Abstract
Huntington's disease (HD) is the most common inherited neurodegenerative disease and is characterized by uncontrolled excessive motor movements and cognitive and emotional deficits. The mutation responsible for HD leads to an abnormally long polyglutamine (polyQ) expansion in the huntingtin (Htt) protein, which confers one or more toxic functions to mutant Htt leading to neurodegeneration. The polyQ expansion makes Htt prone to aggregate and accumulate, and manipulations that mitigate protein misfolding or facilitate the clearance of misfolded proteins tend to slow disease progression in HD models. This article will focus on HD and the evidence that it is a conformational disease.
Inherited polyQ mutations cause huntingtin to misfold and aggregate. This may overload the cell's chaperone network so other metastable proteins misfold, producing a complex loss-of-function phenotype that leads to neurodegeneration.
HUNTINGTON'S DISEASE: A BRIEF HISTORY
In 1872 George Huntington, a young physician from Connecticut, published one of the first descriptions of the disorder that would come to bear his name (Huntington 1872). The publication, one of only two produced in his entire career, was based on families under the care of his father who was also a physician. The neurological disorder was dominantly inherited and characterized by excessive motor movements and neuropsychological deficits. Earlier descriptions of a neurological disorder that is almost certainly Huntington's disease (HD) can be found (Harper 2002; Lund 1860); however, George Huntington's is still considered by many to be the first fairly complete description of HD.
More than a century passed before the underlying genetic mutation in HD was identified (Bates 2005). A consortium of researchers engaged in one of the most ambitious gene hunting efforts of the time. They relied on DNA samples from families in the Lake Maracaibo region of Venezuela, an area with a high density of HD and significant consanguinity. In 1993, the consortium reported the successful discovery of an unstable triplet repeat expansion within IT15 (interesting transcript #15) that cosegregated with coHD (Group 1993). IT15, later called the HUNTINGTIN (HTT) gene, had no significant homology to other genes in the genome and encoded an mRNA of approximately 10 kB and a protein of 350 kDa. A triplet nucleotide repeat, cytosine-adenonsine-guanine (CAG), is found near the 5′ end of the gene's coding region and is translated into a polyglutamine (polyQ) stretch.
HUNTINGTON'S DISEASE: A CLINICAL OVERVIEW OF GENOTYPE–PHENOTYPE CORRELATIONS
Genetic Determinants of Symptom Onset and Disease Progression
With the discovery of HTT, the clinical features of HD could be better defined by investigating genotype-phenotype relationships with greater specificity. A major focus of these early studies was on understanding the relationship between the age when an individual first develops neurological symptoms and the length of the triplet repeat (Duyao et al. 1993; Rubinsztein et al. 1996; Snell et al. 1993) (Fig. 1A). Alleles of HTT with fewer than 35 CAGs were found to pose no risk for causing HD. Individuals harboring alleles with between 36–40 CAGs may or may not develop HD symptoms; those that do tend to become symptomatic late in life. However, those with alleles containing expansions greater than 40 CAGs repeats will eventually develop symptoms of HD if they live long enough. Curiously, individuals who harbor two HD-causing alleles (i.e., homozygous for mutant HTT) appear to develop symptoms about the same age as people with a single allele and the same CAG expansion. However, homozygosity for the CAG mutation has been reported to lead to a more severe clinical course (Squitieri et al. 2003). These observations should be considered cautiously because they are based on very small numbers of people: homozygosity is rare.
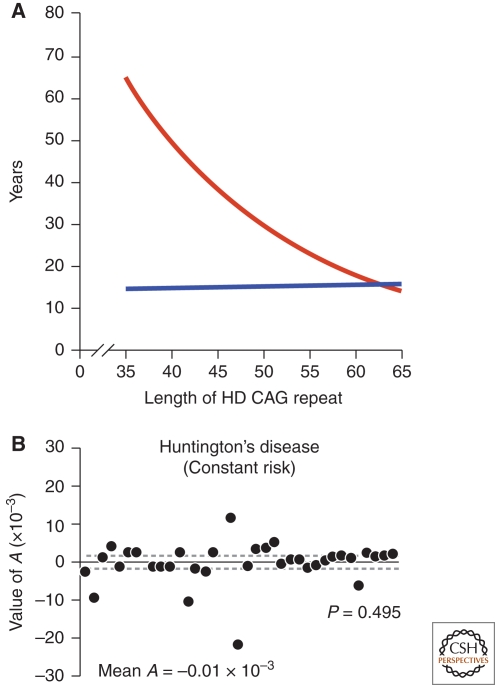
Figure 1.
The role of the CAG expansion in IT-15 on HD pathogenesis. (A) Correlation of HD CAG-repeat length with age at onset. Best-fit curves for age at neurological onset (red) and duration of disease from onset to death (blue), plotted against CAG-repeat length for the expanded mutant allele from Huntington disease (HD) patients. Age at onset is strongly correlated with the CAG-repeat length (r2 = 0.54; p < 0.001), and duration of disease shows no correlation with the CAG-repeat length, suggesting that, after onset of HD, factors independent of the original trigger of pathogenesis determine the rate of progression. Adapted with permission from (Gusella and MacDonald 2006). (B) The kinetics of metabolic decline in Huntington's disease patients is best fit by a constant risk of cell loss. Values of A, the exponent of an equation relating longitudinal metabolic changes to time, do not differ significantly from zero (P = 0.495, Student's t-test). Each point represents the estimated A for an individual patient. Solid line, mean A (mean ± s.d.: −0.01 × 10−3 + 5.3 × 10−3 mM/h1, n = 38 patients) across patient population; dashed lines, 95% confidence interval for the mean value of A (−1.7 × 10−3, 1.7 × 10−3). Adapted with permission from (Clarke et al. 2000).
Interestingly, there is a striking and significant negative relationship between the length of the CAG expansion and the age of symptom onset; the longer the CAG stretch, the earlier symptoms typically appear (Fig. 1A). The most common HD alleles contain 40–50 polyQ. In that range, 50–70% of age of symptom onset appears to be explained by the length of the polyQ stretch, the remainder is determined by other modifying genes and environmental influences (Wexler et al. 2004). At longer polyQ stretches, an even greater proportion of age of symptom onset is explained by the length of the polyQ stretch. Length of the polyQ stretch also appears to influence the progression of pathology, although the link between disease progression and the length of the polyQ stretch appears to be weaker than for age of symptom onset (Brandt et al. 1996; Penney et al. 1997) (Fig. 1A).
The ability to correlate the length of the CAG expansion to clinical phenotype uncovered the molecular basis for genetic anticipation (Telenius et al. 1993; Telenius et al. 1995). Anticipation in HD referred to the puzzling phenomenon whereby inheritance of the mutant HTT allele through the male germ line often led to a more severe clinical course than inheritance through the female germ line. On average, children experienced HD symptoms 8 years earlier than their fathers (Ranen et al. 1995). In those children, the CAG stretch had further expanded. The molecular mechanism responsible for the propensity of the CAG stretch to expand through the male germ line is debated (MacDonald et al. 1993; Telenius et al. 1994; Telenius et al. 1995).
Genetic Determinants of Symptom Profile and Neuropathology
The length of the CAG stretch also affects the types of symptoms. The most common HD alleles (40–50 CAGs) tend to produce classic HD symptoms that manifest during mid-life. The best-known symptoms are excessive uncontrolled jerky motor movements, called chorea, and gait disturbances. Some patients experience dystonia, an increase in muscle tone. Neuropsychiatric symptoms are common, especially depression and anxiety. Some patients report obsessive-compulsive symptoms and effects on cognition. Deficits in executive function appear early; a global dementia often ensues. Unusually long CAG stretches (>50) display symptoms so different that the syndrome is called juvenile HD (JHD), reflecting the earlier onset (Nance and Myers 2001). Unlike adult-onset HD, JHD is associated with a paucity of movements (bradykinesia) and an increased incidence of seizures.
Neuropathological changes are also related to CAG expansion length. The brain region believed to show the earliest changes is the striatum, and within the striatum, medium spiny neurons that contain the neuropeptide enkephalin and express the D2-subtype of dopamine receptor appear to be the most vulnerable (Augood et al. 1996; Ginovart et al. 1997; Le Moine et al. 1990; Marshall et al. 1983; Richfield et al. 1995; Sapp et al. 1995). Subregions of cortex are also significantly affected relatively early (Rosas et al. 2005; Sax et al. 1996). Other regions, such as the cerebellum, are relatively spared (Vonsattel et al. 1985). In JHD, the pathology is more widespread (Myers et al. 1988; Nance and Myers 2001); presumably, as the polyQ expansion increases in length, the mechanisms that restrict the specificity of neurodegeneration to a relatively small subpopulation of neurons in adult onset HD are progressively lost.
The Role of Aging in Huntington's Disease
That an individual harboring a genetic mutation could appear normal for decades before developing a mid-life neurodegenerative disease has puzzled investigators. Some interpret this to mean HD is caused by an underlying biochemical event—the nucleation of a misfolded form of polyQ-expanded HTT or the sufficient accumulation of protein deposits, which takes decades to occur (Chen et al. 2002b). However, it is probably incautious to assume a direct biochemical link between symptoms and biochemistry. In Parkinson's disease, for example, symptoms do not appear until most dopaminergic neurons in the substantia nigra are lost, suggesting that the nervous system has excess capacity before symptoms manifest (Bezard et al. 2003). The nervous system also has powerful cell-autonomous and cell-nonautonomous mechanisms to mitigate the effects of disease-associated mutations (Arrasate et al. 2004; Finkbeiner et al. 2006; Mitra et al. 2009; Palop et al. 2007). Indeed, some adaptive changes may contribute to symptoms under some circumstances (Graybiel 2004; Picconi et al. 2005). Finally, the use of symptoms to detect age of disease onset is somewhat arbitrary. Some clinical tests detect behavioral deficits in patients 15 years before the symptoms appear (Paulsen et al. 2008), and a revision of what constitutes “early-stage” disease may be necessary. Presumably, more sensitive clinical tests could detect deficits earlier.
In 2000, a one-hit model of neurodegeneration was proposed. Clarke et al. carefully counted cells in HD brain sections collected at different disease stages. Surprisingly, they found cell numbers decline exponentially with time with genetic causes of neurodegeneration, such as HD (Clarke et al. 2000) (Fig. 1B). More recently, careful longitudinal live cell measurements in a primary neuron model of HD confirmed and extended the one-hit model by showing that the elevation in the risk of death was proportional to the length of the polyQ expansion beyond the threshold associated with HD (Arrasate et al. 2004; Miller et al. 2010b). The implication of these findings is that the risk that any particular cell would die is relatively constant and independent of the epoch of observation. In other words, the genetic mutation responsible for HD behaves as if it reduces the overall viability of cells by a certain amount, making them more susceptible to apparently random stresses. Although the risk of cell death is constant, cumulative cell loss over time eventually uses up the nervous system's excess capacity and exceeds its mechanisms to cope with cell loss. Network failure results and symptoms manifest (Ying 1996).
What then is the role of aging in HD? Aging is not an absolute requirement. The most common HD alleles (40–50 CAGs) produce symptoms in mid-life. However, longer CAG expansions produce earlier symptoms; the youngest person with HD developed symptoms at age 2. Indeed, electrophysiological abnormalities have been detected in cells derived from mouse models of HD at the embryonic stem cell stage. Aging has been associated with a decline in a variety of pathways relevant to neurodegenerative disease (Beal 1995; Cuervo et al. 2005; Gaczynska et al. 2001; Heydari et al. 1994; Keller et al. 2004; Klivenyi and Vecsei 1997; Massey et al. 2006), especially those that are critical for handling misfolded proteins (Zhou et al. 2003). Thus, aging may further reduce the overall ability of cells to withstand random stresses. The deficits in homeostatic mechanisms caused by aging could overlap or operate independently from those induced by disease-associated forms of HTT, but in either case, aged cells would be predicted to be more susceptible than younger ones. The reader is directed to an excellent article written by Taylor and Dillin (2011) on proteastasis and aging for a fuller account.
HUNTINGTON'S DISEASE AND PROTEOSTASIS
Huntington's Disease: Evidence of a Proteopathy
Having established the CAG expansion as the cause of HD, investigators turned toward understanding how it causes neurodegeneration. The first question was whether neurodegeneration caused by the CAG expansion was mediated at the level of the nucleic acids (DNA and mRNA) that contained them or through the polyQ expansion they encoded in the HTT protein.
CAG expansions might mediate neurodegeneration through abnormal polyQ expansions. Recently, an inducible transgenic mouse model of HD was created with the first exon of HTT (HTTex1), which contains the CAG expansion. In that model, behavioral and pathological deficits appeared when HTTex1 was induced and could be reversed by removing the inducing agent and allowing HTTex1 levels to decrease (Yamamoto et al. 2000). This result excludes a DNA-based mechanism for neurodegeneration. Perhaps the most definitive result was obtained serendipitously years earlier. Hayden and colleagues generated a transgenic mouse that constitutively expressed a human transcript encoding the human HD gene (Goldberg et al. 1996). However, an unplanned stop codon caused the HTT transcript to produce no protein. The animal showed no evidence of a disease-related phenotype, suggesting that a CAG expansion at the DNA or mRNA level was insufficient to produce neurodegeneration.
Others generated a transgenic mouse constitutively expressing the human HD gene but replaced the pure CAG expansion with a mixture of glutamine-encoding CAG and CAA codons (Gray et al. 2008). The mixture did not affect the HTT protein, the polyQ stretch was identical to one encoded by a pure CAG stretch. However, the CAG expansion was less likely to change length during breeding. This mouse, which expressed polyQ-expanded HTT but lacked a pure CAG stretch, had behavioral and pathological features of neurodegeneration that resembled HD. The mix of CAA and CAG codons is significant for another reason. mRNAs containing pure triplet repeat expansions form double-stranded hairpins stabilized by intrastrand base-pairing. The use of alternating CAA and CAG codons disrupts hairpin formation. Because the formation of RNA hairpins has been proposed as a mechanism of toxicity mediated by mRNA-containing triple repeat expansions, the presence of a phenotype despite a manipulation designed to reduce mRNA hairpin formation is consistent with the conclusion that CAG-expansion-mediated neurodegeneration results primarily from the production of proteins that contain polyQ rather than from effects mediated by mRNA. These results strongly implicate proteotoxicity, but some mRNA-mediated toxicity cannot be excluded (Li et al. 2008).
Correlations to Protein Accumulation: HTT Aggregation and Inclusion Body Formation
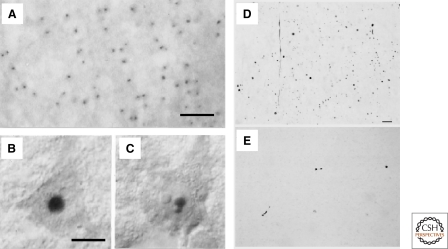
The idea that HD might be a proteopathy arising from abnormal accumulation of mutant HTT got a considerable boost with the generation of the first mouse model of HD (Mangiarini et al. 1996). It expressed a transgene encoding HTTex1 and contained a disease-associated CAG expansion. These mice showed behavioral phenotypes, reduced survival, and gene expression changes that parallel those seen in HD and other HD models based on the full-length HTT protein (Kuhn et al. 2007; Menalled et al. 2009). One neuropathological feature was nuclear accumulations of mutant HTT called intraneuronal nuclear inclusions (INNs) (Davies et al. 1997) (Fig. 2). The appearance of INNs correlated with the onset of behavioral deficits, and the overall burden of INNs correlated with the severity of symptoms. Thus, Davies et al. concluded that INN formation underlies HD. Soon after the observation of INNs were reported in HD models, they were described in HD brains (DiFiglia et al. 1997). Accumulations (or simply inclusion bodies [IBs]) of mutant HTT have also been reported in the cytoplasm, especially the perinuclear area and in dendrites (DiFiglia et al. 1997; Gutekunst et al. 1999). Indeed, dendritic IBs may be more common than those found elsewhere (Gutekunst et al. 1999).
Figure 2.
HD is characterized by abnormal protein deposits containing mutant huntingtin. (A) Huntingtin immunoreactivity in neuronal intranuclear inclusions (hNIIs) and dystrophic neurites in HD brain. Cortex of a juvenile patient shows numerous hNIIs prominently stained. (B and C) Cortical pyramidal neurons in a different juvenile patient shown with Nomarski optics contain one (B) and two (C) hNIIs. A–C, Adapted with permission from (DiFiglia et al. 1997). The nucleolus in each cell is unlabeled. (D–E) Huntingtin aggregates in human postmortem cerebral cortex and striatum from a presymptomatic case. Light micrographs are from the insular cortex (D) and dorsal striatum (E). Large numbers of EM48-immunoreactive aggregates of a wide variety of shapes and sizes are visible in cortex. All of these aggregates are in the neuropil. In contrast, striatal aggregates are exceedingly uncommon. Scale bar, 70 µm. D–E, Adapted with permission from (Gutekunst et al. 1999).
Inclusion Bodies as a Mismatch of Protein Production and Clearance
Abnormal IB accumulations containing mutant HTT, particularly in brain regions affected in HD, provided prima facie evidence that HD is fundamentally associated with protein misfolding and inadequate protein clearance. IBs contain ubiquitin, molecular chaperones, and proteasome subunits, suggesting production of misfolded protein exceeds the ability of cells to clear them (Mitra and Finkbeiner 2008; Sieradzan et al. 1999; Stenoien et al. 1999; Waelter et al. 2001). Generation of an inducible HD mouse model allowed tests of the effects of terminating production of mutant HTT at an age that IBs had already formed and the animal had developed behavioral deficits (Yamamoto et al. 2000). The fact that IBs could be cleared and that behavioral deficits recovered were evidence that neurons must contain cellular mechanisms that metabolize IBs. More recently, experiments using siRNA or antisense oligonucleotides have targeted HTT in several HD models (Boudreau et al. 2009; DiFiglia et al. 2007; Harper et al. 2005). In every case, suppressing mutant HTT production was beneficial, leading to either slowed progression of pathology and behavioral deficits or partial reversal in some cases. Thus, IBs and behavioral deficits appear to result from an imbalance of production and clearance of mutant HTT (Waelter et al. 2001).
How polyQ expansion in HTT yields a mismatch of HTT production and clearance remains unresolved. Discovery of ubiquitin and proteasome subunits in IBs led to investigations of the effect of mutant HTT on proteasome function (Chai et al. 1999). Pharmacological inhibition (Myung et al. 2001) of the proteasome increased IB formation (Wyttenbach et al. 2000), and protein aggregation inhibited proteasome function (Bence et al. 2001; Díaz-Hernández et al. 2006; Holmberg et al. 2004; Venkatraman et al. 2004). The implied feedback between accumulation of aggregation-prone proteins and proteasome inhibition suggested a mechanism (Ardley et al. 2005; Bennett et al. 2005; Ciechanover and Brundin 2003; Ding and Keller 2001; Ding et al. 2002; Gaczynska et al. 2001; Goellner and Rechsteiner 2003; Jana and Nukina 2003; Petrucelli and Dawson 2004): dysregulation of steady-state levels of vast numbers of cellular proteins whose degradation depended on the proteasome. However, other studies reported that the proteasome degrades polyQ-expanded proteins efficiently (Michalik and Van Broeckhoven 2004; Saric et al. 2004). Furthermore, several studies in mouse models of polyQ-induced neurodegeneration detected no proteasome inhibition (Bowman et al. 2005) or contribution of proteasome inhibition to disease progression (Bett et al. 2006).
The dynamics of proteasome function and IB formation have been measured longitudinally in single neurons expressing mutant HTT (Mitra et al. 2009). These studies revealed a transient increase in an artificial proteasome substrate, implying reduced flux through the proteasome, which preceded IB formation. Remarkably, substrate levels fell shortly after IBs formed, suggesting improved flux through the proteasome. Similar findings in vivo were reported recently (Ortega et al. 2010). A transient change in flux through the proteasome associated with polyQ-expanded proteins would likely be undetected using conventional measurements and might explain the conflicting data. However, these findings do not resolve whether the change in flux arises from a direct inhibitory effect of polyQ expanded HTT on the proteasome. Alternatively, polyQ expanded HTT could lead to widespread stresses on cellular protein-fielding machinery and a pleiotropic increase in the load of misfolded proteins to the proteasome, which would compete with the artificial proteasome substrate, increasing its steady-state levels.
Directly measuring the effect of polyQ expansion on cellular clearance of proteins that contain them is difficult. For example, conventional approaches for measuring protein half-life are confounded by aggregation and toxicity from long polyQ expansions. Nevertheless, the available evidence suggests that HTT is normally a long-lived protein (Persichetti et al. 1996). In spinocerebellar ataxia 7, a disease-associated polyQ expansion was associated with overexpression of ataxin-7 (Yoo et al. 2003). The authors suggested that polyQ expansions might stabilize proteins, such as ataxin-7, that contain them, leading to their overproduction. However, steady state levels of HTTex1 in neurons appear inversely correlated to the polyQ length (Arrasate et al. 2004; Miller et al. 2010a). Because different HTTex1 versions were expressed from the identical heterologous promoter, polyQ-expanded HTTex1 might be cleared faster than wild-type versions. Thus, neurons might selectively target proteins with disease-associated polyQ expansions for degradation.
Huntington's Disease, Protein Folding, and the Role of Molecular Chaperones
Scherzinger and colleagues reported polyQ expansions conferred to proteins the propensity to aggregate (Scherzinger et al. 1997) (Fig. 3A,B). Initially, aggregation was reported as an emergent property of polyQ expansions associated with disease. In fact, proteins with sub-threshold polyQ stretches also aggregate (Hollenbach et al. 1999). However, the rate of aggregation dramatically increases as the polyQ expansion grows (Chen et al. 2002a; Georgalis et al. 1998). Moreover, aggregates of HTT with disease-associated polyQ expansions adopt relatively insoluble amyloid-like structures in vitro (Huang et al. 1998; Scherzinger et al. 1997; Scherzinger et al. 1999; Wanker et al. 1999). This relative insolubility helps to distinguish types of inclusions in situ (Arrasate et al. 2004; Kazantsev et al. 1999).
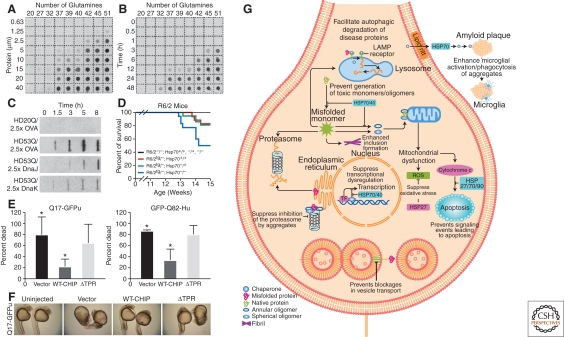
Figure 3.
Aggregation of polyQ expansions is regulated by a network of chaperones and cochaperones. (A) Concentration dependence of polyQ-containing amino-terminal exon 1 peptide fragment of huntingtin (HDex1p) aggregation. Glutathione S-transferase (GST)-HDex1 proteins with polyQ tracts of different lengths were incubated at the indicated concentrations with trypsin for 24 h at 37°C. Aliquots (200 ng) of each protein were then diluted into 0.2 ml of 2% SDS/50 mM dithiothreitol, boiled for 3 min, and filtered through a cellulose acetate membrane. Captured aggregates were detected by incubation with antiAG51 serum (1:1000), followed by incubation with alkaline phosphatase-conjugated antirabbit secondary antibody and the fluorescent substrate AttoPhos. (B) Time course of HDex1p aggregation. The various GST-HDex1 proteins were incubated at a concentration of 20 µM with trypsin. At the indicated times, aliquots (200 ng) of each protein were removed and analyzed by the filter retardation assay as in A. A–B, Adapted with permission from (Scherzinger et al. 1999). (C) Modulation of aggregation of amino-terminal exon 1 fragment of huntingtin (HDexon 1) by Hsp70 and Hsp40 in vitro. Time-dependent formation of SDS-insoluble aggregates of HD20Q and HD53Q (3 µM) in the presence and absence of ovalbumin (OVA) and DnaJ (Hsp40) or DnaK (Hsp70) of Escherichia coli (7.5 µM each) in the absence of ATP as detected in filter-trap assays. Chaperones were added when aggregation was initiated by proteolytic cleavage of GST-HD fusion proteins. OVA served as a nonchaperone control protein. Adapted with permission from Muchowski et al. (2000). (D) Deletion of Hsp70.1 and Hsp70.3 decreases survival in the R6/2 mouse model of HD. Kaplan–Meier survival curve for the indicated genotypes [R6/2−/−; Hsp70+/+ (n = 21), R6/2tg/−; Hsp70+/+ (n = 18), R6/2−/−; Hsp70−/+ (n = 27), R6/2tg/−; Hsp70−/+ (n = 22), R6/2−/−; Hsp70−/− (n = 18), and R6/2tg/−; Hsp70−/− (n = 18)] shows that the absence of Hsp70.1/3 significantly decreased survival of R6/2 mice (log rank: p = 0.033). Nonontransgenic, Hsp70 heterozygous knock-out, or Hsp70 homozygous knock-out mice died during the 14 week time course. Adapted with permission from (Wacker et al. 2009). (E) Carboxy-terminal heat shock protein 70-interacting protein (CHIP) suppresses polyQ toxicity in developing zebrafish. Quantitation of embryo death 24 h after fertilization, after injection at the one-cell stage of Q71-GFPu (left) or GFP-Q82-Htt (right) together with the indicated CHIP plasmids. Death is decreased by coexpression of WT-CHIP but not of mutant CHIP-ΔTPR (deletion of tetratrico peptide repeat). The bars depict the mean and SD of four independent injections for Q71-GFPu (>120 total animals analyzed) and three independent injections for GFP-Q82-Htt (>80 total animals analyzed). *p<0.02, significant difference between groups. (F) Representative bright-field images of 24-h-old survivors from (E) coinjected with Q71-GFPu and the indicated empty vector, WT-CHIP, or mutant CHIP. WT-CHIP partially restores normal embryo length, optical clarity, and development of head, tail, and somite structures. (E–F) Adapted with permission from (Miller et al. 2005). (G) Direct and indirect effects of molecular chaperones on disease protein toxicity. Molecular chaperones might prevent toxicity by blocking inappropriate protein interactions, by facilitating disease protein degradation or sequestration, and by blocking downstream signaling events that lead to neuronal dysfunction and apoptosis. ER, endoplasmic reticulum; ERAD, endoplasmic reticulum-associated degradation; HSP, heat shock protein; LAMP, lysosomal-associated membrane protein; ROS, reactive oxygen species; TF, transcription factor. Adapted with permission from (Muchowski and Wacker, 2005).
Whether HTT forms amyloid in vivo is more controversial. Fibrillar structures have been detected in situ ultrastructurally, and insoluble HTT-containing complexes can be biochemically extracted from brain (Scherzinger et al. 1997; Wanker et al. 1999). However, HD brains show little staining with Congo Red or Thioflavin T, which detect amyloid (DiFiglia et al. 1997). Also, ultrastructural analysis suggests IBs may be substantially amorphous granular material rather than mostly fibrils (DiFiglia et al. 1997; Miller et al. 2010a). Thus, the visible protein accumulations in situ may be mostly less-ordered nonamyloid protein aggregates.
The nomenclature used to describe protein aggregation in cells has sometimes led to confusion, especially in comparison to aggregation of purified proteins. Biochemical techniques can determine with some certainty whether assemblies of aggregated proteins are dimers, trimers, oligomers, or higher ordered species, the ability to make these determinations in cells is much more limited. With light microscopy, it is usually only possible to visualize large deposits, which we prefer to call IBs. Generally, it is not possible to distinguish monomers from dimers, trimers, or oligomers in most cases, so we prefer to avoid using the term “aggregates” altogether. For lack of a better word, we have chosen to describe intracellular forms of aggregation-prone proteins that are not bound up in an IB as “diffuse” to reflect the limitations of light microscopy. Similarly, we think it is prudent to avoid terms such as “soluble” to refer to diffuse pools of aggregation prone protein or “insoluble” to refer to visible protein deposits unless the necessary additional biochemical tests have been performed to make that determination. The mere collection of proteins into a visible protein deposit does not prove the proteins in the IBs are insoluble. Similarly, the “diffuse” pool of proteins might contain a substantial amount of SDS-resistant multimeric species.
Identification of HTT facilitated the creation of cell and animal disease models. HD models in yeast (Krobitsch and Lindquist, 2000; Muchowski et al. 2000; Willingham et al. 2003), Caenorhabditis elegans (Faber et al. 1999; Kim et al. 2002; Satyal et al. 2000), and Drosophila (Jackson et al. 1998) are amenable to unbiased screens for genes that modify HTT toxicity. Because the polyQ expansion is believed to confer a toxic function to HTT and the toxic function might be unrelated to normal function, unbiased screens might reveal critical biological networks that have eluded conventional hypothesis-driven research. In the first screen for genetic enhancers of polyQ toxicity in Drosophila, two molecular chaperones, dHDJ1, a homolog of heat shock protein 40 (HSP40), and dTPR2, a homolog of tetratricopeptide repeat protein 2, were potent suppressors of HTT toxicity (Kazemi-Esfarjani and Benzer 2000). In the first screen for genetic enhancers of the toxicity of an amino-terminal fragment of mutant HTT in yeast, host proteins in the protein folding, stress response, and ubiquitin-dependent protein catabolism pathways were significantly overrepresented among the list of modifiers of mutant HTT toxicity (Willingham et al. 2003). For example, loss of cytoplasmic or ER versions of the molecular chaperone HSP40 significantly enhanced HTT toxicity. Similarly, a screen was performed for genes whose suppression led to the premature appearance of inclusion bodies by a polyQ containing polypetide in Caenorhabditis elegans (Nollen et al. 2004). Five classes of genes were discovered, including those involved in RNA metabolism, protein synthesis, protein folding, protein degradation and protein trafficking.
Before molecular chaperones were implicated in HTT toxicity, they already had been suspected in pathogenesis. Several studies reported the effects of manipulating chaperone function on polyQ aggregation, inclusion formation, and toxicity in vitro and in vivo. Hsp40 and/or Hsp70 suppress polyQ-dependent aggregation and neurodegeneration in a variety of systems (Carmichael et al. 2000; Jana et al. 2000; Kim et al. 2003; Kobayashi et al. 2000; Krobitsch and Lindquist 2000; Muchowski et al. 2000; Wacker et al. 2004; Warrick et al. 1999) (Fig. 3C). A yeast chaperone, Hsp104, without a known mammalian ortholog, disaggregates polyQ and suppresses cell death (Glover and Lindquist 1998; Krobitsch and Lindquist 2000; Parsell et al. 1994; Satyal et al. 2000; Wyttenbach et al. 2000). Other chaperones also disrupt polyQ aggregation and toxicity (Kitamura et al. 2006; Tam et al. 2006). Overexpressing or reducing Hsp70 in mice, for instance, has been associated with suppressing or enhancing polyQ toxicity, respectively (Cummings et al. 2001; Wacker et al. 2009) (Fig. 3D). Additionally, overexpressing Hsp104 in a mouse HD model was associated with reduced protein deposition and better survival (Vacher et al. 2005). The effects of chaperones on neurodegeneration are partly mediated by direct interactions that govern protein misfolding, but probably also involve additional more complex and indirect effects (Muchowski and Wacker 2005) (Fig. 3E).
The Significance of Inclusion Bodies to Neurodegeneration
The correlation of onset and severity of behavioral deficits and IB burden in mouse disease models implicated IBs themselves as the pathogenic species for neurodegeneration (Davies et al. 1997) (Fig. 4). IBs might sequester critical proteins to cause neurodegeneration (Chai et al. 2002; Donaldson et al. 2003; McCampbell et al. 2000; Preisinger et al. 1999). However, several findings did not fit a simple causal connection. In a primary neuron model, IB formation could be reduced by expressing a ubiquitin ligase, but cell death was exacerbated (Saudou et al. 1998). Also, several trophic factors suppressed neurodegeneration induced by mutant HTT while simultaneously increasing IB formation. Orr, Zoghbi and colleagues reported analogous findings from a mouse model of a related polyQ disorder, spinocerebellar ataxia 1 (SCA1) (Klement et al. 1998). They showed that deletion of a self-association domain in SCA1, which reduced IB formation, worsened pathology, and behavioral deficits. Subsequent neuropathology studies implied that IB formation could be dissociated from neurodegeneration (Gutekunst et al. 1999; Kuemmerle et al. 1999; Slow et al. 2005).
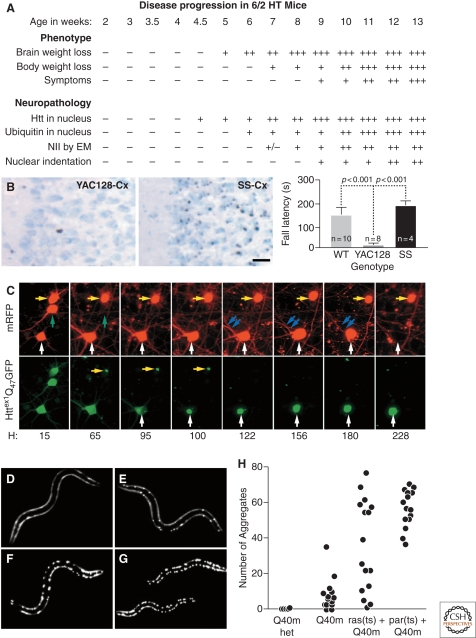
Figure 4.
HD pathogenesis unfolds as a complex temporal dynamic, instigated by polyQ-expanded huntingtin but shaped by a network of cellular adaptive responses. (A) In the R6/2 mouse model of HD, IB formation precedes and correlates to behavioral deficits. Adapted with permission from (Davies et al. 1997). However, the Shortstop mouse model of HD (B) has more IBs (left, micrographs) but fewer behavioral deficits (right, graph) than the YAC128 mouse model of HD. Adapted with permission from (Slow et al. 2005). (C) The paradox could be explained if IB formation behaves as a coping response to more diffuse forms of mutant huntingtin. Here striatal neurons were transfected with mutant huntingtin tagged with GFP and followed longitudinally. Levels of diffuse mutant huntingtin decrease to nearly baseline levels after IB formation and, at the same, begin to show a lower risk of death (i.e., better survival). Adapted with permission from Arrasate et al. (2004). (D–H) Progressive disruption of cellular folding capacity by misfolded proteins. Fluorescent images of representative L2 larvae at the permissive temperature of heterozygous (D) or homozygous (E) Q40 m, ras (ts)+Q40 m (F), and paramyosin (ts)+Q40 m (G) strains. (H) Number of visible aggregates in L2 larvae expressing indicated proteins. ras (ts)+Q40 m in (F) and (G) denotes the fluorescent progeny of an F2 ras (ts) animal expressing Q40 m; these progeny could be either homozygous or heterozygous for Q40 m. Adapted with permission from Gidalevitz et al. (2006).
These reports (Saudou et al. 1998), (Klement et al. 1998) spurred a controversy about IB formation in neurodegeneration (Davies et al. 1998; Floyd and Hamilton 1999; Ross 1997; Rubinsztein et al. 1999; Sisodia 1998). Defining the role of IBs in HD exemplifies a general problem in understanding the significance of changes to neurodegenerative disease (Finkbeiner et al. 2006). How can a disease-relevant phenotype, such as death or dysfunction, be related to an earlier change, such as the formation of an IB? The conventional approach of collecting snapshots of disease progression and making gross correlations is fundamentally flawed. It can mislead investigators to interpret their data exactly opposite to the underlying pathobiology. No wonder the study of neurodegenerative disease is fraught with controversies; the potential for misinterpretation is great.
Inclusion Body Formation as a Regulated Mechanism to Cope with Misfolded Proteins
An automated microscope system provides an innovative solution to this dilemma. It follows individual neurons as they express mutant HTT and show neurodegeneration and analyzes images in an automated user-independent fashion (Arrasate and Finkbeiner 2005; Arrasate et al. 2004). The images record individual neurons until their fate is known, and powerful statistical tools used for clinical studies can be applied to these data to identify factors that predict a particular fate. Importantly, the tool addresses thorny questions in neurodegenerative disease (Orr 2004). Do particular changes, such as IB formation, have prognostic value? Which are the most important? Do they predict a better outcome or a worse one? Which changes are causal and which are effects? What “system” of factors determines neurodegeneration?
This new system allowed us to revisit the role of IBs in HD. Surprisingly, striatal neurons that formed IBs had lower risks of death; they survived better than neurons without IBs (Arrasate et al. 2004). This finding has been replicated by others (Miller et al. 2010a; Mitra et al. 2009; Takahashi et al. 2008; Taylor et al. 2003). The factor that predicted neuronal death was the level of “diffuse” mutant HTT, all forms of mutant HTT other than those bound in an IB. By 24–48 h after IB formation, levels of diffuse mutant HTT fell to nearly baseline levels along with the risk of death (Arrasate et al. 2004; Miller et al. 2010a). A simple model integrated IB formation as a coping response to more toxic forms of diffuse mutant HTT (Fig. 5). IBs were proposed to mitigate the effects of diffuse mutant HTT by sequestering it, thereby reducing its chemical activity and/or possibly refolding it into a more inert form (Arrasate et al. 2004; Miller et al. 2010a).
Figure 5.
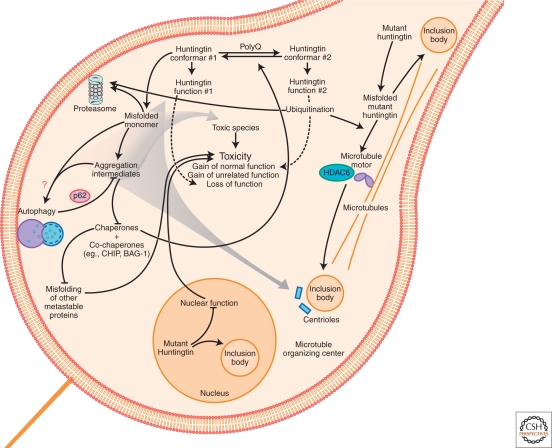
Mechanisms of neurodegeneration induced by protein malfolding in HD. A diagram depicting some of the pathways by which misfolded mutant huntingtin is collected into IBs or cleared. Toxicity could result from direct aberrant interactions between mutant huntingtin and myriad specific effector proteins. Mutant huntingtin could also cause neurodegeneration indirectly by stressing the protein homeostasis system sufficiently that other metastable proteins fail to fold properly and widespread dysfunction of the proteome ensues. Mutant huntingtin induces neurodegeneration predominantly through gain-of-function (GOF) mechanisms, although huntingtin loss-of-function (LOF) may have a role. GOF mechanisms may involve functions unrelated to huntingtin's normal function, but simultaneous GOF and LOF might also result if polyQ expansion increased a normal function of huntingtin to supraphysiological levels at the expense of another normal function of huntingtin.
Why then does IB formation grossly correlate positively with toxicity in some cases and negatively in others? The lesson from single-cell longitudinal work is that inferences made about the significance of pathological changes from gross correlations are unreliable and can be misleading (Table 1). A manipulation that increases IB formation because it increases the load of misfolded protein is going to be associated with greater toxicity because, like any homeostatic response, it typically will be only partially effective at mitigating the perturbation to the equilibrium. In other words, a homeostatic response makes things less bad than they would be without the response but never as good as they would be if the instigating problem were eliminated altogether. On the other hand, a manipulation might somehow accelerate IB formation without increasing the load of misfolded protein. It might act on the process of IB formation itself and downstream of the initial misfolding, leading to even more reduced levels of diffuse misfolded protein than in the absence of the manipulation and, therefore, a reduction in toxicity. In the first case, toxicity and IB formation go in the same direction, and in the second, they go in the opposite direction. Yet, both would be consistent with the simple model that incorporating misfolded proteins into IBs reduces their toxicity (Table 1). The roles of IB formation in pathogenesis may someday be shown to be more complex, but no published findings indicate that it has to be.
Table 1.
As a coping response, inclusion body formation should correlate better with levels of misfolded protein than neurodegeneration.
| Manipulation | Possible examples | Levels of diffuse misfolded protein | Inclusion body formation | Neurodegeneration |
|---|---|---|---|---|
| ↑ Load of misfolded protein | 1. Chaperone deficiency leading to increased production (Wacker et al. 2009) | ↑ | ↑/- | ↑ |
| 2. Inhibition of proteasome (Chai et al. 1999; Mitra et al. 2009; Wyttenbach et al. 2000) or autophagic clearance (Pandey et al. 2007) | ||||
| 3. Oxidative stress (Charvin et al. 2005) | ||||
| ↓ Load of misfolded protein | 1. Suppression of mutant Htt expression (Boudreau et al. 2009; DiFiglia et al. 2007; Harper et al. 2005; Yamamoto et al. 2000) | ↓ | ↓ | ↓ |
| 2. Induction of autophagic clearance (Rideout et al. 2004; Sarkar et al. 2007a; Sarkar et al. 2007b; Yamamoto et al. 2006) | ||||
| ↑ Extent of inclusion body formation | 1. Small-molecule inducers (Bodner et al. 2006) | ↓ | ↑ | ↓ |
| 2. Htt mutations (Slow et al. 2005) | ||||
| 3. NMDAR-dependent synaptic activity (Okamoto et al. 2009) | ||||
| ↓ Extent of inclusion body formation | 1. Intereference with Ubiquitin ligase (Saudou et al. 1998) | ↑ | ↓ | ↑ |
| 2. Mutation of dimerization domain (Klement et al. 1998) | ||||
| 3. Rhes (Subramaniam et al. 2009) | ||||
| ↑ Ability to withstand toxic misfolded protein/↑cellular survival and longevity | 1. Growth factors (Saudou et al. 1998; Yamamoto et al. 2006) | ↑/- | ↑/- | ↓ |
| 2. Activation of prosurvival pathways (Humbert et al. 2002) |
If a manipulation reduces the susceptibility of a cell to the toxicity of misfolded protein, the cell may live longer and may withstand accumulation of higher levels of toxic protein leading to more IBs formation. In such cases, gross neurodegeneration may be reduced although levels of diffuse mutant protein are higher.
Several groups showed IB formation is regulated, at least for IBs in the perinuclear area. In nonneuronal cells, these inclusions or aggresomes are associated with the microtubule-organizing center (MTOC) (Johnston et al. 1998; Lelouard et al. 2004; Shimohata et al. 2002). Indeed, IB formation itself appears to be microtubule dependent, suggesting misfolded proteins are gathered to the MTOC by microtubule-based transport (Muchowski et al. 2002). HDAC6 might function in this process, possibly in some adapter capacity (Kawaguchi et al. 2003) or as part of a complex process involving p62 and ubiquitin proteasome and autophagic mechanisms of protein clearance (Babu et al. 2005; Iwata et al. 2005; Nagaoka et al. 2004; Pandey et al. 2007; Webb et al. 2004; Zatloukal et al. 2002). Activating Ca2+ influx through the NMDA receptor promotes IB formation, suggesting that the process in neurons is regulated by synaptic activity (Okamoto et al. 2009). Intriguingly, pharmacological manipulations of the NMDA receptor that promoted IB formation reduced disease phenotypes. Conversely, Rhes, a protein implicated in HTT sumoylation, mediates mutant HTT-induced cytotoxicity, apparently by interfering with IB formation (Subramaniam et al. 2009). The complex process and regulation of IB formation may also explain why many proteins aggregate readily in a test tube, but form IBs in cells with significantly different propensities.
Neurons also clear IBs in vitro. In a few neurons, IBs cleared spontaneously despite the continued production of mutant HTT (Arrasate et al. 2004). Because proteins must be unfolded to be degraded in proteasomes, other protein clearance pathways might clear IBs. Macroautophagy can, in principle, clear aggregation-prone proteins (Ravikumar and Rubinsztein 2004; Shacka et al. 2008). In this process, double-membrane organelles called autophagosomes engulf cellular cargo and fuse with lysosomes to form autophagolysosomes, which then degrade the organelle's contents (Kim and Klionsky 2000; Meijer and Codogno 2004). HTT expression is associated with up-regulated autophagy (Kegel et al. 2000; Petersén et al. 2001; Rogers and Leavitt 2004), and autophagy degrades polyQ-expanded proteins (Qin et al. 2003; Ravikumar et al. 2002; Yamamoto et al. 2006). Induction of autophagy is associated with reductions in IBs, but it is controversial whether IBs are engulfed in toto and targeted to autophagosomes. Autophagy might reduce IBs by clearing IBs precursors and eventually depleting them of mutant HTT (Fortun et al. 2003; Rideout et al. 2004; Sarkar et al. 2007b; Wong et al. 2008a).
AN INTEGRATED VIEW OF PROTEIN MISFOLDING AND NEURODEGENERATION IN HUNTINGTON'S DISEASE
A Network for Protein Homeostasis
Reductionist approaches to mechanisms of neurodegeneration in HD have been helpful for uncovering important disease-related molecular and cellular changes. However, the initial insights have often been revised after additional factors were analyzed. Including temporal dynamics and simultaneous measurements of HTT levels, IB formation, and fate, for example, revealed that proteasome inhibition occurred transiently and was time-linked to IB formation, and that IB formation led to reduced levels of diffuse forms of mutant HTT and improved survival (Arrasate et al. 2004; Mitra et al. 2009). Now excellent evidence links the molecular chaperone system, ubiquitin proteasome pathway, and autophagic protein clearance pathway. These are mediated by specific molecules (e.g., HDAC6, BAG1, and CHIP) in some cases, and the proteasome and autophagic pathways communicate (Al-Ramahi et al. 2006; Jana and Nukina 2003; Lüders et al. 2000; Massey et al. 2006; Miller et al. 2005; Pandey et al. 2007). The new view is that cells maintain healthy proteins through an adaptable network of interconnected pathways that sense misfolded proteins and either refold them or target them for clearance.
This idea was shown convincingly in C. elegans. Morimoto and colleagues (Gidalevitz et al. 2006) examined effects of expression of a polyQ expansion fused to a fluorescent protein on the folding of other unrelated proteins that harbored temperature-sensitive (ts) mutations. At low temperatures, these proteins remain folded and functioned normally. At slightly elevated temperatures, they misfolded and aggregated. Remarkably, polyQ expansions with lengths close to or slightly above the threshold for human disease led to misfolding of the ts mutant proteins at normal temperatures, even though the two proteins did not interact directly. The likeliest explanation is that the polyQ proteins competed with the ts mutant proteins for access to the chaperone system. The capacity of the chaperone system, which was sufficient to prevent aggregation of either misfolded protein alone, was exceeded with both. If these findings apply to humans, the implications are profound. The human chaperone system may have evolved to match the average load of metastable proteins in humans. If so, any polymorphisms affecting protein stability could be genetic modifiers of protein misfolding diseases, whether or not those polymorphisms affect function. Moreover, small sequence differences in proteins among species, which are often ignored in disease models, might be important if they affect protein stability.
From a network perspective, a cogent model might combine the dynamics of IB formation, proteasome inhibition, and IB clearance. A protein prone to misfold, such as mutant HTT, presumably would be sensed initially by the chaperone system. If the load exceeds the capacity of the cell's chaperone system, it may deprive other endogenous metastable proteins of the chaperones they need for proper folding and lead to a substantial increase in the amount of catastrophically misfolded proteins requiring clearance. The increased load would lead to a “traffic jam” of proteins requiring proteasome-dependent degradation and might be detected experimentally as an accumulation of an artificial proteasome substrate. The stress on the chaperone system and protein clearance machinery could signal the cell that emergency measures are needed to avoid apoptosis, including the induction of IB formation and the longer-term up-regulation of protein clearance pathways, including the ubiquitin proteasome pathway and the autophagic clearance pathway. To the extent that IB formation results in homotypic aggregation of a protein and quantitative reduction in its levels through storage in inert structures, it would reduce the nonnative surfaces requiring chaperone protection. The resulting stress relief on the chaperone system would reduce the load of misfolded proteins requiring proteasome degradation. Experimentally, this might look like improved flux of an artificial proteasome substrate. If IB formation was part of a broader cellular coping response, which involved an up-regulation of autophagy and proteasome function, levels of mutant HTT might remain low after IB formation and some IBs might spontaneously be cleared.
How Does Protein Misfolding Lead to Neurodegeneration?
Several lines of evidence indicate that HD is fundamentally a proteopathy and that measures to mitigate protein misfolding or to promote the clearance of misfolded proteins reduce behavioral deficits and neuropathological abnormalities. Still, the fundamental question remains. How do misfolded proteins lead to neurodegeneration? The fact that the polyQ expansion confers to HTT (1) the propensity to aggregate and form IBs and (2) one or more toxic functions led early on to the idea that those two properties were directly linked: aggregation somehow mediated toxicity in HD (Thakur et al. 2004).
The early focus on the role of IB formation and amyloid led to the concept of a toxic protein species, a particular form of aggregated HTT that was envisioned to be responsible for a disproportionate amount of neuronal toxicity. Initially, that species was proposed to be IBs (Becher et al. 1998; Davies et al. 1997; DiFiglia et al. 1997; Yang et al. 2002). Toxicity was proposed to arise secondarily from sequestration of other critical proteins (Donaldson et al. 2003; McCampbell et al. 2000; Preisinger et al. 1999). The amyloid hypothesis of neurodegeneration has occupied a special place in the field for decades (Tanzi and Bertram, 2005; Wirths et al. 2004), although it has decreased on hard times now. Sequestration of key proteins by IBs is insufficient to explain the observed dysfunction (Bennett et al. 2005; Yu et al. 2002). Moreover, functional amyloids exist in mammals and appear to serve useful functions, indicating that amyloid formation per se is not toxic (Fowler et al. 2006).
More recently, attention shifted to submicroscopic molecular assemblies called oligomers (Behrends et al. 2006; Glabe and Kayed, 2005; Kayed et al. 2003; Nagai et al. 2003; Nandi, 1996; Onodera et al. 1997; Sánchez et al. 2003; Sathasivam et al. 2010; Takahashi et al. 2008; Wacker et al. 2004; Wong et al. 2008b) or malfolded monomers (Nagai et al. 2007; Wang et al. 2006; C Peters-Libeu, E Rutenber, J Miller, et al. unpubl.). How oligomers or malfolded monomers are toxic and trigger neurodegeneration remains a matter of speculation. Tools to resolve these questions are limited. No tools detect specific protein species in situ and link functions to particular proteins, especially in the context of and as distinct from other submicroscopic species. HTT appears to adopt many conformations, even as a monomeric protein, and an equilibrium likely exists among the conformers and assemblies. Proteins cannot be produced with a particular homogeneous conformation and remain in that pure conformation after they are delivered to cells. Despite these limitations, oligomers might cause toxicity by creating pores in membranes (Kayed et al. 2004) or by binding to specific cellular proteins, such as receptors, and altering their function (Trommer et al. 2005; Walsh et al. 2002). Monomers may also bind to transcription factors or other targets and degrade their function (Schaffer et al. 2004).
Because the effects of protein misfolding might be mediated through a proteostasis network, the concept of a toxic species may require revision. Chaperones recognize nonnative folds in proteins through unfolded-polypeptide-binding domains, which prefer to bind short sequences (∼5–7 amino acids) enriched in hydrophobic residues (Gething 1996). From the perspective of the cell's molecular chaperone system, it may be more productive to think of “toxic folds” (or “unfolds”) rather than “toxic species” (Ignatova and Gierasch 2006; Poirier et al. 2005). By the same logic, the most stressful forms of aggregation-prone proteins would be predicted to be those that deliver the largest dose of binding sites for chaperones per protein and are the most difficult for the cell to clear. The key unanswered question, therefore, is whether certain monomeric, oligomeric, or aggregated forms of HTT are particularly effective at binding chaperones and stressing the proteostasis system. Because protein aggregation often buries hydrophobic surfaces of polypeptides so they are unavailable to chaperones, it is possible, in principle, to understand how malfolded monomers might confer the greatest toxicity per protein and how aggregation might mitigate toxicity under some circumstances. Stress on the proteostasis system from mutant HTT could reduce cellular folding capacity sufficiently that myriad other metastable proteins misfold, producing a complex loss-of-function phenotype and eventual neurodegeneration.
Finally, enhancing a normal function might explain how polyQ expansions confer toxicity (Duvick et al. 2010; Kratter and Finkbeiner 2010; Nedelsky et al. 2010). Proteins containing polyQ expansions might adopt two or more conformations, and each might be linked to a specific function. Different conformers might exist in equilibrium, but disease-associated polyQ expansions shift this equilibrium so that one conformation and function (e.g., gene transcription, protein trafficking, etc.) is effectively overexpressed. In this model, neurodegeneration effectively is the result of “too much of a good thing.” It has been challenging to test this hypothesis for HD—HTT functions are poorly defined. Nevertheless, structural studies suggest that the polyQ expansion is flexible (Klein et al. 2007; Masino et al. 2002) and that it can adopt multiple conformations (Kim et al. 2009). For SCA1, SBMA, and SCA7, a disease-associated polyQ expansion might affect normal functions that contribute to disease (Bowman et al. 2007; Duvick et al. 2010; Kratter and Finkbeiner 2010; Lam et al. 2006; Lim et al. 2008; Nedelsky et al. 2010; Yoo et al. 2003).
CONCLUDING REMARKS
After 140 years, many questions remain about HD. The genetic mutation that causes all HD leads to an abnormal expansion of a polyQ tract in the HTT protein. In turn, the polyQ expansion confers to HTT one or more toxic functions and the propensity to aggregate and accumulate intracellularly. Cells contain complex interconnected molecular networks to prevent proteins from misfolding and to correct or degrade those that become misfolded. Those networks might sense mutant HTT as a threat and initiate beneficial coping responses, but ultimately these responses only delay HD. How a polyQ expansion in mutant HTT induces neurodegeneration is a mystery, but monomeric or aggregated protein species might form structures that confer particular toxic functions, and sufficient nonnative folds intracellularly might overwhelm the molecular chaperone system and cause a collapse of the cellular proteostasis system or promote too much of one of the normal functions of HTT. Regardless, efforts to pharmacologically reduce HTT misfolding or mutant HTT levels by inducing proteasome function or autophagy could have therapeutic potential.
Footnotes
Editors: Richard I. Morimoto, Jeffery W. Kelly, and Dennis J. Selkoe
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Al-Ramahi I, Lam YC, Chen H-K, de Gouyon B, Zhang M, Pérez AM, Branco J, de Haro M, Patterson C, Zoghbi HY, et al. 2006. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J Biol Chem 281: 26714–26724 [DOI] [PubMed] [Google Scholar]
- Ardley HC, Hung C-C, Robinson PA 2005. The aggravating role of the ubiquitin–proteasome system in neurodegeneration. FEBS Lett 579: 571–576 [DOI] [PubMed] [Google Scholar]
- Arrasate M, Finkbeiner S 2005. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci 102: 3840–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810 [DOI] [PubMed] [Google Scholar]
- Augood SJ, Faull RLM, Love DR, Emson PC 1996. Reduction in enkephalin and substance P messenger RNA in the striatum of early grade Huntington's disease: A detailed cellular in situ hybridization study. Neuroscience 72: 1023–1036 [DOI] [PubMed] [Google Scholar]
- Babu JR, Geetha T, Wooten MW 2005. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem 94: 192–203 [DOI] [PubMed] [Google Scholar]
- Bates GP 2005. History of genetic disease: The molecular genetics of Huntington disease—a history. Nat Rev Genet 6: 766–773 [DOI] [PubMed] [Google Scholar]
- Beal MF 1995. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol 38: 357–366 [DOI] [PubMed] [Google Scholar]
- Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, Ross CA 1998. Intranuclear neuronal inclusions in Huntington's disease and Dentatorubral and Pallidoluysian atrophy–Correlation between the density of inclusions and IT-15 CAG triplet repeat length. Neurobiol Dis 4: 387–397 [DOI] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Böttcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. 2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23: 887–897 [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292: 1552–1555 [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Bence NF, Jayakumar R, Kopito RR 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell 17: 351–365 [DOI] [PubMed] [Google Scholar]
- Bett JS, Goellner GM, Woodman B, Pratt G, Rechsteiner M, Bates GP 2006. Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington's disease mice: Exclusion of proteasome activator REGγ as a therapeutic target. Hum Mol Genet 15: 33–44 [DOI] [PubMed] [Google Scholar]
- Bezard E, Gross CE, Brotchie JM 2003. Presymptomatic compensation in Parkinson's disease is not dopamine-mediated. Trends Neurosci 26: 215–221 [DOI] [PubMed] [Google Scholar]
- Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, McLean PJ, Young AB, Housman DE, Kazantsev AG 2006. Pharmacological promotion of inclusion formation: A therapeutic approach for Huntington's and Parkinson's disease. Proc Natl Acad Sci 103: 4246–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL 2009. Nonallele-specific silencing of mutant and wild-type huntingtin shows therapeutic efficacy in Huntington's disease mice. Mol Ther 17: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AB, Lam YC, Jafar-Nejad P, Chen H-K, Richman R, Samaco RC, Fryer JD, Kahle JJ, Orr HT, Zoghbi HY 2007. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet 39: 373–379 [DOI] [PubMed] [Google Scholar]
- Bowman AB, Yoo S-Y, Dantuma NP, Zoghbi HY 2005. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin—proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum Mol Genet 14: 679–691 [DOI] [PubMed] [Google Scholar]
- Brandt J, Bylsma FW, Gross R, Stine OC, Ranen N, Ross CA 1996. Trinucleotide repeat length and clinical progression in Huntington's disease. Neurology 46: 527–531 [DOI] [PubMed] [Google Scholar]
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC 2000. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease. Proc Natl Acad Sci 97: 9701–9705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL 1999. Evidence for proteasome involvement in polyglutamine disease: Localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet 8: 673–682 [DOI] [PubMed] [Google Scholar]
- Chai Y, Shao J, Miller VM, Williams A, Paulson HL 2002. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc Natl Acad Sci 99: 9310–9315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvin D, Vanhoutte P, Pages C, Borrelli E, Caboche J 2005. Unraveling a role for dopamine in Huntington's disease: The dual role of reactive oxygen species and D2 receptor stimulation. Proc Natl Acad Sci 102: 12218–12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ferrone FA, Wetzel R 2002b. Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci 99: 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Berthelier V, Hamilton JB, O'Nuallain B, Wetzel R 2002a. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry 41: 7391–7399 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P 2003. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 40: 427–446 [DOI] [PubMed] [Google Scholar]
- Clarke G, Collins RA, Leavitt BR, Andrews DF, Hayden MR, Lumsden CJ, McInnes RR 2000. A one-hit model of cell death in inherited neuronal degenerations. Nature 406: 195–199 [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A 2005. Autophagy and aging: The importance of maintaining “clean” cells. Autophagy 1: 131–140 [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Sun Y, Opal P, Anatalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY 2001. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet 10: 1511–1518 [DOI] [PubMed] [Google Scholar]
- Davies SW, Beardsall K, Turmaine M, DiFiglia M, Aronin N, Bates GP 1998. Are neuronal intranuclear inclusions the common neuropathology of triplet-repeat disorders with polyglutamine-repeat expansions? Lancet 351: 131–133 [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90: 537–548 [DOI] [PubMed] [Google Scholar]
- Díaz-Hernández M, Valera AG, Morán MA, Gómez-Ramos P, Alvarez-Castelao B, Castaño JG, Hernández F, Lucas JJ 2006. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J Neurochem 98: 1585–1596 [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277: 1990–1993 [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et al. 2007. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci 104: 17204–17209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Keller JN 2001. Proteasomes and proteasome inhibition in the central nervous system. Free Radic Biol Med 31: 574–584 [DOI] [PubMed] [Google Scholar]
- Ding Q, Lewis JJ, Strum KM, Dimayuga E, Bruce-Keller AJ, Dunn JC, Keller JN 2002. Polyglutamine expansion, protein aggregation, proteasome activity, and neural survival. J Biol Chem 277: 13935–13942 [DOI] [PubMed] [Google Scholar]
- Donaldson KM, Li W, Ching KA, Batalov S, Tsai C-C, Joazeiro CAP 2003. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc Natl Acad Sci 100: 8892–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J, Giesler G, Zoghbi H, Orr HT 2010. SCA1-like disease in mice expressing wild type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron 67: 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M 1993. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet 4: 387–392 [DOI] [PubMed] [Google Scholar]
- Faber PW, Alter JR, MacDonald ME, Hart AC 1999. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci 96: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S, Cuervo AM, Morimoto RI, Muchowski PJ 2006. Disease-modifying pathways in neurodegeneration. J Neurosci 26: 10349–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd JA, Hamilton BA 1999. Intranuclear inclusions and the ubiquitin-proteasome pathway: Digestion of a red herring? Neuron 24: 765–766 [DOI] [PubMed] [Google Scholar]
- Fortun J, Dunn WA Jr, Joy S, Li J, Notterpek L 2003. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J Neurosci 23: 10672–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW 2006. Functional amyloid formation within mammalian tissue. PLoS Biol 4: e6 ( 0100–0107). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaczynska M, Osmulski PA, Ward WF 2001. Caretaker or undertaker? The role of the proteasome in aging. Mech Aging Dev 122: 235–254 [DOI] [PubMed] [Google Scholar]
- Georgalis Y, Starikov EB, Hollenbach B, Lurz R, Scherzinger E, Saenger W, Lehrach H, Wanker EE 1998. Huntingtin aggregation monitored by dynamic light scattering. Proc Natl Acad Sci 95: 6118–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M-J 1996. Molecular chaperones: Clasping the prize. Curr Biol 6: 1573–1576 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474 [DOI] [PubMed] [Google Scholar]
- Ginovart N, Lundin A, Farde L, Halldin C, Backman L, Swahn CG, Pauli S, Sedvall G 1997. PET study of the pre- and post-synaptic dopaminergic markers for the neurodegenerative process in Huntington's disease. Brain 120: 503–514 [DOI] [PubMed] [Google Scholar]
- Glabe CG, Kayed R 2005. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology, S74–S78 [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S 1998. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Goellner GM, Rechsteiner M 2003. Are Huntington's and polyglutamine-based ataxias proteasome storage diseases? Int J Biochem Cell Biol 35: 562–571 [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Kalchman MA, Metzler M, Nasir J, Zeisler J, Graham R, Koide HB, O'Kusky J, Sharp AH, Ross CA, et al. 1996. Absence of disease phenotype and intergenerational stability of the CAG repeat in transgenic mice expressing the human Huntington disease transcript. Hum Mol Genet 5: 177–185 [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu X-H, Tao J, Yamazaki I, Li S-H, Sun YE, et al. 2008. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci 28: 6182–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM 2004. Network-level neuroplasticity in cortico-basal ganglia pathways. Parkinsonism Relat Disord 10: 293–296 [DOI] [PubMed] [Google Scholar]
- Group THsDCR 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983 [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME 2006. Huntington's disease: Seeing the pathogenic process through a genetic lens. Trends Biochem Sci 31: 533–540 [DOI] [PubMed] [Google Scholar]
- Gutekunst C-A, Li S-H, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li X-J 1999. Nuclear and neuropil aggregates in Huntington's disease: Relationship to neuropathology. J Neurosci 19: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper P 2002. Huntington's disease: A historical background. In Huntington's Disease (ed. Bates G., Harper P.S., Jones L.), pp. 3–37 Oxford, Oxford University Press [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL 2005. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci 102: 5820–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari AR, Takahashi R, Gutsmann A, You S, Richardson A 1994. Hsp70 and aging. Experientia 50: 1092–1098 [DOI] [PubMed] [Google Scholar]
- Hollenbach B, Scherzinger E, Schweiger K, Lurz R, Lehrach H, Wanker EE 1999. Aggregation of truncated GST-HD exon 1 fusion proteins containing normal range and expanded glutamine repeats. Philos Trans R Soc Lond B Biol Sci 354: 991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI 2004. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J 23: 4307–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel J-P, MacDonald ME, Gusella JF 1998. Amyloid formation by mutant huntingtin: Threshold, progressivity and recruitment of normal polyglutamine proteins. Somat Cell Mol Genet 24: 217–233 [DOI] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelières FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F 2002. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves huntingtin phosphorylation by Akt. Dev Cell 3: 1–20 [DOI] [PubMed] [Google Scholar]
- Huntington G 1872. On chorea. In The Medical and Surgical Reporter: A Weekly Journal, (ed. Butler SW), pp. 317–321 Philadelphia [Google Scholar]
- Ignatova Z, Gierasch LM 2006. Extended polyglutamine tracts cause aggregation and structural perturbation of an adjacent β barrel protein. J Biol Chem 281: 12959–12967 [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 280: 40282–40292 [DOI] [PubMed] [Google Scholar]
- Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, Faber PW, MacDonald ME, Zipursky SL 1998. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21: 633–642 [DOI] [PubMed] [Google Scholar]
- Jana NR, Nukina N 2003. Recent advances in understanding the pathogenesis of polyglutamine diseases: Involvement of molecular chaperones and ubiquitin-proteasome pathway. J Chem Neuroanat 26: 95–101 [DOI] [PubMed] [Google Scholar]
- Jana NR, Tanaka M, Wang G-H, Nukina N 2000. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: Their role in suppression of aggregation and cellular toxicity. Hum Mol Genet 9: 2009–2018 [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR 1998. Aggresomes: A cellular response to misfolded proteins. J Cell Biol 143: 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao T-P 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486–489 [DOI] [PubMed] [Google Scholar]
- Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG 2004. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J Biol Chem 279: 46363–46366 [DOI] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D 1999. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci 96: 11404–11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science 287: 1837–1840 [DOI] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M 2000. Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. J Neurosci 20: 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q 2004. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol 36: 2376–2391 [DOI] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ 2000. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem 69: 303–342 [DOI] [PubMed] [Google Scholar]
- Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I 2009. Secondary structure of Huntingtin amino-terminal region. Structure 17: 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Nollen EAA, Kitagawa K, Bindokas VP, Morimoto RI 2002. Polyglutamine protein aggregates are dynamic. Nat Cell Biol 4: 826–831 [DOI] [PubMed] [Google Scholar]
- Kim S, Nollen EAA, Kitagawa K, Bindokas VP, Morimoto RI 2003. Polyglutamine protein aggregates are dynamic. Nat Cell Biol 4: 826–831 [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack C-G, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K 2006. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol 8: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Klein FAC, Pastore A, Masino L, Zeder-Lutz G, Nierengarten H, Oulad-Abdelghani M, Altschuh D, Mandel J-L, Trottier Y 2007. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: Towards a length-dependent toxicity gradient. J Mol Biol 371: 235–244 [DOI] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT 1998. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95: 41–53 [DOI] [PubMed] [Google Scholar]
- Klivenyi P, Vecsei L 1997. Neurodegeneration: Aging and dementia. Etiopathogenic role of electron transport disorders. Therapeutic possibilities. Orv Hetil 138: 331–335 [PubMed] [Google Scholar]
- Kobayashi Y, Kume A, Li M, Doyu M, Hata M, Ohtsuka K, Sobue G 2000. Chaperones Hsp70 and Hsp40 suppress aggregate formation and apoptosis in cultured neuronal cells expressing truncated androgen receptor protein with expanded polyglutamine tract. J Biol Chem 275: 8772–8778 [DOI] [PubMed] [Google Scholar]
- Kratter IH, Finkbeiner S 2010. PolyQ disease: Too many Q's, too much function? Neuron 67: 897–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S 2000. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci 97: 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle S, Gutekunst C-A, Klein AM, Li X-J, Li S-H, Beal F, Hersch SM, Ferante RJ 1999. Huntingtin aggregates may not predict neuronal death in Huntington's disease. Ann Neurol 46: 842–849 [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha J-HJ, et al. 2007. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet 16: 1845–1861 [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, et al. 2006. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127: 1335–1347 [DOI] [PubMed] [Google Scholar]
- Le Moine C, Normand E, Guitteny AF, Fouque B, Teoule R, Bloch B 1990. Dopamine receptor gene expression by enkephalin neurons in rat forebrain. Proc Natl Acad Sci 87: 230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H, Ferrand V, Marguet D, Bania J, Camosseto V, David A, Gatti E, Pierre P 2004. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol 164: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM 2008. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453: 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY 2008. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452: 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J, Demand J, Höhfeld J 2000. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem 275: 4613–4617 [DOI] [PubMed] [Google Scholar]
- Lund JC 1860. Chorea Sti Viti I Stersdalen. In Uddrag af Distriktsoege J C Lunds Medicinalberetning for 1860, pp. 137–138 (Beretning om Sundhedstilstandenm, Norway: ). [Google Scholar]
- MacDonald ME, Barnes G, Srinidhi J, Duyao MP, Ambrose CM, Myers RH, Gray J, Conneally PM, Young A, Penney J 1993. Gametic but not somatic instability of CAG repeat length in Huntington's disease. J Med Genet 30: 982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87: 493–506 [DOI] [PubMed] [Google Scholar]
- Marshall PE, Landis DM, Zalneraitis EL 1983. Immunocytochemical studies of substance P and leucine-enkephalin in Huntington's disease. Brain Res 289: 11–26 [DOI] [PubMed] [Google Scholar]
- Masino L, Kelly G, Leonard K, Trottier Y, Pastore A 2002. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett 513: 267–272 [DOI] [PubMed] [Google Scholar]
- Massey AC, Zhang C, Cuervo AM 2006. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol 73: 205–235 [DOI] [PubMed] [Google Scholar]
- McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, et al. 2000. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet 9: 2197–2202 [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P 2004. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol 36: 2445–2462 [DOI] [PubMed] [Google Scholar]
- Menalled L, El-Khodor BF, Patry M, Suárez-Fariñas M, Orenstein SJ, Zahasky B, Leahy C, Wheeler V, Yang XW, MacDonald M, et al. 2009. Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiol Dis 35: 319–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik A, Van Broeckhoven C 2004. Proteasome degrades soluble expanded polyglutamine completely and efficiently. Neurobiol Dis 16: 202–211 [DOI] [PubMed] [Google Scholar]
- Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S 2010a. Quanitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into Huntington‘s disease molecular pathogenesis. J Neurosci 30: 10541–10550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR, Paulson HL 2005. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci 25: 9152–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Finkbeiner S 2008. The ubiquitin-proteasome pathway in Huntington's disease. The Scientific World Journal 8: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Tsvetkov AS, Finkbeiner S 2009. Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in Huntington's disease. J Biol Chem 284: 4398–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL 2005. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6: 11–22 [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Ning K, D'Souza-Schorey C, Fields S 2002. Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc Natl Acad Sci 99: 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU 2000. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci 97: 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RH, Vonsattel JP, Stevens TJ, Cupples LA, Richardson EP, Martin JB, Bird ED 1988. Clinical and neuropathologic assessment of severity in Huntington's disease. Neurology 38: 341–347 [DOI] [PubMed] [Google Scholar]
- Myung J, Kim KB, Crews CM 2001. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev 21: 245–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Fujikake N, Ohno K, Higashiyama H, Popiel HA, Rahadian J, Yamaguchi M, Strittmatter WJ, Burke JR, Toda T 2003. Prevention of polyglutamine oligomerization and neurodegeneration by the peptide inhibitor QBP1 in Drosophila. Hum Mol Genet 12: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T 2007. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol 14: 332–340 [DOI] [PubMed] [Google Scholar]
- Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N 2004. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem 91: 57–68 [DOI] [PubMed] [Google Scholar]
- Nance MA, Myers RH 2001. Juvenile onset Huntington's disease—Clinical and research perspectives. Ment Retard Dev Disabil Res Rev 7: 153–157 [DOI] [PubMed] [Google Scholar]
- Nandi PK 1996. Protein conformation and disease. Vet Res 27: 373–382 [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP 2010. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron 67: 936–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EAA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RHA 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci 101: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, Zaidi R, Clemente A, Kaul M, Graham RK, et al. 2009. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med 15: 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera O, Burke JR, Miller SE, Hester S, Tsuji S, Roses AD, Strittmatter WJ 1997. Oligomerization of expanded-polyglutamine domain fluorescent fusion proteins in cultured mammalian cells. Biochem Biophys Res Commun 238: 599–605 [DOI] [PubMed] [Google Scholar]
- Orr HT 2004. Neuron protection agency. Nature 431: 747–748 [DOI] [PubMed] [Google Scholar]
- Ortega Z, Díaz-Hernández M, Maynard CJ, Hernández F, Dantuma NP, Lucas JJ 2010. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J Neurosci 30: 3675–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Roberson ED, Wang J, Thwin MT, Bien-Ly N, Yoo J, Ho KO, Yu G-Q, Kreitzer A, et al. 2007. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer's disease. Neuron 55: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447: 859–863 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478 [DOI] [PubMed] [Google Scholar]
- Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, MacDonald M, Beglinger LJ, et al. 2008. Detection of Huntington's disease decades before diagnosis: The Predict-HD study. J Neurol Neurosurg Psychiatry 79: 874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney JB Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH 1997. CAG repeat number governs the development rate of pathology in Huntington's disease. Ann Neurol 41: 689–692 [DOI] [PubMed] [Google Scholar]
- Persichetti F, Carlee L, Faber PW, McNeil SM, Ambrose CM, Srinidhi J, Anderson M, Barnes GJ, Gusella JF, MacDonald ME 1996. Differential expression of normal and mutant Huntington's disease gene alleles. Neurobiol Dis 3: 183–190 [DOI] [PubMed] [Google Scholar]
- Petersén A, Larsen KE, Behr GG, Romero N, Przedborski S, Brundin P, Sulzer D 2001. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum Mol Genet 10: 1243–1254 [DOI] [PubMed] [Google Scholar]
- Petrucelli L, Dawson TM 2004. Mechanism of neurodegenerative disease: Role of the ubiquitin proteasome system. Ann Med 36: 315–320 [DOI] [PubMed] [Google Scholar]
- Picconi B, Pisani A, Barone I, Bonsi P, Centonze D, Bernardi G, Calabresi P 2005. Pathological synaptic plasticity in the striatum: Implications for Parkinson's disease. Neurotoxicology 26: 779–783 [DOI] [PubMed] [Google Scholar]
- Poirier MA, Jiang H, Ross CA 2005. A structure-based analysis of huntingtin mutant polyglutamine aggregation and toxicity: Evidence for a compact β-sheet structure. Hum Mol Genet 14: 765–774 [DOI] [PubMed] [Google Scholar]
- Preisinger E, Jordan BM, Kazantsev A, Housman D 1999. Evidence for a recruitment and sequestration mechanism in Huntington's disease. Philos Trans R Soc Lond B Biol Sci 354: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z-H, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, Yoder J, Aronin N, DiFiglia M 2003. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet 12: 3231–3244 [DOI] [PubMed] [Google Scholar]
- Ranen NG, Stine OC, Abbott MH, Sherr M, Codori A-M, Franz ML, Chao NI, Chung AS, Pleasant N, Callahan C, et al. 1995. Anticipation and instability of IT-15 (CAG)n repeats in parent-offspring pairs with Huntington disease. Am J Hum Genet 57: 593–602 [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Rubinsztein DC 2004. Can autophagy protect against neurodegeneration caused by aggregate-prone proteins? Neuroreport 15: 2443–2445 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC 2002. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Richfield EK, Maguire-Zeiss KA, Vonkeman HE, Voorn P 1995. Preferential loss of preproenkephalin versus preprotachykinin neurons from the striatum of Huntington's disease patients. Ann Neurol 38: 852–861 [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Lang-Rollin I, Stefanis L 2004. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol 36: 2551–2562 [DOI] [PubMed] [Google Scholar]
- Rogers D, Leavitt B 2004. HD aggregates accelerate autophagy. Clin Genet 66: 293–294 [Google Scholar]
- Rosas HD, Hevelone ND, Zaleta AK, Greve DN, Salat DH, Fischl B 2005. Regional cortical thinning in preclinical Huntington disease and its relationship to cognition. Neurology 65: 745–747 [DOI] [PubMed] [Google Scholar]
- Ross CA 1997. Intranuclear neuronal inclusions: A common pathogenic mechanism for glutamine-repeat neurodegenerative diseases. Neuron 19: 1147–1150 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Wyttenbach A, Rankin J 1999. Intracellular inclusions, pathological markers in disease caused by expanded polyglutamine tracts? J Med Genet 36: 265–270 [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman J-J, Chotai K, Connarty M, Craufurd D, Curtis A, et al. 1996. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am J Hum Genet 59: 16–22 [PMC free article] [PubMed] [Google Scholar]
- Sánchez I, Mahlke C, Yuan J 2003. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature 421: 373–379 [DOI] [PubMed] [Google Scholar]
- Sapp E, Ge P, Aizawa H, Bird E, Penney J, Young AB, Vonsattel J-P, DiFiglia M 1995. Evidence for a preferential loss of enkephalin immunoreactivity in the external globus pallidus in low grade Huntington's disease using high resolution image analysis. Neuroscience 64: 397–404 [DOI] [PubMed] [Google Scholar]
- Saric T, Graef CI, Goldberg AL 2004. Pathway for degradation of peptides generated by proteasomes: A key role for thimet oligopeptidase and other metallopeptidases. J Biol Chem 279: 46723–46732 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC 2007a. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J Biol Chem 282: 5641–5652 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, et al. 2007b. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol 3: 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, Finkbeiner S, Paganetti P, Muchowski PJ, Wilson S, Bates GP 2010. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington's disease. Hum Mol Genet 19: 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal S, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI 2000. Polyglutamine aggregates alter protein folding homeostasis in Caenorrhabditis elegans. Proc Natl Acad Sci 97: 5750–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME 1998. Huntingtin acts in the nucleus to induce apoptosis, but death does not correlate with the formation of intranuclear inclusions. Cell 95: 55–66 [DOI] [PubMed] [Google Scholar]
- Sax DS, Powsner R, Kim A, Tilak S, Bhatia R, Cupples LA, Myers RH 1996. Evidence of cortical metabolic dysfunction in early Huntington's disease by single-photon-emission computed tomography. Mov Disord 11: 671–677 [DOI] [PubMed] [Google Scholar]
- Schaffer G, Breuer P, Boteva R, Behrends C, Tzetkov K, Stripple N, Hayer-Hartl M, Hartl F-U 2004. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol Cell 15: 95–105 [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE 1997. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90: 549–558 [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates GP, Lehrach H, Wanker EE 1999. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for Huntington's disease pathology. Proc Natl Acad Sci 96: 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacka JJ, Roth KA, Zhang J 2008. The autophagy-lysosomal degradation pathway: Role in neurodegenerative disease and therapy. Front Biosci 13: 718–736 [DOI] [PubMed] [Google Scholar]
- Shimohata T, Sato A, Burke JR, Strittmatter WJ, Tsuji S, Onodera O 2002. Expanded polyglutamine stretches form an ‘aggresome’. Neurosci Lett 323: 215–218 [DOI] [PubMed] [Google Scholar]
- Sieradzan KA, Mechan AO, Jones L, Wankers EE, Nukina N, Mann DMA 1999. Huntington's disease intranuclear inclusions contain truncated, ubiquitinated huntingtin protein. Exp Neurol 156: 92–99 [DOI] [PubMed] [Google Scholar]
- Sisodia SS 1998. Nuclear inclusions in glutamine repeat disorders: Are they pernicious, coincidental or beneficial? Cell 95: 1–4 [DOI] [PubMed] [Google Scholar]
- Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, et al. 2005. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci 102: 11402–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ 1993. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet 4: 393–397 [DOI] [PubMed] [Google Scholar]
- Squitieri F, Gellera C, Cannella M, Mariotti C, Cislaghi G, Rubinsztein DC, Almqvist EW, Turner D, Bachoud-Lévi A-C, Simpson SA, et al. 2003. Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain 126: 946–955 [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN, Marcelli M, Weigel NL, Mancini MA 1999. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet 8: 731–741 [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, Snyder SH 2009. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science 324: 1327–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O 2008. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet 17: 345–356 [DOI] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J 2006. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol 8: 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L 2005. Twenty years of the Alzheimer's disease amyloid hypothesis: A genetic perspective. Cell 120: 545–555 [DOI] [PubMed] [Google Scholar]
- Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH 2003. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet 12: 749–757 [DOI] [PubMed] [Google Scholar]
- Taylor RC, Dillin A 2011. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol doi:10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenius H, Almqvist E, Kremer B, Spence N, Squitieri F, Nichol K, Grandell U, Starr E, Benjamin C, Castaldo I 1995. Somatic mosaicism in sperm is associated with intergenerational (CAG)n changes in Huntington disease. Hum Mol Genet 4: 189–195 [DOI] [PubMed] [Google Scholar]
- Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, Zeisler J, Adam S, Greenberg C, Ives EJ, Clarke LA 1994. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet 6: 409–414 [DOI] [PubMed] [Google Scholar]
- Telenius H, Kremer HP, Theilmann J, Andrew SE, Almqvist E, Anvret M, Greenberg C, Greenberg J, Lucotte G, Squitieri F 1993. Molecular analysis of juvenile Huntington disease: The major influence on (CAG)n repeat length is the sex of the affected parent. Hum Mol Genet 2: 1535–1540 [DOI] [PubMed] [Google Scholar]
- Thakur AK, Yang W, Wetzel R 2004. Inhibition of polyglutamine aggregate cytotoxicity by a structure-based elongation inhibitor. FASEB J 18: 923–925 [DOI] [PubMed] [Google Scholar]
- Trommer BL, Shah C, Yun SH, Gamkrelidze G, Pasternak ES, Blaine Stine W, Manelli A, Sullivan P, Pasternak JF, LaDu MJ 2005. ApoE isoform-specific effects on LTP: Blockade by oligomeric amyloid-b1–42. Neurobiol Dis 18: 75–82 [DOI] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC 2005. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum Mol Genet 14: 3425–3433 [DOI] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL 2004. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell 14: 95–104 [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr 1985. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol 44: 559–577 [DOI] [PubMed] [Google Scholar]
- Wacker JL, Huang SY, Steele AD, Aron R, Lotz GP, Nguyen Q, Giorgini F, Roberson ED, Lindquist S, Masliah E, et al. 2009. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. J Neurosci 29: 9104–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ 2004. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol 11: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 12: 1393–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ 2002. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416: 535–539 [DOI] [PubMed] [Google Scholar]
- Wang X, Vitalis A, Wyczalkowski MA, Pappu RV 2006. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins 63: 297–311 [DOI] [PubMed] [Google Scholar]
- Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H 1999. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol 309: 375–386 [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HYE, Gray-Board GL, Chai Y, Paulson HL, Bonini NM 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet 23: 425–428 [DOI] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Rubinsztein DC 2004. Microtubule disruption inhibits autophagosome-lysosome fusion: Implications for studying the roles of aggresomes in polyglutamine diseases. Int J Biochem Cell Biol 36: 2541–2550 [DOI] [PubMed] [Google Scholar]
- Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayan J, et al. 2004. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci 101: 3498–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ 2003. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or α-synuclein. Science 302: 1769–1772 [DOI] [PubMed] [Google Scholar]
- Wirths O, Multhaup G, Bayer TA 2004. A modified β-amyloid hypothesis: Intraneuronal accumulation of the β-amyloid peptide–the first step of a fatal cascade. J Neurochem 91: 513–520 [DOI] [PubMed] [Google Scholar]
- Wong SLA, Chan WM, Chan HYE 2008b. Sodium dodecyl sulfate-insoluble oligomers are involved in polyglutamine degeneration. FASEB J 22: 3348–3357 [DOI] [PubMed] [Google Scholar]
- Wong ESP, Tan JMM, Soong W-E, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim K-L 2008a. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet 17: 2570–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, Rubinsztein DC 2000. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc Natl Acad Sci 97: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R 2000. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101: 57–66 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE 2006. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 172: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Dunlap JR, Andrews RB, Wetzel R 2002. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet 11: 2905–2917 [DOI] [PubMed] [Google Scholar]
- Ying W 1996. A new hypothesis of neurodegenerative diseases: The deleterious network hypothesis. Med Hypotheses 47: 307–313 [DOI] [PubMed] [Google Scholar]
- Yoo S-Y, Pennesi ME, Weeber EJ, Xu B, Atkinson R, Chen S, Armstrong DL, Wu SM, Sweatt JD, Zoghbi HY 2003. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron 37: 383–401 [DOI] [PubMed] [Google Scholar]
- Yu Z-X, Li S-H, Nguyen H-P, Li X-J 2002. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum Mol Genet 11: 905–914 [DOI] [PubMed] [Google Scholar]
- Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H 2002. p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol 160: 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cao F, Wang Z, Yu Z-X, Nguyen H-P, Evans J, Li S-H, Li X-J 2003. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J Cell Biol 163: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]