Abstract
Our concept of biological membranes has markedly changed, from the fluid mosaic model to the current model that lipids and proteins have the ability to separate into microdomains, differing in their protein and lipid compositions. Since the breakthrough in crystallizing membrane proteins, the most powerful method to define lipid-binding sites on proteins has been X-ray and electron crystallography. More recently, chemical biology approaches have been developed to analyze protein–lipid interactions. Such methods have the advantage of providing highly specific cellular probes. With the advent of novel tools to study functions of individual lipid species in membranes together with structural analysis and simulations at the atomistic resolution, a growing number of specific protein–lipid complexes are defined and their functions explored. In the present article, we discuss the various modes of intramembrane protein–lipid interactions in cellular membranes, including examples for both annular and nonannular bound lipids. Furthermore, we will discuss possible functional roles of such specific protein–lipid interactions as well as roles of lipids as chaperones in protein folding and transport.
Lipids can act as a glue that holds oligomeric membrane proteins together. They may also act as chaperones that facilitate folding and insertion of these proteins and ensure they adopt the correct topology.
Our concept of biological membranes has markedly changed in the last two decades, from the fluid mosaic model (Singer and Nicolson 1972), in which the membrane was thought to be formed by a homogenous lipid fluid phase with proteins embedded, to the current model that lipids and proteins are not homogenously distributed, but have the ability to separate into microdomains, differing in their protein and lipid compositions. A well established example of domains are lipid rafts (see Box 1 for definitions). Raft domains are described as dynamic domain structures enriched in cholesterol, sphingolipids, and membrane proteins (Brown and London 1998; Simons and Ikonen 1997) that have an important role in different cellular processes (Lingwood and Simons 2010). Formation of domains within cellular membranes has been extensively investigated over the past years leading to various models that differ in the primary forces involved in the formation and the recruitment of surrounding membrane components into such domains.
BOX 1. Definitions.
Annular Lipids/Lipid Shell
An annular lipid shell is formed when selected lipid classes or molecular species bind preferentially to the hydrophobic and/or hydrophilic surfaces of a membrane protein. Per definition these lipids show markedly reduced residence times at the protein–lipid interface as compared to bulk lipids.
Bulk Lipids
Lipids within the membrane that diffuse rapidly in the bilayer plane and show a low residence time at the protein–lipid interface following random collisions. Typical diffusion coefficients for bulk lipids in a liquid disordered phase are in the range of DL = 7×10−12 m2/sec (DOPC) (Filippov et al. 2003).
Hydrophobic Mismatch
A term to describe any deviation from the compatibility of the hydrophobic surface of membrane proteins (their TMDs) to the vertically and laterally encountered hydrophobic surfaces of the lipid bilayer in biological membranes. In the case of a hydrophobic mismatch, the resulting energy penalty may cause the recruitment of a suitable local lipid environment, the deformation of the membrane and/or in conformational changes of the protein to achieve a status of hydrophobic match (for advanced reading, see Killian 1998).
Lateral Pressure Field/Profile of Membranes
Biological membranes can be considered as the “solvent” for membrane proteins that are embedded in them. The lateral pressure profile (Ω(z)) describes the force or pressure that is exerted by the membrane on the matter residing inside it. This pressure is modulated by different extents of lipid–lipid interactions and asymmetries across and within the bilayer, which in turn results in varying lateral pressures that may locally correspond to several hundreds of atmospheres.
Lipid Rafts
Sterol and sphingolipid-dependent microdomains that form a network of lipid–lipid, protein–protein, and protein–lipid interactions; involved in the compartmentalization of processes such as signaling within biological membranes.
Liquid-Disordered Phase (Id)
A predominantly fluid phase of lipids, characterized by a high degree of mobility (cis-gauche flexibility of acyl chains; lateral diffusion) and a high content of short and/or unsaturated fatty acyl chains.
Liquid-Ordered Phase (Io)
A liquid crystalline phase (that displays physical properties of both liquids and of solid crystals), characterized by a high degree of acyl chain order (“packing”), a reduced lateral mobility of lipid and protein molecules, and a reduction in the elasticity of the membrane as a result of specific interactions between sterols and phospholipids containing long, saturated acyl chains and/or glycosphingolipids.
Microdomains
Membrane compartments of distinct lipid and protein composition that may modulate the enzymatic functions of membrane proteins.
Molecular Lipid Species
Individual members of a lipid class that differ in their fatty acid composition.
Nonannular Lipids
Lipids that specifically interact with membrane proteins are neither bulk lipids, nor do they belong to the shell/annulus of lipids that surround the membrane protein. These nonannular lipids often reside within membrane protein complexes, in which they may fulfill diverse functions ranging from structural building blocks to allosteric effectors of enzymatic activity (see text). Nonannular lipids bind to distinct hydrophobic sites of membrane proteins or membrane protein complexes.
According to one model, membrane domains can form by specific protein–protein interactions (Douglass and Vale 2005). This model is based on single-molecule microscopy experiments. In these studies, single fluorophores were chemically attached to specific proteins, and the dynamics of individual proteins was tracked by monitoring the fluorescent probe. In this kind of set up, a dynamic behavior of lipids is not assessed. Here, proteins involved in signaling processes are trapped within interconnected microdomains created by specific protein–protein interactions, probably involving additional scaffolding proteins. The proteins of such domains can exchange with the surrounding membrane area at individual kinetics, some components are immobile over minutes, and others can diffuse rapidly.
Another model emphasizes the importance of lipid–lipid interactions, initiating the formation of subdomains of defined lipid compositions. Transmembrane proteins then can be attracted to such subdomains via various specific interactions with lipids. The resulting lipid–protein complexes then eventually coalesce to form larger lipid–protein assemblies (Anderson and Jacobson 2002).
The idea of lipid-dependent domain formation is inherent to the biophysical properties and therefore to the complex lipid composition of cellular membranes that include up to a thousand lipids that vary in structure (van Meer et al. 2008). This wide range of lipid species has been proposed to facilitate the “solvation” of membrane proteins. Taken into account the sum of lipid species present in a cellular membrane, it is important to understand the different interactions and affinities within the bilayer between different lipids. Molecular dynamics simulations have been successfully employed to investigate lipid interactions between different lipid species and found specific interactions of various lipid classes and molecular species (Hofsass et al. 2003; Niemela et al. 2004, 2006, 2009; Pandit et al. 2004; Zaraiskaya and Jeffrey 2005; Bhide et al. 2007). These results are supported and expanded by recent data from our group that suggest a specific order of interactions of sphingomyelin species with cholesterol in membranes (A.M. Ernst, F. Wieland, and B. Brügger, unpubl.). At low cholesterol concentrations, some sphingomyelin species preferentially interact with cholesterol, whereas others prefer their kin. At higher cholesterol concentrations, all sphingomyelin species investigated display an increased affinity for the sterol. These findings open the possibility of differentiated pathways of self-assembly of microdomains, dependent on molecular lipid species.
In the present article the various modes of intramembrane protein–lipid interactions in cellular membranes (Fig. 1) will be discussed. This includes possible functional roles of such specific protein–lipid interactions.
Figure 1.
Intramembrane protein–lipid interactions within a cell membrane. (A) Bulk lipids; (B) annular lipids; (C) nonannular lipids/lipid ligands. For details see text.
METHODS USED TO ANALYZE PROTEIN–LIPID INTERACTIONS
Crystallization of membrane proteins (Knapp et al. 1985) is one of the most powerful methods to define lipid-binding sites on proteins (Hunte and Richers 2008). Crystallography has the potential to revealing complete structural insight at atomic resolution, although in many cases lipid classes and molecular species cannot be determined unequivocally.
For structures in which crystals are not available, nuclear magnetic resonance (NMR) methods come into play, specifically solid state NMR of multiple protein/bilayer stacks that provides various modes to assess the orientation of a TMD toward lipids within the membrane. Current analytical methods to assess protein–lipid interactions are listed in Box 2, and explained below.
BOX 2. Selected Methods to Study Protein–Lipid Interactions.
X-Ray Crystallography
X-ray crystallography is the method of choice to obtain high-resolution structures of membrane proteins, with the number of protein structures at high resolution (<3 Å) rapidly increasing. However, crystals of membrane proteins are always obtained from detergent solutions, which do not reflect their native lipid environment. Lipid molecules present in protein–lipid assemblies might not survive the purification procedure, or if copurified and crystallized appear not sufficiently ordered in the crystal to allow for their unambiguous characterization. In many cases lipid polar groups are not resolved and the acyl chains show unusual conformations, raising the possibility of inadequate refinement.
Electron Crystallization
Electron crystallization is increasingly used to solve membrane protein structures with atomic resolution from two-dimensional (2D) crystals in a lipid environment (for recent reviews see (Raunser and Walz 2009; Reichow and Gonen 2009). 2D crystals (typically with a thickness of only one protein per array) can be produced of the purified protein in detergent by dialysis in the presence of lipids in a specific ratio to the protein, to allow membrane formation and 2D crystallization of the stabilized protein. This technique was successfully employed to solve the structure of bacteriorhodopsin at 3 Å resolution (Kimura et al. 1997; Mitsuoka et al. 1999) and of aquaporin at 1.9 Å resolution (Gonen et al. 2005), including individual lipid molecules. In the case of aquaporin a complete lipid shell around one aquaporin tetramer was resolved.
NMR Spectroscopy
NMR spectroscopy is an important analytical technique to obtain molecular structures in solution. However, transmembrane proteins in bilayers are not amenable to solution NMR. Technical improvements like magic-angle spinning (MAS) and cross polarization (CP) approaches allow to analyze transmembrane proteins by solid-state NMR with a resolution close to that obtained in solution NMR experiments. In contrast to solution NMR, solid state NMR can be employed to study molecules larger than 100 kDa (Tycko 2001).
Atomic Force Microscopy (AFM)
AFM provides a three-dimensional surface profile of the biological sample. Samples do not require any specific treatment. Notably, AFM can be performed conveniently in ambient air or in a liquid environment. Only few micrograms of sample are necessary. The lateral resolution is several nanometers, and the height resolution about 0.1 nm. High quality AFM profiles are taken at low time resolution (min), making dynamic processes less tractable. Unlike NMR or X-ray crystallography, AFM does not provide insight into changes down to the level of secondary protein structure.
IR Spectroscopy
The technique of FT-IR (Fourier Transform-Infrared spectroscopy) has the potential to measure lipid acyl chain configuration, phospholipid head group-ion interactions (Dluhy et al. 1983), and protein secondary structure in a single experiment. No extrinsic probe molecules are required that could perturb the properties of the system under investigation (Taylor and Smith 1981). Relatively small amounts of material (in the microgram range) may be examined in a variety of physical states, such as bilayer vesicles or monolayer films on an IR substrate or aqueous surface (Dluhy and Cornell 1985). Interpretation of IR data is often difficult because of water vapor that interferes with protein amide I and II bands, and hampered by the complexity of most biological samples.
Electron Spin Resonance (ESR) and Electron Paramagnetic Resonance (EPR)
With these methods one can, at high sensitivity and speed, estimate the number of lipids bound to the protein, and, for example, determine the residence time of one lipid in a protein–lipid complex. The method depends on large amounts of protein and on the introduction into the sample molecule of reporter groups (spin labels) that cannot report fine details of order and mobility gradients.
Differential Scanning Calorimetry (DSC)
DSC is a useful tool to study molecular interactions of membrane proteins, including formation of membrane domains and lipid hydrocarbon chain order. Basically, this technique monitors differences in energy required to maintain the sample and the reference at the same temperature. The protein of interest can be reconstituted at various molar concentrations into liposomes of a defined lipid composition, to monitor (e.g., phase transitions within the membrane or heat-denaturation of the protein). High amounts of material (lipid and protein), and long analysis times (∼30 min) are required.
Molecular Dynamics Simulations
In silico approaches are of increasing importance for analyzing the molecular mechanisms of protein–lipid interactions (Lindahl and Sansom 2008). The atomistic simulations of molecular mechanics used to be limited by the calculation times on the available computer systems. This has been partially overcome by the use of parallelized super computers and a variety of optimized simulation software, which enable simulations in atomistic details for systems. The simulation of molecular dynamics (MD) allows the analyses of the interactions of atoms and macromolecules for a short period of time by the established laws of physics, and can hence be considered as an animation of Newtonian mechanics. Unlike NMR and X-ray based approaches, the motions and interactions of proteins and lipids can be monitored in atomistic detail with high temporal and spatial resolution. To model even larger systems or to analyze longer simulation times, systems can be simplified and simulated at “coarse-grained” resolution. Coarse-graining refers to combining neighboring atoms into a single interaction site, leading to a significant reduction in the number of particles and interactions in the MD simulation, enabling the simulations of processes beyond 1 microsecond, although with a loss in molecular detail (Ding et al. 2003; Paci et al. 2002; Smith and Hall 2001). The major limitation in simulating the MD of macromolecules is the number of particles that need to be analyzed, with an increasing particle number resulting in exponentially increasing computational times. Although MD simulations have been successfully employed to study lipid–lipid and protein–lipid interactions (reviewed in Niemela et al. 2009), further improvements in the hardware of computer systems might be necessary to broaden the access to this approach.
More recently, chemical biology approaches are being developed to analyze protein–lipid interactions. Such methods have the advantage of providing highly specific cellular probes.
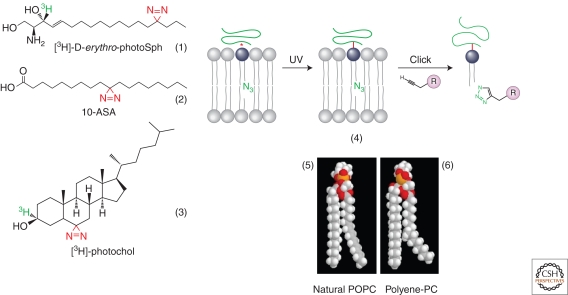
One method to analyze specificity of cellular intramembrane protein–lipid interactions is based on a combination of radioactive labeling with covalent photo-crosslinking. Tritium-labeled membrane lipids or precursors thereof that carry a photoactivatable group (e.g., see structures 1–3 in Fig. 2) are fed to cells, and their conversion into membrane lipids is monitored by thin layer chromatography and autoradiography. Lipids are photo-activated by irradiation of the cells, and after immunoprecipitation of the protein of interest, covalently cross-linked lipids are visualized by autoradiography after gel electrophoresis (Thiele et al. 2000; Haberkant et al. 2008).
Figure 2.
Novel lipid tools to study protein–lipid interactions. For details see text. (Structure 4 is adapted from Haberkant and van Meer [2009] and reprinted with permission from de Gruyter © 2009.)
A combination of covalent cross-linking by photo-labeling of one site of a lipid interacting with a protein, and labeling at another site for a sensitive readout of this interaction, has proven powerful. Here readout is based on introducing a click chemistry group that can then react with a second molecule, that is, biotin for detection with avidin. Such approaches are reviewed in Haberkant and van Meer (2009) and Moses and Moorhouse (2007), and typical chemistry is depicted in Figure 2 (scheme 4). For cellular labeling, such a bifunctional lipid is fed to cells, and, after photo-activation by irradiation to trigger covalent cross-linking, the cells are broken to perform the click reaction, with subsequent quantifying of the readout molecule.
Fluorescence quenching has been successfully used to assess in vitro intramembrane protein–lipid interactions (Leto et al. 1980; East and Lee 1982; Markello et al. 1985; Gonzalez-Manas et al. 1992; Powl et al. 2005). Halogenated (mainly bromylated) membrane phospholipids together with nonlabeled lipids are reconstituted with membrane proteins into proteoliposomes. Binding constants for transmembrane-lipid complexes are then deduced by fluorometrically monitoring resonance energy transfer from aromatic intramembrane amino acid residues (usually tryptophan residues) to the dark acceptor halogenide.
Another approach for in vitro and live cell analysis of protein–lipid interactions is based on a novel class of fluorescent lipids with a conjugated pentanyl-group (Kuerschner et al. 2005). These fatty acyl analogs have a structure highly similar to their endogenous counterparts (Fig. 2, structures 5 and 6), and, unlike conventional fluorescently tagged lipids (e.g., NBD- and BODIPY-derivatives) their lipid derivatives distribute within the cell together with their physiological kins. Pentanyl lipids have been employed to localize various lipid classes to intracellular compartments in live cells (Kuerschner et al. 2005). Notably, these fluorophores are able to serve as Förster resonance energy transfer (FRET) acceptors from protein-tryptophanyl residues exclusively in a hydrophobic environment as donor. Therefore, they are highly suited as probes for intramembrane protein–lipid interactions in reconstituted liposomal systems, and have been used in vitro to confirm a specific lipid molecular species–protein interaction found in live cells (F.-X. Contreras, F. Wieland, and B. Brügger, unpubl.).
DEFINING PROTEIN–LIPID INTERACTIONS
For membrane proteins, different types of interactions with lipid molecules can be distinguished by the relative “residence” time of a particular lipid at the protein–lipid interface (Lee 2003). If a lipid displays a low degree of interaction with the transmembrane domain (TMD) of the protein, it is considered a “bulk” lipid, defined by its fast exchange rate with lipids in close proximity (Fig. 1).
The residence time can be mediated by specific interactions with the lipid polar head group, by hydrophobic matching to the lipids' hydrocarbon chains, or both (discussed below). Such interactions lead to a significant reduction of exchange rates with the “bulk” lipids and the formation of an annulus or shell of (“annular”) lipids that surround the membrane protein. For large, multiple transmembrane-spanning proteins, the composition of this shell is not necessarily homogenous, because the interactions depend on the local architecture of the membrane protein and its compatibility with the various lipids. Individual lipids of a shell may vary in their residence time (for a review see Anderson and Jacobson 2002).
Lipids with even lower exchange rates are denominated as nonannular lipids (Fig. 1). These lipids often reside within large membrane protein complexes with a large number of subunits or within proteins that contain multiple transmembrane domains.
ANNULAR PROTEIN–LIPID INTERACTIONS
As described above, the major distinction between annular and bulk lipids is based on their exchange rates at the protein–lipid interface of membrane proteins. The composition of a lipid annulus or shell around TMDs is dictated by their local architecture. This may result in various specificities for lipid classes (and molecular species), at various interfaces with the same membrane protein complex. Atomistic molecular dynamics simulations suggest that membrane proteins, together with their adjacent lipids, form a dynamic protein–lipid complex with up to 50–100 lipids (Niemela et al. 2010). Within this lipid shell, the diffusion rates, and hence the exchange rates with the bulk lipids, are found to be significantly reduced. Accepting that membranes are “more mosaic than fluid” (Engelman 2005), it becomes difficult to tell apart an actively recruited annulus from lipids in preexisting liquid-ordered microdomains, in which the lateral mobility of lipids is reduced.
Impact of Membrane Properties and Membrane Protein Architecture on Interactions at the Protein–Lipid Interface
Several mechanisms rule the interactions of membrane proteins with distinct lipids: (1) hydrophobic thickness of the lipids, (2) the lateral pressure field of the membrane, (3) the distribution of charges at the protein–lipid interface, and (4) from the protein side the presence of specifically localized amino acid side chains.
The hydrophic thickness of a lipid bilayer defines the distance between opposing head groups of the inner and outer leaflet, typically between 35 and 55 Å. The hydrophic thickness is determined mainly by the lipid composition of the bilayer.
An important factor that influences the structure and dynamics of membrane proteins is the lateral pressure profile of membranes. It describes, similar to the buoyant force that is effective on matter embedded in fluids, the influence of a membrane as a solvent on a membrane protein as the matter dissolved in it. The highest pressure is at the interfacial region between hydrophobic and hydrophilic areas, at this location reaching peak values that correspond to pressures of hundreds of atmospheres. This force is further modulated by the degree of order of the surrounding lipids as well as the degree of hydrophobic mismatch at the protein–lipid interface and across the bilayer (Cantor 1999a,b; Gaines 1966).
Hydrophobic mismatch describes any deviation from an ideal hydrophobic compatibility of a transmembrane domain with its surrounding lipids. The above mechanisms are illustrated with the example of mechanosensitive channels. Mechanosensitive channels are found in both prokaryotes (MscL) and eukaryotes (TREK-1, TRAAK), and belong to a class of membrane proteins that are regulated by their local membrane environment, and for which the importance of binding of anionic annular lipids to a “hot spot” of positively charged amino acid residues was shown (Powl et al. 2008a,b). Alterations of the local lateral pressure fields were proposed as the molecular mechanism that provides the mechanical force to shift the channels from an open to a closed state. This was shown by activation of the channels on addition of nonbilayer-forming phospholipids to cylindrical, bilayer-forming ones (Perozo et al. 2002).
As another example, the peptide antibiotic gramicidin (gA) of Bacillus brevis can only form channels if the hydrophobic length of the surrounding lipid acyl chains is exactly (hydrophobically) matched to its potassium-conducting conformation (Koeppe and Anderson 1996; Girshman et al. 1997; Mobashery et al. 1997).
An example of how the distribution of charges at the protein–lipid interface gives rise to the selectivity of membrane proteins for distinct polar moieties of lipids is given with the peripheral antenna complex LHII of Rhodobacter sphaeroides. Here, the enrichment of phosphatidylethanolamine (PE) in the boundary phase is thought to be mediated by specific spectrin-like PE-binding sites (Liu et al. 2004; Kwa et al. 2008).
The availability of an increasing number of membrane protein structures solved by X-ray crystallography (Hunte and Richers 2008; White 2009; Byrne and Iwata 2002) has enormously contributed to our understanding of protein–lipid interactions. In some of these structures, lipids tightly bound to the transmembrane domains have been observed (see Tables 1 and 2). These lipids appear in the electron density map as elongated structures mainly oriented perpendicular to the membrane plane. Correspondingly, the majority of the bound lipids are reproducibly copurified with the protein. Only few crystal structures containing an inner shell of annular lipids have been completely characterized. These annular lipids, bound to the surface of the protein, mediate between the protein and the bulk lipids, and seem to play a major role in the orientation of the membrane-spanning domain within the bilayer. In the yeast cytochrome bc1 complex, phospholipids of the matrix leaflet (Lange et al. 2001) and phospholipids present in the intermembrane leaflet (Palsdottir et al. 2003) have been used to determine the hydrophobic thickness of the annulus surrounding the protein complex. Here, the distance between the phosphodiester groups of two oppositely oriented phospholipids (36 Å) is in good agreement with the published thickness measure for a dioleoyl-phosphatidylcholine (DOPC) bilayer (38 Å) (Lewis and Engelman 1983). Another example of an annular shell is a bilayer of up to 18 tightly bound lipids that covers 80% of the surface of the trimeric seven-transmembrane-span protein bacteriorhodopsin (Belrhali et al. 1999; Luecke et al. 1999).
Table 1.
Examples of nonannular lipids
| Protein | Method | Lipid | Function | Selected references |
|---|---|---|---|---|
| β2-adrenergic receptor) | X-ray crystallography | Chol | Protein fold (putative) | Hanson et al. 2008 |
| Caveolin-1 | SDS-PAGE, in vitro oligomerization | Chol | oligomeric state | Monier et al. 1995; Murata et al. 1995 |
| Cytochrome bc1 complex | X-ray crystallography | PI, CL | Stability and integrity of the complex, enzymatic activity | Gomez and Robinson 1999; Lange et al. 2001 |
| Cytochrome c oxidase | X-ray crystallography | PG | Oxygen transfer (putative) | Shinzawa-Itoh et al. 2007 |
| K+-channel | X-ray crystallography | anionic PLs | Potassium conductance | Valiyaveetil et al. 2002; Marius et al. 2008 |
| Metabotropic glutamate receptor | Agonist binding kinetics, lipid mass spectrometry | Erg | Allosteric effector, targeting to sterol-rich microdomains | Eroglu et al. 2003 |
| Mitochondrial ADP/ATP carrier | X-ray crystallography | CL | Oligomeric state | Nury et al. 2005 |
| Nitrate reductase A | X-ray crystallography | PG | Oligomeric state/protein fold (“building blocks”) | Bertero et al. 2003 |
| Oxytocin receptor | Agonist binding kinetics | Chol | Allosteric effector | Gimpl et al. 2002a |
| Peripheral-type benzodiazepine receptor | Ligand-dependent cholesterol uptake and release kinetics, mutational analyses | Chol | Cholesterol transport and compartmentali-zation (putative) | Li and Papadopoulos 1998; Jamin et al. 2005 |
| Plasma membrane Ca2+-ATPase | Transport & fluorescence assays | Cer DAG | Allosteric effector | Perez-Gordones et al. 2009 |
| Rhodopsin | In vitro photolysis assays, differential scanning calorimetry (DSC) | Chol | Metarhodopsin formation, reprotonation (photocycle) | Mitchell et al. 1990; Bennet et al. 2008 |
| Serotonin1a receptor | Agonist binding kinetics | Chol | Allosteric effector | Pucadyil and Chattopadhyay 2004 |
| Vacuolar-type Na+-ATPase | X-ray crystallography | DPPG, DPG | Oligomeric state/protein fold (“building blocks”) | Murata et al. 2005 |
For details, see text and Ernst et al. 2010.
Table 2.
Examples of annular lipids
| Protein | Method | Lipid | Function | Selected references |
|---|---|---|---|---|
| Aquaporin | Electron crystallography, X-ray crystallography | various | Protein structure | Recently reviewed in Andrews et al. 2008 |
| Bacteriorhodopsin | X-ray crystallography, electron crystallography | various | Protein fold (“building blocks”), reprotonation (photocycle) | Luecke et al. 1999; recently reviewed in Raunser and Walz 2009 |
| Gramicidin (gA) | Capacitance measurements, solid-state NMR | di-18:2-PC, lyso-PLs | Conformational state (conductance) | Koeppe and Anderson 1996; Mobashery et al. 1997; Girshman et al. 1997; Martinac and Hamill 2002 |
| Large-conductance mechanosensitive channel (E. coli; MscL) | Patch clamp analyses, electron paramagnetic resonance (EPR) | lyso-PLs | Conformational state (conductance) | Vasquez et al. 2008; for a review see Powl et al. 2005 |
| Light-harvesting complex II (LHII) | X-ray crystallography, lipid mass spectrometry | PE, PG | Photosynthetic membrane biogenesis | Liu et al. 2004; Kwa et al. 2008 |
| Na,K-ATPase | Electron spin resonance spectroscopy (ESR) | PS, CL | Enzymatic activity (antiport of Na+ and K+) | Reviewed in Esmann et al. 2006 |
| Outer membrane protein F (E. coli; OmpF) | Förster resonance Energy transfer (FRET) | di-14:1-PLs | oligomeric state (trimeric complex) | O'Keeffe et al. 2000 |
| Sarcoplasmatic Ca2+-ATPase (SERCA) | Transport and fluorescence assays, phosphoresence anisotropy | di-18:1-PC, PE | rate of dephosphorylation, modulation of the phosphorylation domain | Starling et al. 1995; Hunter et al. 1999a,b |
For details, see text and Ernst et al. 2010.
Finally, a well-established example of an annular shell surrounding a transmembrane domain is present in the X-ray structure of the membrane rotor ring of the Na+-ATPase from Enterococcus hirae, in which the internal hydrophobic ring surface is covered in both leaflets by 10 molecules of 1,2-dipalmitoyl-phosphatidyglycerol (DPPG) and 10 molecules of 1,2-dipalmitoyl-glycerol (DPG), respectively (Murata et al. 2005). This highly defined structure exemplifies a problem in classifying protein–lipid interactions: Although the lipid molecules form a ring around the transmembrane domains, and therefore represent annuli, the shell is tightly bound to the protein by specific interfaces with the individual lipids on the inner hydrophobic surface of the oligomeric assembly, causing very low exchange rates. Such ring-shaped assemblies are usually characteristic of annular lipids.
In most of the X-ray structures published, lipid–protein interactions are mainly stabilized by polar interactions between the lipid head group and specific amino acids. The majority of the tightly bound lipids are generally stabilized by at least two polar interactions between the phosphodiester group and a set of molecules generally combining a positive charge and a polar amino acid. Nevertheless, the observed binding domains are nonlinear and can even consist of a set of amino acids localized to different subunits, as observed for the yeast cytochrome bc1 complex (Palsdottir et al. 2003). Aromatic amino acids are very often involved in lipid stabilization. Tyrosine residues, present in the interfacial region, interact with the lipid phosphodiester group either alone (via ion pair or hydrogen bond) or in combination with positively charged amino acids. Likewise, tryptophan residues are mainly localized in the interfacial region, with the indole group pointing toward the center of the bilayer (Deisenhofer and Michel 1989). Hydrogen bonds are frequently observed between the indole nitrogen atom and the lipid phosphodiester group (Ridder et al. 2000). Furthermore, a stabilization of the lipid acyl chains has been observed by a lamellar orientation of the indole ring, providing a mechanism for the anchoring of the transmembrane domain to the first shell of annular lipids. In addition to the previous statement, lipid head groups are also stabilized by multiple interactions within the protein (Lange et al. 2001). Nonpolar interactions between the hydrophobic lipid acyl chains and the transmembrane domain stabilize the binding (Luecke et al. 1999; Lange et al. 2001).
NONANNULAR PROTEIN–LIPID INTERACTIONS
Lipids as Structural Building Blocks and Allosteric Effectors of Membrane Proteins
Most of the nonannular protein–lipid interactions have been identified by X-ray-crystallographic approaches (see the following examples). Frequently, as identified in the vacuolar-type (V-type) sodium ATPase of Enterococcus hirae (Murata et al. 2005), lipids that reside within oligomeric membrane protein assemblies act as “molecular glue,” strengthening the contacts of the subunits. Another example, in which lipids were identified as structural building blocks of protein assemblies, are caveolae. Caveolae and caveolae-like domains are found at plasma membranes of higher eukaryotes and are morphologically heterogeneous (Parton et al. 1994; Scherer et al. 1997). They have been suggested to play crucial roles in signal transduction acting as signaling platforms. Today, the role of caveolin in signaling is far from being completely resolved because in cells lacking caveolin the distribution of signaling proteins is not affected (Parton and Simons 2007). The major caveolae proteins are members of the caveolin protein family. Caveolae show a unique lipid composition, predominantly consisting of cholesterol, sphingomyelin, and the ganglioside GM1 (Smart et al. 1995; Schnitzer et al. 1996). Regulation of caveolin-1 expression was shown to be tightly connected to cellular cholesterol levels (Bist et al. 1997). Caveolin-1 binds to cholesterol with a 1:1 stoichiometry, resulting in a complex that even resists treatment with SDS (Murata et al. 1995). Cholesterol is able to promote the oligomerization of caveolin-1 subunits in microsomes (Monier et al. 1995), underlining its role as a structural building block of caveolae. Another example for a nonannular lipid-binding site is found in KcsA, a potassium channel of Streptomyces lividans. This tetrameric complex selects for anionic phospholipids in its core, in which they strongly influence the ability of the protein complex to conduct potassium (Valiyaveetil et al. 2002). Li and Papadopoulos identified a cholesterol-binding motif in the peripheral-type benzodiazepine receptor (PBR) (Li and Papadopoulos 1998), a motif also found in caveolin-1. This consensus motif consists of hydrophobic, aromatic and positively charged amino acids: L/V-(X)1–5-Y-(X)1–5-R/K (cholesterol recognition/interaction amino acid consensus [CRAC]).
A similar motif was found responsible for the tight interaction of nonannular cholesterol molecules within the β2-adrenergic receptor (Hanson et al. 2008), a seven transmembrane-containing G-protein-coupled receptor (GPCR). Here two cholesterol molecules are bound in a shallow surface groove, with four out of the seven TMDs contributing to this lipid-binding structure. Strikingly, a relaxed version of this motif, W/Y-(X)1-3-I/V/L-(X)1-7-K/R, termed the cholesterol-consensus motif (CCM), was identified in a large number of GPCRs. The precise function of cholesterol as a nonannular lipid in these receptors is controversial, however. Two models have been suggested how the presence of the lipid could modulate the receptor's structure and function: (1) directly, via a conformational change following interaction with cholesterol, and (2) indirectly, because of an alteration of membrane biophysical properties (Gimpl et al. 2002a,b). Either one (or a combination of both) might mediate targeting of these receptors to distinct membrane domains and/or modulate their affinity for ligands. Other GPCRs, such as rhodopsin, a photoreceptor present in retinal rod cells, are modulated by cholesterol as well. The ordering effect of cholesterol on acyl chains of lipids adjacent to transmembrane domains was shown to affect the formation of metarhodopsin, an intermediate state that is in equilibrium with the light-activated receptor (Mitchell et al. 1990; Bennett and Mitchell 2008). In addition to their role as “nonannular” structural building blocks within GPCRs, sterols are discussed to function as allosteric effectors. For example, Pucadyil and Chattopadhyay showed for the hippocampal serotonin1A receptor (another GPCR with a CCM) that ligand-binding affinity and G-protein coupling was affected by cholesterol (Pucadyil and Chattopadhyay 2004). This was also observed for other GPCRs, for example, the metabotropic glutamate receptor of Drosophila melanogaster (DMGluRA) (Eroglu et al. 2003) and the oxytocin receptor (Gimpl et al. 2002b), in which the presence of sterols was shown to shift these receptors to a high-affinity agonist-binding state. In the case of DMGluRa, this protein–lipid interaction was further shown to target the receptor to sterol-rich microdomains.
In addition, other lipid classes were reported to regulate as nonannular lipids enzymatic functions of membrane proteins, such as diacylglycerol that activates the Ca2+-ATPase (PMCA) (Perez-Gordones et al. 2009). In summary, these and other examples (see Tables 1 and 2) emphasize the importance of nonannular lipids within membrane protein complexes in modulating their architecture (structure/conformation), localization (targeting to distinct membrane domains), and enzymatic functions.
ROLE OF LIPIDS AS CHAPERONES
It has been postulated that protein–lipid interactions can be critical for correct insertion, folding, and topology of membrane proteins in a way similar to protein chaperones (van Klompenburg et al. 1997; Dowhan and Bogdanov 2009). Here, lipids assuming protein chaperone-like functions are defined as lipochaperones. This term must not be confused with the expression lipid chaperones, defining proteins that keep soluble or transport individual lipid molecules, such as lipid transport proteins (Hanada et al. 2003; Ile et al. 2006; Wirtz 2006) and fatty acid binding proteins (Storch and McDermott 2009). As a prominent lipochaperone function, membrane lipids can rule the topology of membrane proteins. X-ray data show that the majority of lipids tightly bound to membrane proteins via head groups are localized in the electronegative side of the membrane. In addition to sequential protein–protein interactions within the translocon, specific interactions between negatively charged phospholipids and positively charged amino acids help guiding membrane protein orientation (von Heijne 2006; Rapoport 2007). This is in agreement with the asymmetric lipid distribution of biological membranes, in which anionic lipids are localized in the cytoplasmic leaflet. This preferential localization is consistent with the “positive-inside” rule described previously (von Heijne and Gavel 1988; von Heijne 1989), in which membrane proteins facing the negative side of the membrane are generally enriched in arginine and lysine residues. For example, addition or removal of positively charged amino acids in E. coli leader peptidase Lep completely changed the membrane proteins' orientation, which is influenced by the extent of anionic lipids (van Klompenburg et al. 1997). In the case of the secondary transporter LacY (lactose permease) it has been shown that PE is necessary for its cellular function, assembly, and folding (Bogdanov and Dowhan 1998, 1999; Bogdanov et al. 1999). This zwitterionic phospholipid is not required for membrane insertion because LacY, a 12 TMD protein, can be inserted in E. coli cells lacking PE, but its active transport function is disturbed. This lack of activity in the absence of PE is because of a different membrane topological organization. Under these conditions, some transmembrane domains and hydrophilic domains are topologically inverted with respect to the bilayer. However, addition of PE after membrane insertion restores the topological orientation, facilitates the LacY structural maturation, and reestablishes its transport activity. Therefore, PE seems to play an active role in controlling membrane protein topology and assembly (Bogdanov et al. 2002). Further indication that the topological organization of lacY is regulated by the membrane phospholipid composition comes from reconstitution of lacY in liposomes of different lipid composition, in which only in presence of PE active transport of lactose by LacY was achieved. Interestingly, in liposomes containing another zwitterionic lipid (PC), the membrane topology of the C6 domain was the same as that observed in presence of PE, however, no active transport was detected. In these in vitro studies, the presence of zwitterionic lipids is sufficient to drive and facilitate membrane topology. In addition, other properties of PE are required to sustain active transport (Wang et al. 2002). One important difference between PC and PE is that the latter can form hydrogen bonds or exchange protons, which will help the cotransport of a proton along with lactose to couple substrate uptake with the proton electrochemical potential across the bilayer. Likewise, a requirement of PE for proper topological organization and function has been described for other secondary transporters (e.g., phenylalanine permease [PheP] or γ-aminobutyric acid [GABA]) (Zhang et al. 2003, 2005).
The number of diseases related to lipids that interfere with protein folding is growing over the years (Kuznetsov and Nigam 1998). In Alzheimer's disease the specific molecular initiator of the disease is still unresolved. However, biochemical studies indicate that soluble amyloid-β (Aβ) oligomers, mainly dimers, are the key intermediates in the manifestation of the disease (for a recent review see Li et al. 2010). The formation of Aβ aggregates in the brain of patients with Alzheimer disease correlates with a specific peptide–lipid interaction. Several studies showed that Aβ peptides strongly interact with the monosialoganglioside GM1 (Yanagisawa et al. 1995; Wakabayashi et al. 2005). Aβ specifically recognizes GM1 clusters at the cell surface, and following GM1-Aβ interaction, Aβ undergoes a conformational change from a α-helix-rich structure to a β-sheet-rich structure and serve as a seed for Aβ fibrillogenesis (Matsuzaki et al. 2010). In this case, GM1 functions as an “anti”-lipochaperone activity in facilitating aggregation of Aβ peptides.
Expanding the term lipochaperones beyond roles in the folding and topology of transmembrane proteins, lipids might assist the transport of proteins to their target membrane. Within its transport pathway a newly synthesized or cycling TMD is faced with membranes composed of varying lipid classes and molecular species. These compositions determine the physicochemical characteristics of a membrane as well as its bilayer thickness. As a consequence, a given TMD may be confronted with unfavorable membrane environments, mainly because of a lack of hydrophobic match. Specific binding of lipid molecular species to a transmembrane domain can minimize such hydrophobic mismatch (Hite et al. 2010), which will decrease the corresponding energy penalty. Varying local energy states of a TMD, because of locally different lipid compositions, would then allow efficient protein sorting. In eukaryotic cells, transmembrane domains of plasma membrane proteins in the average are five amino acids longer than proteins localized to the Golgi (Bretscher and Munro 1993; Sharpe et al. 2010). Thus, plasma membrane proteins that travel within the Golgi have TMDs too long to be accommodated by the membrane, and therefore their helices will either tilt within the bilayer, or their annular lipids expand to increase hydrophobic thickness. In both cases bilayer stress will be increased. Such unfavorable situation could be ameliorated by lipid molecular species tightly bound to the transmembrane domain to reduce the hydrophobic mismatch. Based on the fact that each organelle has a specific lipid composition, a state of minimal energy will contribute to a preferential distribution of the membrane protein to its target membrane.
Additional functions can be envisaged for interactions between specific lipid molecular species and TMDs. Employing novel methods to analyze protein–lipid interactions, we have observed highly specific protein–lipid molecular species interactions that regulate the oligomerization of TMDs (F-X Contreras, F Wieland, and B Brügger, unpubl.). Furthermore, binding of specific lipids to a membrane receptor can regulate its activity directly as allosteric effectors (as outlined above), or by targeting to specific membrane domains, or both. Likewise, a membrane protein may be kept in its resting state by interaction with a specific lipid molecular species, therefore decreasing unspecific interactions with other proteins. Following ligand association, a conformational change will release the lipid, leading to the association of the protein with other proteins to form an active complex (U Coskun, M Grzybek, and K Simons, pers. commun.).
In summary, such mechanisms involving specific intramembrane protein–lipid interactions are reminiscent of chaperone functions.
OPEN PROBLEMS
A major obstacle for rapid progress in our understanding of the physiology of protein–lipid interactions is the lack of molecular and cell biology-based tools to probe interactions at the level of lipid molecular species specificity. Unlike analyses of protein function by targeted knockdown or knockout of individual proteins, or introduction of mutations, no equivalent methods exist to manipulate lipid molecular species individually in live cells. Although qualitative and quantitative lipid analysis has seen rapid progress (Brügger et al. 1997; Shevchenko and Simons 2010), mass spectrometric analysis on the protein side, to determine TMDs or derivatives thereof, is still challenging, hampering the characterization of TMD-lipid complexes. The fundamental question is still open whether protein TMDs exist that are simply dissolved in the hydrophobic phase of the membrane, with no specificity for any lipid whatsoever, or if there is some specificity in each TMD for one or more of the manifold lipid species that make up a membrane. A systems biology approach may help to answer such questions: To this end, a high throughput system would be needed to screen all physiological TMDs for their interactions with a comprehensive set of membrane lipid building blocks. Technically, such a screen may be assisted by the availability for read out of fluorescent detection methods as outlined above.
Protein–lipid interactions and their physiological relevance have now found the interest of a wide community of molecular cell biologists, chemists, physicists, and pharmacologists, and synergy derived from their combined activities will further help spurring this field in the molecular life sciences.
Footnotes
Editor: Kai Simons
Additional Perspectives on The Biology of Lipids available at www.cshperspectives.org
REFERENCES
- Anderson RG, Jacobson K 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296: 1821–1825 [DOI] [PubMed] [Google Scholar]
- Andrews S, Reichow SL, Gonen T 2008. Electron crystallography of aquaporins. IUBMB Life 60: 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belrhali H, Nollert P, Royant A, Menzel C, Rosenbusch JP, Landau EM, Pebay-Peyroula E 1999. Protein, lipid and water organization in bacteriorhodopsin crystals: A molecular view of the purple membrane at 1.9 Å resolution. Structure 7: 909–917 [DOI] [PubMed] [Google Scholar]
- Bennett MP, Mitchell DC 2008. Regulation of membrane proteins by dietary lipids: Effects of cholesterol and docosahexaenoic acid acyl chain-containing phospholipids on rhodopsin stability and function. Biophys J 95: 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NC 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol 10: 681–687 [DOI] [PubMed] [Google Scholar]
- Bhide SY, Zhang Z, Berkowitz ML 2007. Molecular dynamics simulations of SOPS and sphingomyelin bilayers containing cholesterol. Biophys J 92: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bist A, Fielding PE, Fielding CJ 1997. Two sterol regulatory element-like sequences mediate up-regulation of caveolin gene transcription in response to low density lipoprotein free cholesterol. Proc Natl Acad Sci 94: 10693–10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W 1998. Phospholipid-assisted protein folding: Phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J 17: 5255–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W 1999. Lipid-assisted protein folding. J Biol Chem 274: 36827–36830 [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Heacock PN, Dowhan W 2002. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J 21: 2107–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Umeda M, Dowhan W 1999. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem 274: 12339–12345 [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S 1993. Cholesterol and the Golgi apparatus. Science 261: 1280–1281 [DOI] [PubMed] [Google Scholar]
- Brown DA, London E 1998. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol 14: 111–136 [DOI] [PubMed] [Google Scholar]
- Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci 94: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B, Iwata S 2002. Membrane protein complexes. Curr Opin Struct Biol 12: 239–243 [DOI] [PubMed] [Google Scholar]
- Cantor RS 1999a. The influence of membrane lateral pressures on simple geometric models of protein conformational equilibria. Chem Phys Lipids 101: 45–56 [DOI] [PubMed] [Google Scholar]
- Cantor RS 1999b. Lipid composition and the lateral pressure profile in bilayers. Biophys J 76: 2625–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J, Michel H 1989. Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. EMBO J 8: 2149–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Borreguero JM, Buldyrey SV, Stanley HE, Dokholyan NV 2003. Mechanism for the α-helix to β-hairpin transition. Proteins 53: 220–228 [DOI] [PubMed] [Google Scholar]
- Dluhy RA, Cornell DG 1985. In situ measurement of the infrared spectra of insoluble monolayers at the air–water interface. J Phys Chem 89: 3195–3197 [Google Scholar]
- Dluhy RA, Mendelsohn R, Casal HL, Mantsch HH 1983. Interaction of dipalmitoylphosphatidylcholine and dimyristoylphosphatidylcholine-d54 mixtures with glycophorin. A Fourier transform infrared investigation. Biochemistry 22: 1170–1177 [DOI] [PubMed] [Google Scholar]
- Douglass AD, Vale RD 2005. Single-molecule microscopy reveals plasma membrane microdomains created by protein–protein networks that exclude or trap signaling molecules in T cells. Cell 121: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W, Bogdanov M 2009. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem 78: 515–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- East JM, Lee AG 1982. Lipid selectivity of the calcium and magnesium ion dependent adenosinetriphosphatase, studied with fluorescence quenching by a brominated phospholipid. Biochemistry 21: 4144–4151 [DOI] [PubMed] [Google Scholar]
- Engelman DM 2005. Membranes are more mosaic than fluid. Nature 438: 578–580 [DOI] [PubMed] [Google Scholar]
- Esmann M, Marsh D 2006. Lipid–protein interactions with the Na,K-ATPase. Chem Phys Lipids 141: 94–104 [DOI] [PubMed] [Google Scholar]
- Ernst AM, Contreras FX, Brügger B, Wieland F 2010. Determinants of specificity at the protein–lipid interface in membranes. FEBS Lett 584: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Eroglu C, Brügger B, Wieland F, Sinning I 2003. Glutamate-binding affinity of Drosophila metabotropic glutamate receptor is modulated by association with lipid rafts. Proc Natl Acad Sci 100: 10219–10224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov A, Oradd G, Lindblom G 2003. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J 84: 3079–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines GL 1966. Insoluble monolayers at liquid–gas interfaces. Interscience Publishers, New York [Google Scholar]
- Gimpl G, Burger K, Fahrenholz F 2002a. A closer look at the cholesterol sensor. Trends Biochem Sci 27: 596–599 [DOI] [PubMed] [Google Scholar]
- Gimpl G, Wiegand V, Burger K, Fahrenholz F 2002b. Cholesterol and steroid hormones: Modulators of oxytocin receptor function. Prog Brain Res 139: 43–55 [DOI] [PubMed] [Google Scholar]
- Girshman J, Greathouse DV, Koeppe RE II, Andersen OS 1997. Gramicidin channels in phospholipid bilayers with unsaturated acyl chains. Biophys J 73: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez B Jr, Robinson NC 1999. Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry 38: 9031–9038 [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T 2005. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Manas JM, Lakey JH, Pattus F 1992. Brominated phospholipids as a tool for monitoring the membrane insertion of colicin A. Biochemistry 31: 7294–7300 [DOI] [PubMed] [Google Scholar]
- Haberkant P, van Meer G 2009. Protein–lipid interactions: Paparazzi hunting for snap-shots. Biol Chem 390: 795–803 [DOI] [PubMed] [Google Scholar]
- Haberkant P, Schmitt O, Contreras FX, Thiele C, Hanada K, Sprong H, Reinhard C, Wieland FT, Brugger B 2008. Protein–sphingolipid interactions within cellular membranes. J Lipid Res 49: 251–262 [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature 426: 803–809 [DOI] [PubMed] [Google Scholar]
- Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC 2008. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Li Z, Walz T 2010. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J 29: 1652–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsass C, Lindahl E, Edholm O 2003. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys J 84: 2192–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Richers S 2008. Lipids and membrane protein structures. Curr Opin Struct Biol 18: 406–411 [DOI] [PubMed] [Google Scholar]
- Hunter GW, Bigelow DJ, Squier TC 1999a. Lysophosphatidylcholine modulates catalytically important motions of the Ca-ATPase phosphorylation domain. Biochemistry 38: 4604–4612 [DOI] [PubMed] [Google Scholar]
- Hunter GW, Negash S, Squier TC 1999b. Phosphatidylethanolamine modulates Ca-ATPase function and dynamics. Biochemistry 38: 1356–1364 [DOI] [PubMed] [Google Scholar]
- Ile KE, Schaaf G, Bankaitis VA 2006. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol 2: 576–583 [DOI] [PubMed] [Google Scholar]
- Jamin N, Neumann JM, Ostuni MA, Vu TK, Yao ZX, Murail S, Robert JC, Giatzakis C, Papadopoulos V, Lacapere JJ 2005. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol Endocrinol 19: 588–594 [DOI] [PubMed] [Google Scholar]
- Killian JA 1998. Hydrophobic mismatch between proteins and lipids in membranes. Biochim Biophys Acta 1376: 401–415 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y 1997. Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature 389: 206–211 [DOI] [PubMed] [Google Scholar]
- Knapp EW, Fischer SF, Zinth W, Sander M, Kaiser W, Deisenhofer J, Michel H 1985. Analysis of optical spectra from single crystals of Rhodopseudomonas viridis reaction centers. Proc Natl Acad Sci 82: 8463–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppe RE II, Anderson OS 1996. Engineering the gramicidin channel. Annu Rev Biophys Biomol Struct 25: 231–258 [DOI] [PubMed] [Google Scholar]
- Kuerschner L, Ejsing CS, Ekroos K, Shevchenko A, Anderson KI, Thiele C 2005. Polyene-lipids: A new tool to image lipids. Nat Methods 2: 39–45 [DOI] [PubMed] [Google Scholar]
- Kuznetsov G, Nigam SK 1998. Folding of secretory and membrane proteins. N Engl J Med 339: 1688–1695 [DOI] [PubMed] [Google Scholar]
- Kwa LG, Wegmann D, Brugger B, Wieland FT, Wanner G, Braun P 2008. Mutation of a single residue, β-glutamate-20, alters protein–lipid interactions of light harvesting complex II. Mol Microbiol 67: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C 2001. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J 20: 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury PT Jr 1997. Structural neurology: Are seeds at the root of neuronal degeneration? Neuron 19: 1151–1154 [DOI] [PubMed] [Google Scholar]
- Lee AG 2003. Lipid–protein interactions in biological membranes: A structural perspective. Biochim Biophys Acta 1612: 1–40 [DOI] [PubMed] [Google Scholar]
- Leto TL, Roseman MA, Holloway PW 1980. Mechanism of exchange of cytochrome b5 between phosphatidylcholine vesicles. Biochemistry 19: 1911–1916 [DOI] [PubMed] [Google Scholar]
- Lewis BA, Engelman DM 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol 166: 211–217 [DOI] [PubMed] [Google Scholar]
- Li H, Papadopoulos V 1998. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 139: 4991–4997 [DOI] [PubMed] [Google Scholar]
- Li S, Shankar GM, Selkoe DJ 2010. How do soluble oligomers of amyloid β-protein impair hippocampal synaptic plasticity? Front Cell Neurosci 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E, Sansom MS 2008. Membrane proteins: Molecular dynamics simulations. Curr Opin Struct Biol 18: 425–431 [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K 2010. Lipid rafts as a membrane-organizing principle. Science 327: 46–50 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W 2004. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK 1999. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol 291: 899–911 [DOI] [PubMed] [Google Scholar]
- Marius PM, Zagnoni M, Sandison ME, East JM, Morgan H, Lee AG 2008. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys J 94: 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markello T, Zlotnick A, Everett J, Tennyson J, Holloway PW 1985. Determination of the topography of cytochrome b5 in lipid vesicles by fluorescence quenching. Biochemistry 24: 2895–2901 [DOI] [PubMed] [Google Scholar]
- Martinac B, Hamill OP 2002. Gramicidin A channels switch between stretch activation and stretch inactivation depending on bilayer thickness. Proc Natl Acad Sci 99: 4308–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K, Kato K, Yanagisawa K 2010. Aβ polymerization through interaction with membrane gangliosides. Biochim Biophys Acta 1801: 868–877 [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Straume M, Miller JL, Litman BJ 1990. Modulation of metarhodopsin formation by cholesterol-induced ordering of bilayer lipids. Biochemistry 29: 9143–9149 [DOI] [PubMed] [Google Scholar]
- Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y 1999. The structure of bacteriorhodopsin at 3.0 Å resolution based on electron crystallography: Implication of the charge distribution. J Mol Biol 286: 861–882 [DOI] [PubMed] [Google Scholar]
- Mobashery N, Nielsen C, Andersen OS 1997. The conformational preference of gramicidin channels is a function of lipid bilayer thickness. FEBS Lett 412: 15–20 [DOI] [PubMed] [Google Scholar]
- Monier S, Parton RG, Vogel F, Behlke J, Henske A, Kurzchalia TV 1995. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell 6: 911–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses JE, Moorhouse AD 2007. The growing applications of click chemistry. Chem Soc Rev 36: 1249–1262 [DOI] [PubMed] [Google Scholar]
- Murata M, Peranen J, Schreiner R, Wieland F, Kurzchalia TV, Simons K 1995. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci 92: 10339–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE 2005. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science 308: 654–659 [DOI] [PubMed] [Google Scholar]
- Niemela P, Hyvonen MT, Vattulainen I 2004. Structure and dynamics of sphingomyelin bilayer: Insight gained through systematic comparison to phosphatidylcholine. Biophys J 87: 2976–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemela PS, Hyvonen MT, Vattulainen I 2006. Influence of chain length and unsaturation on sphingomyelin bilayers. Biophys J 90: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemela PS, Hyvonen MT, Vattulainen I 2009. Atom-scale molecular interactions in lipid raft mixtures. Biochim Biophys Acta 1788: 122–135 [DOI] [PubMed] [Google Scholar]
- Niemela PS, Miettinen MS, Monticelli L, Hammaren H, Bjelkmar P, Murtola T, Lindahl E, Vattulainen I 2010. Membrane proteins diffuse as dynamic complexes with lipids. J Am Chem Soc 132: 7574–7575 [DOI] [PubMed] [Google Scholar]
- Nury H, Dahout-Gonzalez C, Trezeguet V, Lauquin G, Brandolin G, Pebay-Peyroula E 2005. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett 579: 6031–6036 [DOI] [PubMed] [Google Scholar]
- O'Keeffe AH, East JM, Lee AG 2000. Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys J 79: 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci E, Vendruscolo M, Karplus M 2002. Validity of Go models: Comparison with a solvent-shielded empirical energy decomposition. Biophys J 83: 3032–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsdottir H, Lojero CG, Trumpower BL, Hunte C 2003. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem 278: 31303–31311 [DOI] [PubMed] [Google Scholar]
- Pandit SA, Jakobsson E, Scott HL 2004. Simulation of the early stages of nano-domain formation in mixed bilayers of sphingomyelin, cholesterol, and dioleylphosphatidylcholine. Biophys J 87: 3312–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Simons K 2007. The multiple faces of caveolae. Nat Rev Mol Cell Biol 8: 185–194 [DOI] [PubMed] [Google Scholar]
- Parton RG, Joggerst B, Simons K 1994. Regulated internalization of caveolae. J Cell Biol 127: 1199–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gordones MC, Lugo MR, Winkler M, Cervino V, Benaim G 2009. Diacylglycerol regulates the plasma membrane calcium pump from human erythrocytes by direct interaction. Arch Biochem Biophys 489: 55–61 [DOI] [PubMed] [Google Scholar]
- Perozo E, Kloda A, Cortes DM, Martinac B 2002. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol 9: 696–703 [DOI] [PubMed] [Google Scholar]
- Powl AM, East JM, Lee AG 2008a. Anionic phospholipids affect the rate and extent of flux through the mechanosensitive channel of large conductance MscL. Biochemistry 47: 4317–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powl AM, East JM, Lee AG 2008b. Importance of direct interactions with lipids for the function of the mechanosensitive channel MscL. Biochemistry 47: 12175–12184 [DOI] [PubMed] [Google Scholar]
- Powl AM, Carney J, Marius P, East JM, Lee AG 2005. Lipid interactions with bacterial channels: Fluorescence studies. Biochem Soc Trans 33: 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Chattopadhyay A 2004. Cholesterol modulates ligand binding and G-protein coupling to serotonin1A receptors from bovine hippocampus. Biochim Biophys Acta 1663: 188–200 [DOI] [PubMed] [Google Scholar]
- Rapoport TA 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Raunser S, Walz T 2009. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys 38: 89–105 [DOI] [PubMed] [Google Scholar]
- Reichow SL, Gonen T 2009. Lipid–protein interactions probed by electron crystallography. Curr Opin Struct Biol 19: 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder AN, Morein S, Stam JG, Kuhn A, de Kruijff B, Killian JA 2000. Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry 39: 6521–6528 [DOI] [PubMed] [Google Scholar]
- Scherer PE, Lewis RY, Volonte D, Engelman JA, Galbiati F, Couet J, Kohtz DS, van Donselaar E, Peters P, Lisanti MP 1997. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J Biol Chem 272: 29337–29346 [DOI] [PubMed] [Google Scholar]
- Schnitzer JE, Oh P, McIntosh DP 1996. Role of GTP hydrolysis in fission of caveolae directly from plasma membranes. Science 274: 239–242 [DOI] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Simons K 2010. Lipidomics: Coming to grips with lipid diversity. Nat Rev Mol Cell Biol 11: 593–598 [DOI] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, et al. 2007. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J 26: 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E 1997. Functional rafts in cell membranes. Nature 387: 569–572 [DOI] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL 1972. The fluid mosaic model of the structure of cell membranes. Science 175: 720–731 [DOI] [PubMed] [Google Scholar]
- Smart EJ, Ying YS, Anderson RG 1995. Hormonal regulation of caveolae internalization. J Cell Biol 131: 929–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Hall CK 2001. Assembly of a tetrameric α-helical bundle: Computer simulations on an intermediate-resolution protein model. Proteins 44: 376–391 [DOI] [PubMed] [Google Scholar]
- Starling AP, East JM, Lee AG 1995. Effects of phospholipid fatty acyl chain length on phosphorylation and dephosphorylation of the Ca2+-ATPase. Biochem J 310: 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J, McDermott L 2009. Structural and functional analysis of fatty acid-binding proteins. J Lipid Res 50: S126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MG, Smith IC 1981. Reliability of nitroxide spin probes in reporting membrane properties: A comparison of nitroxide- and deuterium-labeled steroids. Biochemistry 20: 5252–5255 [DOI] [PubMed] [Google Scholar]
- Thiele C, Hannah MJ, Fahrenholz F, Huttner WB 2000. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol 2: 42–49 [DOI] [PubMed] [Google Scholar]
- Tycko R 2001. Biomolecular solid state NMR: Advances in structural methodology and applications to peptide and protein fibrils. Annu Rev Phys Chem 52: 575–606 [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, Zhou Y, MacKinnon R 2002. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 41: 10771–10777 [DOI] [PubMed] [Google Scholar]
- Van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B 1997. Anionic phospholipids are determinants of membrane protein topology. EMBO J 16: 4261–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW 2008. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez V, Sotomayor M, Cordero-Morales J, Schulten K, Perozo E 2008. A structural mechanism for MscS gating in lipid bilayers. Science 321: 1210–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G 1989. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341: 456–458 [DOI] [PubMed] [Google Scholar]
- von Heijne G 2006. Membrane-protein topology. Nat Rev Mol Cell Biol 7: 909–918 [DOI] [PubMed] [Google Scholar]
- von Heijne G, Gavel Y 1988. Topogenic signals in integral membrane proteins. Eur J Biochem 174: 671–678 [DOI] [PubMed] [Google Scholar]
- Wakabayashi M, Okada T, Kozutsumi Y, Matsuzaki K 2005. GM1 ganglioside-mediated accumulation of amyloid β-protein on cell membranes. Biochem Biophys Res Commun 328: 1019–1023 [DOI] [PubMed] [Google Scholar]
- Wang X, Bogdanov M, Dowhan W 2002. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J 21: 5673–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH 2009. Biophysical dissection of membrane proteins. Nature 459: 344–346 [DOI] [PubMed] [Google Scholar]
- Wirtz KW 2006. Phospholipid transfer proteins in perspective. FEBS Lett 580: 5436–5441 [DOI] [PubMed] [Google Scholar]
- Yanagisawa K, Odaka A, Suzuki N, Ihara Y 1995. GM1 ganglioside-bound amyloid β-protein (A β): A possible form of preamyloid in Alzheimer's disease. Nat Med 1: 1062–1066 [DOI] [PubMed] [Google Scholar]
- Zaraiskaya T, Jeffrey KR 2005. Molecular dynamics simulations and 2H NMR study of the GalCer/DPPG lipid bilayer. Biophys J 88: 4017–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W 2003. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem 278: 50128–50135 [DOI] [PubMed] [Google Scholar]
- Zhang W, Campbell HA, King SC, Dowhan W 2005. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the γ-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem 280: 26032–26038 [DOI] [PubMed] [Google Scholar]