC6 inhibition results in increased transfection efficiency Mol Ther 161841–1847. [DOI] [PubMed] [Google Scholar]
- Verderame M, Alcorta D, Egnor M, Smith K., and, Pollack R. Cytoskeletal F-actin patterns quantitated with fluorescein isothiocyanate-phalloidin in normal and transformed cells. Proc Natl Acad Sci USA. 1980;77:6624–6628. doi: 10.1073/pnas.77.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eynard N, Rols MP, Ganeva V, Galutzov B, Sabri N, Teissie J. Electrotransformation pathways of prokaryotic and eukaryotic cells: recent developments. Bioelectrochem Bioenerg. 1997;44:103–110. [Google Scholar]
- Rols MP, Dahhou F, Mishra KP., and, Teissié J. Control of electric field induced cell membrane permeabilization by membrane order. Biochemistry. 1990;29:2960–2966. doi: 10.1021/bi00464a011. [DOI] [PubMed] [Google Scholar]
- Teissie J., and, Rols MP. Manipulation of cell cytoskeleton affects the lifetime of cell membrane electropermeabilization. Ann N Y Acad Sci. 1994;720:98–110. doi: 10.1111/j.1749-6632.1994.tb30438.x. [DOI] [PubMed] [Google Scholar]
- Soldati T., and, Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Doherty GJ., and, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Conner SD., and, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Bellve KD, Leonard D, Standley C, Lifshitz LM, Tuft RA, Hayakawa A.et al. (2006Plasma membrane domains specialized for clathrin-mediated endocytosis in primary cells J Biol Chem 28116139–16146. [DOI] [PubMed] [Google Scholar]
- Rols MP, Femenia P., and, Teissie J. Long-lived macropinocytosis takes place in electropermeabilized mammalian cells. Biochem Biophys Res Commun. 1995;208:26–35. doi: 10.1006/bbrc.1995.1300. [DOI] [PubMed] [Google Scholar]
- Satkauskas S, Bureau MF, Mahfoudi A., and, Mir LM. Slow accumulation of plasmid in muscle cells: supporting evidence for a mechanism of DNA uptake by receptor-mediated endocytosis. Mol Ther. 2001;4:317–323. doi: 10.1006/mthe.2001.0465. [DOI] [PubMed] [Google Scholar]
- Antov Y, Barbul A, Mantsur H., and, Korenstein R. Electroendocytosis: exposure of cells to pulsed low electric fields enhances adsorption and uptake of macromolecules. Biophys J. 2005;88:2206–2223. doi: 10.1529/biophysj.104.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5:463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P.et al. (2010Membrane curvature controls dynamin polymerization Proc Natl Acad Sci USA 1074141–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D.et al. (2007Shiga toxin induces tubular membrane invaginations for its uptake into cells Nature 450670–675. [DOI] [PubMed] [Google Scholar]
- Ondrej V, Lukásová E, Falk M., and, Kozubek S. The role of actin and microtubule networks in plasmid DNA intracellular trafficking. Acta Biochim Pol. 2007;54:657–663. [PubMed] [Google Scholar]
- Dauty E., and, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm: a new barrier for non-viral gene delivery. J Biol Chem. 2005;280:7823–7828. doi: 10.1074/jbc.M412374200. [DOI] [PubMed] [Google Scholar]
- Ross JL, Ali MY., and, Warshaw DM. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA. Coupling actin dynamics and membrane dynamics during endocytosis. Curr Opin Cell Biol. 2002;14:76–81. doi: 10.1016/s0955-0674(01)00297-6. [DOI] [PubMed] [Google Scholar]
- Bausinger R, von Gersdorff K, Braeckmans K, Ogris M, Wagner E, Bräuchle C.et al. (2006The transport of nanosized gene carriers unraveled by live-cell imaging Angew Chem Int Ed Engl 451568–1572. [DOI] [PubMed] [Google Scholar]
- Gouin E, Welch MD., and, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M.et al. (2002Visualization of the intracellular behavior of HIV in living cells J Cell Biol 159441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest T, Barnard S., and, Baines JD. Active intranuclear movement of herpesvirus capsids. Nat Cell Biol. 2005;7:429–431. doi: 10.1038/ncb1243. [DOI] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP., and, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Ewers H, Rajamäki ML, Day PM, Schiller JT., and, Helenius A. Human papillomavirus type 16 entry: retrograde cell surface transport along actin-rich protrusions. PLoS Pathog. 2008;4:e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Northup N, Marga F, Huber T, Byfield FJ, Levitan I.et al. (2007The effect of cellular cholesterol on membrane-cytoskeleton adhesion J Cell Sci 120Pt 132223–2231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
z-stack projection of a CHO cell 10 minutes after the electric field exposure in the presence of plasmid DNA.
Time series of a CHO cell, expressing EGFP-actin, after application of the electric field in the presence of plasmid DNA.