Abstract
Ras GTPases are best known for their ability to serve as molecular switches regulating cell growth, differentiation and survival. Gene mutations that result in expression of constitutively active forms of Ras proteins have been clearly linked to oncogenesis in animal models and humans. However, over the past two decades, evidence has gradually accumulated to support a paradoxical role for Ras proteins in the initiation of cell death pathways. The balance between the opposing functions of Ras in cell proliferation/survival versus cell death can be critical for determining the overall fate of the cancer cell. In this review we will survey the body of literature that points to the ability of activated Ras proteins to tip the scales toward cell death under conditions where cancer cells encounter adverse environmental conditions or are subjected to apoptotic stimuli. In some cases the consequences of Ras activation are mediated through interactions with known effectors and well defined apoptotic death pathways. However, in other cases it appears that Ras operates by triggering novel non-apoptotic death mechanisms that are just beginning to be characterized. Understanding the details of these pathways, and the various factors that go into changing the nature of Ras signaling from pro-survival to pro-death, could potentially set the stage for the development of novel therapeutic approaches aimed at manipulating the pro-death Ras effector pathways in cancers.
Keywords: Ras, GTPase, Apoptosis, Non-Apoptotic, Cell Death, Review, Cancer
2. Introduction
The ras genes encode monomeric GTPases that function as molecular switches in signal transduction pathways regulating cell proliferation, differentiation and survival in mammalian cells (1,2). The original members of the Ras protein family include H-Ras, N-Ras, and the splice variants, K-Ras4A and K-Ras4B (3,4). The sequences and structural features of these proteins are highly conserved, except for their carboxyl-terminal domains and post-translational lipid modifications (5,6). The Ras family has now expanded to include the related R-Ras subgroup, which includes R-Ras (R-Ras1)(7), TC21 (R-Ras2) (8) and M-Ras (R-Ras3) (9). Similar to the original Ras family members, introduction of activating mutations into members of the R-Ras subgroup can transform mammalian cells (10,11). The R-Ras proteins can also interact with many of the same effectors as H, K, and N-Ras (12). However, the R-Ras proteins are generally viewed as being functionally distinct from H, K, and N-Ras, with activation of R-Ras pathways reported to promote integrin activation, cell adhesion and cell migration (13–15) rather than cell proliferation.
Widespread interest in Ras proteins stems primarily from the knowledge that mutations in ras genes can be oncogenic (16,17). The ras mutations typically found in human cancers result in amino acid substitutions that reduce intrinsic GTPase activity or impair interaction with GTPase-activating proteins (GAPs) (2,17,18). Consequently, mutations such as the common G12→V and Q61→L are regarded as `activating' because they result in chronic unregulated stimulation of Ras effector pathways. Activating mutations in ras genes occur widely in spontaneous and experimentally induced tumors in rodents (19,20) and in approximately 30% of human malignancies(21). Mutations in specific Ras family members are commonly associated with particular types of tumors; e.g., K-Ras in colon (22,23) and pancreatic cancers (24,25), H-Ras in bladder carcinoma (26), and N-Ras in myeloid leukemia (27). With the spotlight on the oncogenic activity of Ras, the vast majority of published studies concerning these proteins have understandably focused on their roles in stimulating cell growth and promoting cell survival. Certainly, the accumulated body of evidence leaves little doubt that the latter can be important consequences of Ras activation. Nevertheless, throughout the past 10–15 years, reports have occasionally appeared suggesting that activation of Ras proteins can have detrimental effects on mammalian cells, ultimately leading to death by apoptosis or other mechanisms (28). The identification of specific Ras effectors that have pro-apoptotic functions (29–31) lends credence to the idea that the death-promoting effects of Ras may be physiologically significant under some circumstances. Even in cases where the negative effects of Ras on cell viability are observed under somewhat artificial experimental conditions (e.g., ectopic overexpression of constitutively active Ras proteins), understanding the relevant signaling pathways could lead to the identification of new molecular targets that might be manipulated to trigger cell death in cancer cells.
The intent of this review is to provide an overview of examples from the literature where activation of endogenous Ras or ectopic expression of activated Ras mutants have been shown to cause apoptotic or non-apoptotic cell death in mammalian cells. We will not discuss the evidence linking Ras to cellular senescence (permanent cell cycle arrest) which is not in itself a cell death pathway (32,33). The scope of the review will be limited to the “classical” H, K, and N-Ras proteins, where connections to cell death pathways are much better established than for the R-Ras proteins. Except for cases where overlap exists with Ras cell death signaling pathways, it will not be possible to consider the large number of Ras-related GTPases (34), which fall into distinct subfamilies involved in regulation of cytoskeletal architecture and cell motility (Rho/Rac), vesicular trafficking (Rab), and nucleocytoplasmic transport (Ran), all of which could potentially influence cell survival. Although a brief overview of the signaling functions of Ras proteins will be included to set the stage for discussion of Ras-mediated cell death pathways, we will not attempt to cover in depth the extensive body of information available on Ras signaling networks or the roles of Ras and its effectors in cell growth and tumorigenesis, which have been the subjects of many previous reviews.
3. Overview of Ras Signaling
Three decades of intense study have yielded a tremendous amount of information about the guanine nucleotide dependent structural conformations of Ras proteins (35–37), the identities and functions of the guanine nucleotide exchange factors (GEFs) and GAPs that control the Ras nucleotide state (38–40), and the complex downstream signaling pathways regulated by Ras GTPases (41–43). From these studies a general picture has emerged wherein Ras proteins are viewed as key intermediates in the transmission of signals from cell surface receptors to intracellular effectors (Fig. 1). The latter may have direct effects on cellular processes that take place in the cytoplasm (e.g., actin organization, endocytosis), or may influence nuclear transcription factors that regulate genes important for cell cycle progression, differentiation, or survival.
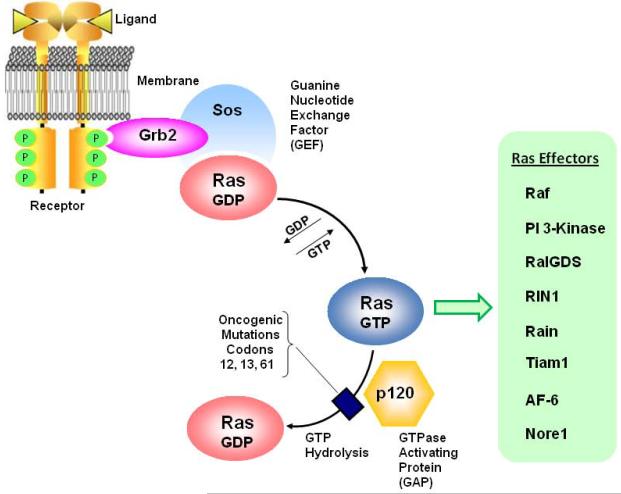
Figure 1.
Ras proteins function as GTP-dependent molecular switches. According to the most widely recognized paradigm for Ras activation, binding of ligands to cell surface receptors initiates tyrosine phosphorylation on cytoplasmic domains. Adapter proteins (e.g., Grb2) containing SH2 domains bind to the p-Tyr and recruit Ras GEFs (e.g. Sos) which promote dissociation of GDP and binding of GTP to the Ras protein. In its active GTP-bound conformation, the Ras protein may associate with a number of different effectors, initiating signaling pathways that can affect cell proliferation, cell survival, differentiation, cytoskeletal organization, vesicular trafficking, or cell death. Normal Ras proteins revert to their inactive GDP state through their intrinsic GTPase activity and interaction with GAPs (e.g., p120) that increase the rate of GTP hydrolysis. The common oncogenic mutations found in Ras proteins interfere with GTP hydrolysis, resulting in proteins that remain “stuck” in the active GTP state and continue to transmit signals to effector pathways in an unregulated manner.
Ras is considered to be active when bound to GTP, and inactive when bound to GDP. In mammalian cells endogenous Ras proteins are predominantly in the GDP state, and activation is transient (44). In one well established paradigm for Ras signaling (Fig. 1), binding of ligand (e.g., EGF) to a receptor tyrosine kinase on the plasma membrane stimulates receptor phosphorylation, with the resulting phosphotyrosines serving as docking sites for adaptor proteins such as Grb2. These adaptors, in turn, recruit guanine nucleotide exchange factors (e.g., Sos) that stimulate conversion of inactive Ras-GDP to active Ras-GTP (45–47). From studies of the three-dimensional structure of Ras, it is now clear that binding of GTP triggers conformational changes in two flexible domains that promote interactions with various effectors; switch I (also termed the effector domain) and switch II (37,48,49). Signals are terminated when bound GTP is hydrolyzed to GDP, with the intrinsic GTPase activity of the Ras protein being accelerated through interaction with members of the Ras GAP family (50,51).
The spectrum of known Ras effectors has grown steadily over the years and it is now clear that Ras occupies a central position in a complex signaling network (41,43,52). The most thoroughly characterized Ras effectors are the Raf kinases (53,54) and the p110 catalytic subunit of type-I phosphatidylinositide 3-kinase (PI 3-Kinase) (55). Interaction of activated Ras with Raf initiates the Raf → MEK → ERK kinase cascade. Activated ERK1/2 (p42/44 MAP kinase) can then phosphorylate many substrates, including kinases that are important for control of translation (e.g., p90Rsk) and transcription factors that control genes involved in cell cycling (e.g., Elk1, Fos, Myc) (56,57). Ras activation of PI 3-Kinase triggers production of the second messenger, PI(3,4,5)P3, leading to downstream activation of Akt (Protein Kinase B). Akt-mediated phosphorylation inhibits some proteins that promote programmed cell death (Bad, FoxO), and stimulates others (Mdm2) that promote cell survival (58).
Many additional Ras effectors have now been identified. These include, RAIN, an endomembrane receptor for Ras (59), NORE1, a pro-apoptotic tumor suppressor (60), and AF-6, a mediator of membrane-cytoskeleton interactions (61,62). Ras also engages in cross-talk with other GTPase signaling pathways involved in regulating actin reorganization and/or endocytic trafficking. This can occur through interactions with GEFs like RalGDS, for RalA and RalB (63), RIN1, for Rab5 (64), and Tiam1, for Rac (65).
Adding to the complexity of Ras signaling, accumulating evidence indicates that the different forms of Ras are not all equivalent in terms of their effector interactions. For example, H-Ras is a more potent activator of PI 3-Kinase than K-Ras (66), while K-Ras is comparatively more effective than H-Ras in stimulating activation of Raf-1 (66,67) and Rac1 (68). Signaling specificity also is dictated to some extent by differential localization of the Ras isoforms in discrete plasma membrane microdomains (69,70) or distinct intracellular membrane compartments (e.g., endosomes, Golgi) (6,71), where the activated GTPase may encounter unique sets of effectors. Thus, as we strive to understand the multiplicity of different, and sometimes conflicting, biological effects that have been reported in conjunction with Ras activation, it is important to bear in mind that the signaling output in response to any given stimulus may vary significantly, depending on both the spectrum of Ras effectors and the particular Ras isoforms that predominate in a particular cell type.
4. Induction of Apoptosis by Ras
The prevailing view that activating mutations in Ras proteins uniformly lead to unregulated cell proliferation, resistance to programmed cell death and more aggressive phenotypes in human tumors has been called into question by a studies evaluating the prognostic importance of N- and K-ras gene mutations in acute myeloid leukemia (72) and H-Ras expression in neuroblastoma (73). Surprisingly, these studies showed a positive correlation of activating ras mutations or increased Ras expression with improved survival. These observations are consistent with numerous studies with cultured cells demonstrating that ectopic expression of activated Ras can trigger cell death under some circumstances, or that activation of endogenous Ras can be an essential step in the process whereby cells initiate death pathways in response to growth factor deprivation or other pharmacological or environmental insults. With some exceptions noted later in this review, such studies have focused mainly on connections between Ras and the most widely recognized form of programmed cell death, apoptosis. In this section, we will provide an overview of early studies which indicated that, in addition to its well-established pro-survival role, Ras can have paradoxical pro-apoptotic effects under specific conditions.
In mammalian cells apoptosis is manifested by several diagnostic morphological characteristics, including cytoplasmic shrinkage, condensation of nuclear chromatin, blebbing of the cell membrane, and eventual fragmentation of the cell into apoptotic bodies (74). At the biochemical level, one of the key features of apoptosis is endonuclease cleavage of DNA at internucleosomal sites, yielding discrete fragments that can be detected as a “ladder” on an electrophoresis gel. The molecular pathways involved in triggering and executing the apoptotic cell death program have been studied extensively and have been the subject of excellent reviews (75,76). Very briefly, apoptosis can be initiated through either an extrinsic pathway or an intrinsic pathway. In the extrinsic pathway, death-promoting ligands like tumor necrosis factor α (TNFα) or FasL bind to their respective receptors on the cell surface and initiate a signaling cascade leading to activation of a cysteine protease, caspase 8, and cleavage of the pro-apoptotic Bcl-2 family member, Bid, to form tBid. The latter promotes release of cytochrome c from the mitochondria, facilitating assembly of the apoptosome complex and activation of a caspase cascade commencing with caspase 9, and culminating with the so called `executioner' caspases (e.g., caspase 3, caspase 7). The terminal caspases work on various substrates in the nucleus and elsewhere in the cell to effect the degenerative changes typical of apoptosis. The intrinsic apoptotic pathway differs from the extrinsic pathway in that it is triggered by a general insult rather than a specific ligand-receptor interaction. For instance, growth factor withdrawal, DNA damage, loss of cell-matrix attachment, and altered calcium flux from the endoplasmic reticulum are all capable of inducing the intrinsic apoptotic pathway. Signals are transmitted to the caspase machinery through one of several well defined mechanisms (e.g., activation of p53, suppression of anti-apoptotic Bcl-2 or Bcl-XL), ultimately resulting in increased association of pro-apoptotic Bcl-2 family members (e.g., Bax, Bak) with the mitochondrial membrane, release of cytochrome c, and propagation of the downstream caspase cascade.
Early reports of Ras being able to stimulate apoptosis instead of promoting cell survival came from experiments in which activated forms of Ras were ectopically expressed in fibroblasts. In one study, Kauffmann-Zeh et al. (77) found that although expression of constitutively active PI 3-Kinase could block c-Myc-induced apoptosis, overexpression of Ras(V12) had the opposite effect, actually enhancing the pro-apoptotic effect of c-Myc. This suggested that in cases where Ras is hyperactive, pro-apoptotic signals may override the pro-survival signals mediated through the PI 3-Kinase effector pathway. In a study that appeared shortly thereafter, Mayo et al. (78) reported that all three forms of Ras with oncogenic mutations were able to trigger p53-independent apoptosis, provided that endogenous NFκB signaling was suppressed. Evidence for a role for Ras in apoptosis has since been extended by numerous studies demonstrating that activation of endogenous Ras is correlated with or required for apoptotic death in a variety of cell types exposed to different pro-apoptotic stimuli. These include, treatment of Jurkat cells with Fas (79), withdrawal of cytokines from T-cells (80,81), treatment of osteosarcoma and neuroblastoma cells with cisplatin (82), treatment of osteosarcoma cells with a protein kinase C inhibitor, chelerythrine (83), and exposure of fibroblasts to cycloheximide, ceramide or mitomycin C (84).
As noted for Ras signaling in general, the specific consequences of Ras activation for apoptosis may vary, depending on the predominant form of Ras expressed in a particular cell type. For instance, H-Ras and K-RasB have opposite effects on radiation-induced apoptosis in fibroblasts, with K-Ras promoting apoptosis and H-Ras having a protective effect (85). Similarly, opposing roles for K-Ras versus H-Ras have been observed in endometrial cells subjected to serum deprivation (86). Further complicating matters, the nature of the posttranslational modifications of Ras can have a significant impact on its pro-apoptotic behavior. For example, treatment of cells expressing N-Ras or K-RasB with a farnesyltransferase inhibitor seems to switch Ras from a stimulator of cell proliferation to an inducer of apoptosis by promoting alternative prenylation of the carboxyl-terminal cysteine (normally farnesylated) with a geranylgeranyl group (87). One possible explanation for the switch to pro-apoptotic behavior might be that altering the lipid modification on Ras could affect its subcellular compartmentalization and/or change its range of potential effector interactions. Phosphorylation also appears to play a key role in enhancing the pro-apoptotic function of KRasB. Specifically, Bivona et al. (88) have demonstrated that protein kinase C (PKC)-mediated phosphorylation of serine-181 in the unique polybasic region near the carboxyl-terminus of KRasB stimulates apoptosis. A mechanism for this effect is suggested by their observation that phosphorylation correlates with translocation of the active GTPase from the plasma membrane to intracellular compartments, including the mitochondria, where Ras interacts with members of the Bcl-2 family (discussed below).
5. Mechanisms of Ras-Induced Apoptosis
5.1 Replication Stress
Early theories concerning the possible mechanism of Ras-induced cell death centered on the concept that loss of cell viability could be associated with sustained mitogenic signaling and aberrant cell cycle progression, leading to replication stress. Along these lines, Miranda et al. (89) initially observed that abnormal expression of wt or mutant H-Ras in HeLa cells leads to accumulation of giant multinucleated cells with chromosomal abnormalities, consistent with a mitotic death mechanism (mitotic catastrophe). Recent evidence indicates that cell death resulting from sustained mitotic arrest proceeds via classical apoptosis pathways triggered by phosphorylation and inactivation of the anti-apoptotic Bcl-2 family member, Mcl-1 (90,91). Other investigators, working with rat fibroblasts (92) or normal thyroid cells (93) have observed different effects of activated Ras on cell cycling, with arrest in the G1 phase or the G2/M phase, preceding apoptosis. In contrast to the many examples where cells transformed by Ras exhibit elevated levels of cyclin D1 (94–97), in these studies the cell cycle perturbations triggered by Ras were accompanied by decreased levels of cyclin D1 (92,93). Moreover, in thyroid cells, acute expression of Ras(V12) also activated the ATR kinase, a sensor of DNA damage (98). Together, these findings support a model wherein some types of cells, challenged by Ras-driven dysregulation of cell cycling, may respond by activating `replication stress' checkpoints, where cells with genetic damage are held until they can be eliminated by apoptosis. It remains unclear why this response has been observed in some types of cells but not others.
5.2 The ERK Pathway
The pleiotrophic effects of activated Ras have made it extremely challenging to pinpoint all of the specific Ras signaling pathways that might contribute to its pro-apoptotic effects under various conditions where Ras-induced cell death has been observed. Despite abundant evidence that Ras activation of the Raf → MEK →ERK pathway plays an important role in stimulating cell proliferation (99–101) and promoting cell survival (102), there is also a case for involvement of this pathway in the pro-apoptotic effects of Ras. Experiments utilizing Ras constructs with partial loss-of-function mutations in the effector domain, which selectively reduce interaction with Raf, demonstrated that interference with Ras signaling to ERK could blunt the induction of apoptosis in response to serum deprivation in fibroblasts (77). Similar conclusions were reached in studies applying MEK inhibitors to block downstream signals from Raf in fibroblasts exposed to a PKC inhibitor (83) or in thyroid cells in which death was induced by genes of the RET/PTC family (103). It is now clear from numerous studies (reviewed in references 104,105) that activation of ERK plays a broad role in the apoptotic response of many types of normal and transformed cells. Although a direct link to activation of Ras has not been established in all cases, there is abundant evidence that activation of ERK1/2 can be important for the response of tissues and cells to death receptor ligands (TNFα, TRAIL), oxidative stress, nutrient deprivation, or DNA-damage caused by drugs or radiation.
Proposed mechanisms linking ERK activation to induction of apoptosis include stimulation of the extrinsic apoptotic pathway through interactions with regulatory proteins of the death receptor → caspase 8 pathway (106,107), direct perturbation of mitochondrial function (108,109), and regulation of transcription factors that control expression of death receptors (110) or proteins involved in modulating the intrinsic apoptotic pathway (e.g., p53, Bax, Bcl-2) (111–113). In addition to its role in apoptosis, ERK has also been implicated as a regulator of nonapoptotic cell death pathways, which will be discussed later in this review. These studies underscore the current challenge of reconciling how activation of ERK can be involved in mediating the pro-apoptotic effects of Ras under certain conditions, while the preponderance of evidence over the past two decades points to a role for Ras-mediated ERK activation in promoting mitogenesis and cell survival. The solution to the puzzle most likely will depend on understanding parallel Ras signaling pathways that could alter the consequences of Ras →ERK signaling. For instance, shifts in the balance of Ras signaling from the pro-survival PI 3-Kinase pathway to the pro-death effector pathways (e.g., Nore1, discussed in the following sections) certainly could play important roles in modifying the net outcome of Ras activation of ERK (28,114). As noted earlier, such differences in the transmission of signals to the different branches of the Ras network might arise from environmental cues (growth factors, nutrients, extracellular matrix interactions), the relative abundance of the specific Ras isoforms in a particular cell type, differential subcellular localization of the Ras proteins, or the variations in the expression levels of the Ras effectors.
5.3 The Rac Pathway
In addition to the aforementioned PI 3-Kinase, Raf, and Nore1 signaling pathways, there have been some hints that the net effect of Ras activation on cell survival versus cell death may also depend on the activity of parallel pathways controlled by other Ras-related GTPases like Rac. The discovery of Rac GEFs like Tiam1, which contains a Ras binding domain and can thereby link activation of Ras to activation of Rac (65,115), suggests that crosstalk between these pathways could be important for some Ras-mediated functions, possibly including the promotion of apoptosis. In support of this notion, one study showed that interference with Rac activation by co-expressing a dominant negative Rac mutant with activated Ras could block the pro-apoptotic activity of Ras (116). However, another study came to the opposite conclusion, demonstrating that induction of apoptosis by Ras was blocked by active Rac (117). In both studies the effects of Rac on cell survival were thought to be mediated by activation of NFκB. The accumulated evidence that NFκB can be pro-apoptotic or anti-apoptotic, depending on the nature of the apoptotic stimulus, its transcriptional targets, and the cell type (118,119), might offer an explanation for these contradictory findings.
Among the many known functions of Rac GTPases, one of the first to be elucidated was the stimulation of free radical production via the NADPH oxidase complex in phagocytic cells (120,121). It has since been established that activated Rac can also stimulate the production of reactive oxygen species (ROS) through other mechanisms in a variety of non-phagocytic cell types (122–125). It thus becomes possible to envision a mechanism whereby Ras activation could affect cell viability by modulating the intracellular production of ROS, possibly via activation of Rac. In support of this concept, Gulbins et al. (79) reported that induction of apoptosis in the Jurkat T-cell leukemia cell line by Fas involves rapid activation of Ras and synthesis of ROS. Introduction of dominant-negative Ras into the cells reduced the production of ROS and also prevented apoptosis. Whether or not the Ras-induced production of ROS may induce apoptosis by activating NFκB remains unclear because of the aforementioned uncertainties about the pro-survival versus pro-death roles of NFκB, and because of a lack of consensus regarding the exact role of endogenous ROS in regulating the activity of NFκB. Nevertheless, a potential role for ROS in Ras-induced apoptosis is worth further consideration, given the well known connections between elevated ROS and apoptosis (126,127).
Aside from its potential ability to induce cell death through stimulation of ROS production, Ras-mediated activation of Rac1 may also activate stress pathways that can lead to cell death. A number of studies have indicated that interaction of Rac with another of its effectors, the stress-activated kinase, MKK7 (128), can have important consequences for cell viability through its downstream target, JNK1/2 (c-Jun N-terminal Kinase) (129). JNK1/2 is an important activator of the cytochrome c-mediated apoptotic pathway (130). Regulation of JNK1/2 by Rac1 has been implicated in apoptosis induced by Fas or ceramide in Jurkat cells (131) and fibroblasts (132) and by TNFα in intestinal epithelial cells (133,134). A direct connection between expression of constitutively active Ras(V12) and Rac1-mediated activation of JNK1/2 was recently demonstrated by Byun et al. (135), although in that case cell death involved a non-apoptotic mechanism (discussed later in this review).
5.4 The Bcl-2 Proteins
There have been a few intriguing reports suggesting that Ras GTPases may associate directly with proteins of the Bcl-2 family. Since the latter are well known regulators of apoptosis (136–138), such interactions are of obvious interest in terms of understanding the contrasting effects of Ras in promoting cell death or cell survival. Very briefly, the anti-apoptotic members of the family, Bcl-2, Bcl-XL and Mcl-1, can inhibit apoptosis induced by a wide variety of chemical and physical stimuli, principally by preventing the release of cytochrome c from mitochondria and subsequent activation of Apaf-1 and downstream caspases (e.g., caspase-9, caspase-3) (139,140). In contrast, the pro-apoptotic members of the Bcl-2 family, such as Bax and Bak, can stimulate release of mitochondrial cytochrome c and trigger activation of the initiator and executioner caspases (141). Although considerable controversy still exists concerning the exact mechanisms whereby the different Bcl-2 family members exert their unique activities, the key finding that anti-apoptotic Bcl-2 proteins can form heterodimers with the pro-apoptotic members (142,143) has led to a prevailing model in which the balance between cell death and survival is thought to rest on the expression levels, localization, and functional interactions of the pro-apoptotic and anti-apoptotic members of the Bcl-2 family (144).
Early evidence for a connection between Ras and Bcl-2 came from the work of Chen and Faller (145), who showed that Bcl-2 could suppress the ability of constitutively active H-Ras to induce apoptosis when PKC was inhibited in Jurkat cells. Most notably, they also showed that the phosphorylation of Bcl-2 was increased in cells expressing activated H-Ras. The same group subsequently demonstrated that activated H-Ras can be immunoprecipitated with Bcl-2, and that the association increases in conjunction with delivery of an apoptotic stimulus and an increase in the phosphorylation state of Bcl-2 (146). Finally, they showed that that upon apoptotic stimulation, the interaction between Bcl-2 and H-Ras takes place at the mitochondrial membrane (147). In considering the functional consequences of the H-Ras-Bcl-2 interaction, one might suspect that binding to Ras could serve to sensitize cells to apoptosis indirectly, by interfering with the anti-apoptotic function of Bcl-2 and thereby facilitating the pro-apoptotic effects of Bax and Bak. However, Chen et al. (146) found that preventing the phosphorylation of Bcl-2 and decreasing its association with Ras actually made cells more susceptible to apoptosis, consistent with their initial observation that Bcl-2 protects against Ras-induced apoptosis. The implication of this finding is that, at least in lymphoblastoid cells, translocation of H-Ras to the mitochondrial membrane actively delivers an apoptotic signal that can be blocked by Bcl-2. In the case of H-Ras, the exact nature of this apoptotic signal remains unknown.
A somewhat different picture of Ras-mediated apoptosis and interaction with the Bcl-2 family has emerged from the work of Philips and colleagues (88), focusing on K-Ras instead of H-Ras. They discovered that PKC stimulates the pro-apoptotic signaling of K-RasB by phosphorylating S181 in its carboxyl-terminal poly-basic domain. By reducing the net positive charge of this region, the phosphorylation weakens association of K-Ras with the plasma membrane. The ability of the “electrostatic switch” to promote apoptosis appears to be related to the translocation of the phosphorylated K-Ras to the mitochondrial membrane, where it interacts with Bcl-XL. Like Bcl-2, the latter is typically regarded as playing an anti-apoptotic role. However, in this case, the association with K-Ras is postulated to confer a pro-apoptotic function onto Bcl-XL, as evidenced by the finding that the pro-apoptotic effect of K-Ras is eliminated in Bcl-XL null fibroblasts (88).
In addition to direct binding of Ras to members of the Bcl-2 family, Ras can affect the expression of p53 via activation of ERK (111–113). Through this pathway there is the potential for Ras to indirectly influence the expression of p53-regulated genes like Bax and Bcl-2, but because of the complexity of the signaling pathways that converge on p53, the relative importance of Ras signals is not easy to delineate. There is however some clear evidence for the ability of Ras to up-regulate the expression of a specific pro-apoptotic member of the Bcl-2 family, BNIP3, from a study done in RAW 264.7 cells, a mouse leukemia line with macrophage characteristics (148). In this report, cell death triggered by nitric oxide was completely dependent on activation of Ras and increased expression of BNIP3. The latter appeared to be mediated through ERK activation of the transcription factor, HIF-1 (hypoxia-inducible factor 1), which binds to the BNIP3 promoter. Most intriguing was the finding that activation of Ras by NO required nitrosylation of C118. It will be interesting to see if the regulation of BNIP3 and the novel post-translational nitrosylation of Ras turn out to be of general importance for apoptosis in other types of cells.
6. Pro-Apoptotic Effects of Ras Mediated by the RASSF Family
In the quest to identify Ras effector pathways that could provide a clear mechanistic basis for the observed pro-apoptotic activity of the activated GTPase, a major breakthrough came with the identification of the Ras-Association Domain Family (RASSF) of proteins. The latter encompasses 10 members, RASSF1–RASSF10, with additional isoforms identified due to alternative promoter use or splice variations of some of the members (reviewed in ref. 149). All members of this family contain a Ras-association (RA) domain which, as the name implies, may enable the proteins to directly interact with, and potentially modulate the activity of, different members of the Ras family. However, not all proteins with an RA domain are capable of interacting with Ras (150). Overexpressed RASSF1 (151), RASSF2 (152) and NORE1 (Novel Ras Effector 1, also known as RASSF5,) (29) have been shown to be Ras effectors, in that they bind preferentially to Ras in its GTP conformation. However, only RASSF2 (153) and RASSF5 (29) have been demonstrated to take part in this interaction in an endogenous environment.
RASSF1A is a tumor suppressor that is frequently silenced by promoter methylation in human tumors (154). Vos et al. (31) have demonstrated that the direct association of RASSF1A with the Modulator of Apoptosis-1 (MOAP-1) can activate the pro-apoptotic protein, Bax, to induce cell death in 293T cells. The interaction of MOAP-1 and RASSF1A is stimulated by the presence of activated K-Ras, resulting in synergistic enhancement of cell death. The authors demonstrate that K-Ras(V12) stimulates the ability of RASSF1A-MOAP-1 complex to cause translocation of Bax from the cytosol to the mitochondrial membrane, where it can induce apoptosis. When RASSF1 mRNA levels are reduced by siRNA, K-Ras(V12) cannot efficiently activate Bax. A similar effect is seen with the Y40C effector domain mutant of K-Ras, which has impaired ability to induce RASSF1A-MOAP-1 interaction. Interestingly, C65R, a tumor-derived point mutation of RASSF1A, does not interact with MOAP-1 or induce cell death in the presence or absence of activated K-Ras.
RASSF1A has also been shown to interact with the proapoptotic kinase, MST1(30), which could also lead to cell death. However, it is not known if this interaction occurs in response to Ras activation. Based on other studies showing that the stimulation of MST1 may activate the kinases LATs1 and LATs2, which leads to down-regulation of the anti-apoptotic Bcl-2 (155,156), Vos et al. (31) suggest the interesting possibility that RASSF1A may regulate multiple pathways to induce apoptosis, leading to its importance as a tumor suppressor.
RASSF1C is a shorter product than RASSF1A, resulting from alternative splicing combined with transcription from a distinct promoter of the RASSF1 gene (157). RASSF1C has been identified as a mediator of Ras-induced apoptosis when transiently co-expressed with Ras(V12) (151). This appears to occur through activation of the SAPK/JNK pathway (158). Others have reported that RASSF1C never associates directly with Ras(V12) (159). Therefore, the physiological relevance and mechanistic details of the interaction of these proteins will require further investigation.
Like RASSF1A, RASSF2 displays characteristics of both a Ras effector (152) and a tumor suppressor (160). Endogenous RASSF2 has been shown to bind to the prostate apoptosis response protein 4 (PAR-4) (161). The latter is an important tumor suppressor in the prostate (162) and is capable of inducing apoptosis following translocation to the nucleus (163). Association of PAR-4 with RASSF2 allows nuclear localization of PAR-4 and formation of this complex is enhanced upon expression of activated K-Ras (161). In the H441 lung carcinoma cell line, which endogenously expresses activated K-Ras, siRNA-mediated knockdown of K-Ras reduced the interaction of PAR-4 with RASSF2 (161). In addition, knockdown of RASSF2 impaired K-Ras mediated nuclear localization of PAR-4 and TRAIL-induced apoptosis in prostate cancer cells (161). Taken together, these data suggest that K-Ras induced apoptosis, linked to PAR-4, may be a key regulatory step for tumor suppression in the prostate.
NORE1, also known as RASSF5, shares approximately 60% homology with RASSF1 at the amino acid level (164). Khokhlatchev et al. (30) have demonstrated that endogenous NORE1 and the proapoptotic kinase, MST-1, exist as a complex in 293 cells. Activated K-Ras can recruit this complex to the membrane, through its interaction with NORE1, to induce apoptosis. While H- and K-Ras(V12) were both shown to bind to the MST-1-NORE1 complex, only stimulation by K-Ras(V12) resulted in the induction of apoptosis. The authors attribute this functional divergence to the fact that H-Ras(V12) is much more efficient at stimulating the PI 3-Kinase “survival” pathway than K-Ras (66), suggesting that perhaps this imbalance tips the scales away from induction of apoptosis. Studies utilizing H-Ras constructs with amino acid substitutions in the effector domain have provided support for this concept. The interaction of Ras with NORE1 occurs through the effector region of Ras (29), and H-Ras harboring either the T35S or Y40C effector mutations is impaired in NORE1 association. However, the E37G Ras mutant remains capable of associating with NORE1 (30). Interestingly, the E37G mutation impairs the ability of Ras proteins to activate Raf or PI 3-Kinase. The paper by Khokhlatchev et al. (30) demonstrates that while overexpression of H-Ras(V12) does not induce NORE1-mediated apoptotic cell death in 293 cells, H-Ras(V12,G37) is capable of causing such death, potentially due to the inability of this protein to stimulate the PI 3-Kinase pathway for survival. Conversely, the authors reveal that overexpression of PI 3-Kinase can mute the apoptotic effect of activated K-Ras. Together these results emphasize the overall importance of the balance between PI 3-Kinase activation of pro-survival pathways and NORE1 activation of pro-death pathways (30).
Recombinant RASSF4 (165) and RASSF6 (166) have been observed to directly interact with activated Ras, and transient overexpression of these proteins leads to apoptosis that is synergistically enhanced by activated Ras. Additionally, overexpressed RASSF6 was shown to co-immunoprecipitate with MOAP and this interaction is enhanced in the presence of activated K-Ras (166). However, like RASSF1C, further studies on endogenous protein interactions and details of the pathways leading to apoptotic cell death have yet to be reported.
Of the four newer members of the RASSF family, RASSF7–10 (167), only RASSF9 has been shown to bind to Ras proteins (168), but little is known about the cellular effects of this interaction. Although the ability of RASSF7 and -8 to associate with Ras has not yet been investigated, it is interesting that both of these proteins have been reported to be required for cell death by necroptosis, a recently recognized form of non-apoptotic cell death (169).
Of the 10 known members of the RASSF family of proteins, only the NORE1-MST-1 complex has been shown to directly interact with a Ras protein and induce apoptosis under physiological conditions (30). Additionally, the interaction of RASSF2 and PAR-4 was shown to be influenced by endogenous levels of activated Ras in lung carcinoma cells (161). On the other hand, many of the studies described above for the RASSF proteins have been done with overexpressed proteins and further investigations will need to be completed to determine if these proteins can mediate Ras-induced apoptosis in an endogenous cellular environment.
7. Induction of Non-Apoptotic Cell Death by Activated Ras
Although apoptosis is the best characterized form of programmed cell death, there is now considerable evidence to indicate that activation of Ras can be important for inducing non-apoptotic forms of cell death in mammalian cells. Typically, non-apoptotic cell death results in loss of cell viability that does not depend on caspase activation or nuclear DNA fragmentation (170). Autophagy-associated cell death (sometimes referred to as Type-II programmed cell death) is the most widely studied form of non-apoptotic cell death. Macroautophagy (hereafter referred to simply as autophagy) was originally identified as a mechanism for protein degradation under conditions of nutrient deprivation (171,172). Newly formed autophagosomes sequester cytosolic proteins and organelles into structures surrounded by a double membrane. These structures then mature into degradative autolysosomes by acquiring acid hydrolases through fusion with pre-existing lysosomes (173–176). In autophagy-associated cell death, early activation of autophagic pathways occurs without overt DNA fragmentation, nuclear collapse or cytoplasmic shrinkage (177,178). Death is thought to occur as the cell digests its own proteins and organelles beyond the point where it can recover. This type of cell death may occur during embryonic development in connection with tissue remodeling (177,179). It has also received considerable attention in the context of neurodegenerative diseases (180,181) and host-pathogen interactions (182). By far the most extensive investigations of autophagy have focused on its role in the response of cancer cells to environmental stress or DNA damage (183–185), antineoplastic agents (186–189) or ionizing radiation (190). Nevertheless, it remains controversial whether the increased autophagic activity that correlates with cell death is actually a direct cause of the cell's demise. Recent evidence supports the alternative view that accumulation of autophagosomes may signify a survival response intended to overcome environmental stress or rid cells of misfolded proteins or damaged organelles (191–194) and that, if this fails, cell death may ultimately occur by apoptosis, necrosis or other mechanisms. In this regard, several additional forms of non-apoptotic cell death have now been described, based on specific cellular or molecular criteria. These include paraptosis (195,196), oncosis (197–199), necroptosis (200,201), entosis (202), programmed necrosis (203,204), and methuosis (205,206). A detailed discussion of these forms of cell death is beyond the scope of this review. However, in at least one case to be described later (methuosis), Ras has been directly implicated.
The importance of non-apoptotic cell death as a consequence of Ras activation was first noted in glioblastoma and gastric carcinoma cells. Although the incidence of ras mutations ranges from moderate to high in many forms of cancer, the latter tumors rarely exhibit ras mutations (21). More than a decade ago Kitanaka, Kuchino and colleagues (207,208) speculated that a strong negative effect of ras mutations on cell survival could explain their absence in these cancers. When they tested this hypothesis by expressing H-Ras(V12) in glioma and gastric carcinoma cells, they observed a non-apoptotic degenerative process instead of the expected induction of apoptosis. This involved accumulation of cytoplasmic vacuoles, with no chromatin condensation or DNA fragmentation. Based on the presence of autophagosomes and autolysosome-like structures, and the fact that cell death could not be reversed by caspase inhibitors, the authors classified this form of Ras-induced cell death as type-2 physiological degeneration (autophagic degeneration) (207,208). Interestingly, cells already harboring activating mutations in H-Ras (e.g., bladder carcinoma cells), were not susceptible to this form of cell death, ostensibly because they had already developed compensatory mechanisms to overcome the death-inducing effects of H-Ras and permit tumor growth and survival (207).
Further evidence to support an association between Ras and non-apoptotic cell death came from studies in neuroblastoma. Although Ras mutations are uncommon in neuroblastoma, (209), the work of Tanaka et al. (73,210) suggested that high expression levels of H-Ras correlate with a favorable prognosis. Based on their earlier observations in glioma cells, Kitanaka et al. (211) hypothesized that perhaps a high level of H-Ras expression might contribute to neuroblastoma regression by activating a non-apoptotic death program. In a survey of human neuroblastoma specimens from mass-screened patients, they found a high percentage of tumors with focal areas of cellular degeneration that coincided with regions of high H-Ras expression. In contrast, a smaller percentage of tumors from patients with advanced neuroblastoma contained such areas of degeneration. Consistent with a non-apoptotic death mechanism, the degenerating cells did not stain with apoptotic markers for DNA fragmentation or caspase activation. Follow-up studies in cultured cell lines revealed that expression of activated H-Ras could indeed induce a form of caspase-independent cell death with morphological features of autophagic degeneration in neuroblastoma cells (211). Although provocative, these studies were not without controversy, as another group failed to detect autophagic degeneration in neuroblastomas with Ras-positive cells (212). In fact, they found that tumors with the highest frequency of classical caspase-dependent apoptosis surprisingly had the worst prognoses.
8. Mechanisms of Ras-Induced Non-Apoptotic Cell Death
The preceding studies strongly suggested that activated Ras is capable of inducing pathways for macroautophagy in some types of cancer cells, but also raised important questions about whether or not the induction of autophagy per se is responsible for killing the cells. Some progress has been made in defining the molecular pathways that link Ras to the autophagy machinery, and most recently, evidence from our laboratory has suggested that Ras-induced vacuolization and killing of glioma cells and other types of transformed cells may actually involve a novel form of non-apoptotic cell death termed methuosis, which is distinct from autophagy. In this section we will summarize the emerging information in this evolving area of Ras biology.
8.1 Ras and Autophagy
Mechanistic insight into one pathway that can link Ras activation to autophagy came from the work of Codogno and colleagues. They discovered that ERK1/2 dependent phosphorylation of Gα-interacting protein (GAIP), a GAP for the α subunit of the trimeric G-protein, Gi3, stimulates autophagy in colon carcinoma cells in response to amino acid starvation (213). They then went on to establish the importance of the Ras →Raf→MEK →ERK signaling pathway in this process by showing that Ras(V12) could mimic the downstream effects of starvation on GAIP and autophagy (214). However, in these studies the long term effects of autophagy induction by Ras on cell viability were not explored.
A more recently discovered mechanism that appears to link Ras to the induction of autophagy involves activation of Rac1. Byun et al. (215) found that overexpression of activated H- or K-Ras, but not N-Ras, was able to induce a high level of caspase-independent cell death in three different fibroblast cell lines. Cell death was dependent on activation of both PI 3-Kinase and Rac1, although these appeared to work through parallel pathways, since activation of Rac1 did not require activation of PI 3-Kinase. In a follow-up study, the same group reported that Ras-induced death of the fibroblasts was accompanied by increased expression of the autophagosome-associated protein Atg5, and accumulation of autophagosomes (135). The likelihood that autophagy was a death-promoting event rather than a survival response was supported by the observation that inhibitors of autophagy were able to protect the cells from Ras-induced death. The induction of autophagic death by Ras in this case required activation of Rac1 and downstream signaling via the Rac1→MKK7→JNK→ c-Jun pathway, as cells were protected by chemical inhibition or siRNA knockdown of components in this pathway, but not the ERK1/2 or p38 kinase pathways (135).
8.2 Ras and Methuosis
Following the initial reports of the induction of non-apoptotic cell death in glioblastoma cells by activated H-Ras (207,208), our research group became intrigued by the cell death phenotype observed in these cells because some of the features appeared inconsistent with the classification as autophagic cell death. Specifically, although the cells clearly contained autophagosomes, the majority of the cytoplasmic vacuoles in the dying cells were phase lucent empty structures that were morphologically distinct from classical autophagic vacuoles and autolysosomes. Therefore, we undertook a series of studies to define more precisely the origin of the Ras-induced vacuoles and the nature of the Ras signaling mechanisms involved in triggering this unique process. These studies led us to define a novel non-apoptotic, non-autophagic, death pathway that can be induced by overexpression of activated Ras proteins in glioblastoma and other cancer cell lines.
Upon close examination by electron microscopy, the vacuoles induced by H-Ras(V12) in glioblastoma cells did not contain degraded proteins or organelles. Nor were they surrounded by a double membrane characteristic of autophagosomes. Moreover, the vacuoles, which contained the activated Ras protein in their limiting membranes, were not acidic and did not display LC3-II, an autophagosome-specific marker (205) (Fig. 2A). Although autophagy was increased in the cells expressing Ras(V12), siRNA knockdown of the pro-autophagy protein Beclin-1 did not block the accumulation of vacuoles or prevent cell death in cells expressing H-Ras(V12). Taken together, these observations confirmed our initial impression that the phase-lucent vacuoles induced by Ras(V12) expression in glioblastoma cells are not autophagosomes or autolysosomes and that the death mechanism might be distinct from autophagic cell death.
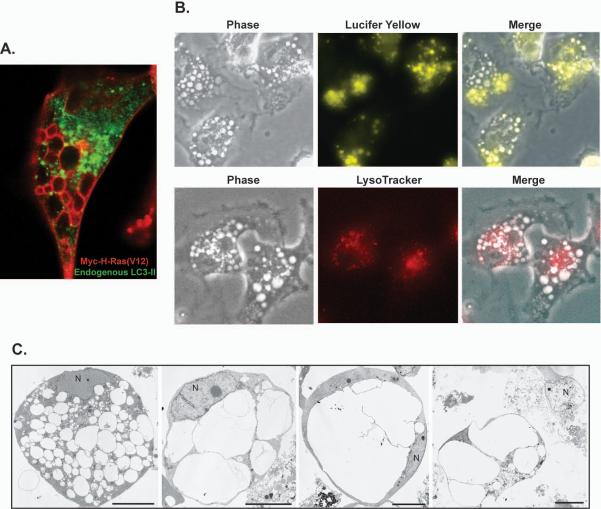
Figure 2.
Ras(V12) induces macropinocytosis and cell death by a non-apoptotic mechanism termed methuosis. A) In glioblastoma cells expressing Ras(V12), LC3-II positive autophagosomes (green) are clearly separate from the large vacuoles circumscribed with myc-tagged H-Ras(V12) (red). B) The vacuoles induced by induced expression of activated Ras in these cells are macropinosomes, as determined in part by their incorporation of the extracellular fluid-phase tracer, Lucifer yellow. The vacuoles are distinct from lysosomes as shown by their exclusion of LysoTracker Red. C) Electron micrographs of detached U251glioblastoma cells after 4–6 days of Ras(V12) expression reveal cells filled with large vacuoles that have coalesced and filled the cytoplasmic space, eventually causing disruption of the cell. Note that the nuclei (N) do not exhibit chromatin condensation typical of apoptosis. Reprinted with permission from Mol Cancer Res 2008;6(6)965–977.
The preponderance of evidence now indicates that the large vacuoles induced by Ras(V12) in glioma cells are in fact derived from macropinosomes (205,206). For example, the Ras-induced vacuoles incorporate fluid-phase tracers (Fig.2B) but they remain distinct from clathrin-coated early endosomes that incorporate transferrin and harbor EEA1, an early endosome marker (205). In combination with the stimulation of macropinocytosis, Ras(V12) appears to induce significant defects in vesicular trafficking, as the vacuoles do not appear to recycle or merge with lysosomes (Fig.2B), although they do acquire the late endosomal markers LAMP1(205) and Rab7 (206) on the surrounding membrane. The accrual of vacuoles eventually fills the cytoplasmic space, leading to cell detachment and cell lysis reminiscent of necrosis (Fig.2C). Throughout this degenerative process, the cells do not undergo DNA fragmentation or nuclear changes typically associated with apoptosis (205) (Fig. 2C), and caspase inhibitors do not prevent death (205,206). Based on the initiating factor of unchecked macropinocytosis (cell drinking), we dubbed this unique form of Ras-induced necrotic cell death, methuosis (methuo, from the Greek; to drink to intoxication).
To elucidate the mechanism of Ras(V12)-induced methuosis, several of the well studied Ras effector pathways were examined. Based on inhibitor studies, as well as the use of constitutively active Raf constructs, we determined that Ras-induced vacuolization was not dependent on the activation of the Raf → MEK →ERK pathway (216). Furthermore, the PI 3-Kinase pathway was not required for vacuole induction, as determined through the use of Ras effector domain mutations or re-introduction of a normal PTEN gene into the U251 glioblastoma cell line, reducing the activity of the PI 3-Kinase pathway (216). A third Ras effector that did not appear to be involved in the formation of vacuoles was RalGDS. Neither Ras effector domain point mutations that prevent interaction with RalGDS nor co-expression of activated Ras with a dominant negative RalA prevented the formation of vacuoles (216). An important clue concerning the Ras effector pathway that was crucial for the induction of vacuolization was provided by the finding that activated Rac1, but not the active form of other small GTPases, like RhoA and Cdc42, could mimic the phenotype induced by Ras(V12) (216).
It has been known for a number of years that activated Ras can induce macropinocytosis (217) and that the latter is linked to activation of Rac (218). However, a relationship between Ras-mediated induction of macropinocytosis and non-apoptotic cell death had not been suspected. In a recent study we defined a requirement for Rac1 activation in methuosis induced by Ras(V12) (206). Expression of activated Ras caused an increase in Rac activation and cell vacuolization in proportion to the relative amount of H-Ras(V12) expressed. Additionally, in the presence of the Rac inhibitor, EHT 1864 (219), Ras-induced vacuolization was blocked. This, combined with evidence that co-expression of dominant-negative Rac1 with activated Ras prevented vacuolization, indicated that activated Rac1 acts downstream from Ras(V12) to induce methuosis (206). It remains to be determined precisely how H-Ras(V12) activates Rac in the context of methuosis, since knockdown of the Rac GEF, Tiam1, did not alter the Ras-induced death phenotype (206).
Downstream from Rac, it appears that the pathway for inducing methuosis involves interaction of activated Rac with components that regulate the trafficking of clathrin-independent endosomes (CIE). Ras(V12)-induced activation of Rac1 leads to a decline in endogenous levels of active Arf6 (206), a GTPase that controls endosome recycling (220). This reduction in Arf6-GTP was shown to be mediated through an interaction between activated Rac1 and an Arf6 GAP, GIT1 (206). Knockdown of GIT1 not only prevented the decrease in active Arf6, but also protected the cells from Rac(V12)- mediated vacuolization and cell death (206). Thus, it appears that interference with CIE recycling, perhaps coupled with additional defects at the step of endosome-lysosome fusion, triggers the accumulation of CIE-derived vacuoles that ultimately disrupt cellular integrity in methuosis. We have observed that Ras(V12) can induce this form of non-apoptotic death in multiple glioma lines (205), as well as in osteosarcoma and human embryonal kidney 293 cells (206). Therefore, it methuosis is not a peculiarity of a single cell line.
9. Perspectives
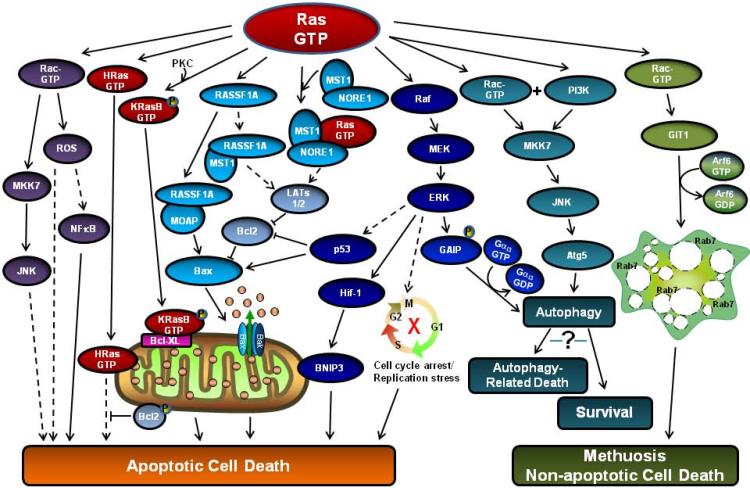
Figure 3 attempts to provide an overview of the growing number of Ras dependent pathways that have the potential to trigger cell death in normal and/or cancer cells. Although the evidence for some specific pathways is still rather sketchy, overall there is strong support for the notion that activation of endogenous Ras proteins can be important for initiation of apoptosis in a variety of normal and transformed cell types in response to environmental stimuli (death receptor ligands, growth factor withdrawal, nutrient deprivation). It is also clear that activation of Ras can play a key role in mediating the apoptotic response of cancer cells to cytotoxic drugs. At the same time, data accumulated over three decades strongly support the conventional view of Ras as an initiator of signals required for stimulation of cell growth and survival in response to growth factors and hormones and during the process of cell transformation. The great challenge has been, and continues to be, defining the specific factors that go into changing Ras from a pro-survival to a pro-death switch. As noted throughout this review, it is important to recognize that not all members of the Ras family are equal with respect to their ability to induce apoptosis. Therefore, the relative expression levels of different Ras proteins in different cell types may be critical for determining the nature of the response to a specific stimulus. Whether Ras activation transmits a pro-apoptotic or a mitogenic signal will also depend to a great extent on the spectrum of effector proteins it encounters within the cell. Some effector proteins, like NORE1, have a clear apoptotic function, while others, like PI 3-Kinase, are more likely to transmit pro-survival signals. The situation with some well known Ras effectors, like Raf, is less certain, as there is good evidence for both mitogenic and pro-apoptotic functions. The recognition that members of the Ras family can undergo a number of unique post-translational modifications (prenylation, carboxylmethylation, palmitylation, phosphorylation, nitrosylation) that may influence their localization within the plasma membrane and other subcellular compartments, coupled with the concept that Ras proteins can actively signal from endomembrane compartments, provides a possible framework for understanding how a given Ras protein might come into contact with a different range of effectors, depending on the physiological circumstances and the specific stimuli impinging on the cell.
Figure 3.
A summary of cell death pathways that may be induced by activated Ras. Supporting evidence and references for each of the indicated pathways are mentioned throughout the text of the review. Dashed arrows indicate that details of the intervening step(s) are unclear, or that the connection with activated Ras has not yet been firmly established.
In addition to the numerous studies demonstrating a role for endogenous Ras activation in apoptotic signaling, there are many examples of studies where apoptosis or non-apoptotic cell death have been triggered in cancer cells by ectopic overexpression of Ras constructs bearing activation mutations, usually V12. The fact that these mutant Ras proteins can induce cell death, despite overwhelming evidence that they are oncogenic, makes it tempting to dismiss the paradoxical pro-death effects as artifacts of Ras overexpression. There is some merit to that argument, since the ability of RasV12 to induce cell death is generally manifested at expression levels that exceed endogenous levels and may therefore not represent true physiological functions of Ras. As an example, the death-promoting effects of ectopic RasV12 observed in glioma cell lines (205,207) contrast sharply with studies showing that Ras plays an important role in glioblastoma tumorigenesis and survival (221–223). Nevertheless, we would argue that information gleaned from the studies with overexpression of Ras at “non-physiological” levels may still be of value in pursuing new avenues for cancer therapy. For example, the prognosis for patients with glioblastoma remains poor, in large part due to the fact that glioblastoma cells harbor genetic mutations that render them resistant to apoptosis induced by conventional therapies (224,225). The discovery of a novel cell death mechanism induced by overexpression of RasV12 (methuosis), which does not depend on activation of the classical mitochondrial apoptotic pathways, could present new opportunities to induce cell death in tumors, provided that one can identify the responsible signaling pathways and find ways to manipulate them pharmacologically. Similar logic would also apply to the definition of autophagic pathways or NORE1-dependent apoptotic pathways stimulated by overexpression of activated Ras.
10. Acknowledgements
The work from the laboratory of the authors was supported by a grant from the National Institutes of Health, R01 CA115495.
11. References
- 1.Barbacid M. Ras genes. Annu.Rev.Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Lowy DR, Willumsen BM. Function and regulation of Ras. Annu.Rev.Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 3.Chang EH, Gonda MA, Ellis RW, Scolnick EM, Lowy DR. Human genome contains four genes homologous to transforming genes of Harvey and Kirsten murine sarcoma viruses. Proc.Natl.Acad.Sci.U.S.A. 1982;79:4848–4852. doi: 10.1073/pnas.79.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall A, Marshall CJ, Spurr NK, Weiss RA. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983;303:396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- 5.Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: Evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 6.Hancock JF. Ras proteins: different signals from different locations. Nat.Rev.Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 7.Lowe DG, Capon DJ, Delwart E, Sakaguchi AY, Naylor SL, Goeddel DV. Structure of the human and murine R-ras genes, novel genes closely related to ras protooncogenes. Cell. 1987;48:137–146. doi: 10.1016/0092-8674(87)90364-3. [DOI] [PubMed] [Google Scholar]
- 8.Chan AM, Miki T, Meyers KA, Aaronson SA. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc.Natl.Acad.Sci.U.S.A. 1994;91:7558–7562. doi: 10.1073/pnas.91.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimmelman A, Tolkacheva T, Lorenzi MV, Osada M, Chan AM. Identification and characterization of R-ras3: a novel member of the RAS gene family with a non-ubiquitous pattern of tissue distribution. Oncogene. 1997;15:2675–2685. doi: 10.1038/sj.onc.1201674. [DOI] [PubMed] [Google Scholar]
- 10.Saez R, Chan AM, Miki T, Aaronson SA. Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene. 1994;9:2977–2982. [PubMed] [Google Scholar]
- 11.Graham SM, Vojtek AB, Huff SY, Cox AD, Clark GJ, Cooper JA, Der CJ. TC21 causes transformation by Raf-independent signaling pathways. Mol Cell Biol. 1996;16:6132–6140. doi: 10.1128/mcb.16.11.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oertli B, Han J, Marte BM, Sethi T, Downward J, Ginsberg M, Hughes PE. The effector loop and prenylation site of R-Ras are involved in the regulation of integrin function. Oncogene. 2000;19:4961–4969. doi: 10.1038/sj.onc.1203876. [DOI] [PubMed] [Google Scholar]
- 13.Hughes PE, Oertli B, Han J, Ginsberg MH. R-Ras regulation of integrin function. Methods Enzymol. 2001;333:163–171. doi: 10.1016/s0076-6879(01)33054-9. [DOI] [PubMed] [Google Scholar]
- 14.Wozniak MA, Kwong L, Chodniewicz D, Klemke RL, Keely PJ. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol Biol.Cell. 2005;16:84–96. doi: 10.1091/mbc.E04-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Hunt DF, Ginsberg MH. RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. J.Cell Biol. 2006;174:877–888. doi: 10.1083/jcb.200603111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang EH, Furth ME, Scolnick EM, Lowy DR. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982;297:479–483. doi: 10.1038/297479a0. [DOI] [PubMed] [Google Scholar]
- 17.Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 18.Krengel U, Schlichting L, Scherer A, Schumann R, Frech M, John J, Kabsch W, Pai EF, Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 19.Conti CJ. Mutations of genes of the ras family in human and experimental tumors. Prog.Clin.Biol.Res. 1992;376:357–378. [PubMed] [Google Scholar]
- 20.Sills RC, Boorman GA, Neal JE, Hong HL, Devereux TR. Mutations in ras genes in experimental tumours of rodents. IARC Sci.Publ. 1999:55–86. [PubMed] [Google Scholar]
- 21.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 22.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 23.Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 24.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 25.Smit VT, Boot AJ, Smits AM, Fleuren GJ, Cornelisse CJ, Bos JL. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res. 1988;16:7773–7782. doi: 10.1093/nar/16.16.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burchill SA, Neal DE, Lunec J. Frequency of H-ras mutations in human bladder cancer detected by direct sequencing. Br.J.Urol. 1994;73:516–521. doi: 10.1111/j.1464-410x.1994.tb07636.x. [DOI] [PubMed] [Google Scholar]
- 27.Bos JL, Verlaan-de Vries M, van der Eb AJ, Janssen JW, Delwel R, Lowenberg B, Colly LP. Mutations in N-ras predominate in acute myeloid leukemia. Blood. 1987;69:1237–1241. [PubMed] [Google Scholar]
- 28.Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22:8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 29.Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J.Biol.Chem. 1998;273:5439–5442. doi: 10.1074/jbc.273.10.5439. [DOI] [PubMed] [Google Scholar]
- 30.Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, Avruch J. Identification of a novel Ras-regulated proapoptotic pathway. Curr.Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 31.Vos MD, Dallol A, Eckfeld K, Allen NP, Donninger H, Hesson LB, Calvisi D, Latif F, Clark GJ. The RASSF1A tumor suppressor activates Bax via MOAP-1. J.Biol.Chem. 2006;281:4557–4563. doi: 10.1074/jbc.M512128200. [DOI] [PubMed] [Google Scholar]
- 32.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 33.Benanti JA, Galloway DA. The normal response to RAS: senescence or transformation? Cell Cycle. 2004;3:715–717. [PubMed] [Google Scholar]
- 34.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 35.Wittinghofer A, Pai EF. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem.Sci. 1991;16:382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 36.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 37.Shima F, Ijiri Y, Muraoka S, Liao J, Ye M, Araki M, Matsumoto K, Yamamoto N, Sugimoto T, Yoshikawa Y, Kumasaka T, Yamamoto M, Tamura A, Kataoka T. Structural basis for conformational dynamics of GTP-bound Ras protein. J.Biol.Chem. 2010;285:22696–22705. doi: 10.1074/jbc.M110.125161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Y, Quilliam LA. Activation of the Ras superfamily of small GTPases. Workshop on exchange factors. EMBO Rep. 2003;4:463–468. doi: 10.1038/sj.embor.embor831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J.Biol.Chem. 2009;284:10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 42.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 43.Cully M, Downward J. SnapShot: Ras Signaling. Cell. 2008;133:1292. doi: 10.1016/j.cell.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Scheele JS, Rhee JM, Boss GR. Determination of absolute amounts of GDP and GTP bound to Ras in mammalian cells: comparison of parental and Rasoverproducing NIH 3T3 fibroblasts. Proc.Natl.Acad.Sci.U.S.A. 1995;92:1097–1100. doi: 10.1073/pnas.92.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 46.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 47.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 48.Pai EF, Krengel U, Petsko GA, Goody RS, Kansch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H- ras p21 at 1.35A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moodie SA, Paris M, Villafranca E, Kirshmeier P, Willumsen BM, Wolfman A. Different structural requirements within the switchII region of the Ras protein for interactions with specific downstream targets. Oncogene. 1995;11:447–454. [PubMed] [Google Scholar]
- 50.Wittinghofer A, Scheffzek K, Ahmadian MR. The interaction of Ras with GTPase-activating proteins. FEBS Lett. 1997;410:63–67. doi: 10.1016/s0014-5793(97)00321-9. [DOI] [PubMed] [Google Scholar]
- 51.Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim.Biophys.Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- 52.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat.Rev.Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 53.Katz ME, McCormick F. Signal transduction from multiple Ras effectors. Curr.Opin.Genet.Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 54.Marshall CJ. Ras effectors. Curr.Opin.Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 56.Davis RJ. Transcriptional regulation by MAP kinases. Mol.Reprod.Dev. 1995;42:459–467. doi: 10.1002/mrd.1080420414. [DOI] [PubMed] [Google Scholar]
- 57.Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J.Biol.Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 58.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitin NY, Ramocki MB, Zullo AJ, Der CJ, Konieczny SF, Taparowsky EJ. Identification and characterization of rain, a novel Ras-interacting protein with a unique subcellular localization. J Biol.Chem. 2004;279:22353–22361. doi: 10.1074/jbc.M312867200. [DOI] [PubMed] [Google Scholar]
- 60.Vos MD, Martinez A, Ellis CA, Vallecorsa T, Clark GJ. The pro-apoptotic Ras effector Nore1 may serve as a Ras-regulated tumor suppressor in the lung. J Biol.Chem. 2003;278:21938–21943. doi: 10.1074/jbc.M211019200. [DOI] [PubMed] [Google Scholar]
- 61.Kuriyama M, Harada N, Kuroda S, Yamamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C, Canaani E, Kaibuchi K. Identification of AF-6 and Canoe as Putative Targets for Ras. J. Biol. Chem. 1996;271:607–610. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, Rehmann H, Price LS, Riedl J, Bos JL. AF6 negatively regulates Rap1-induced cell adhesion. J. Biol. Chem. 2005;280:33200–33205. doi: 10.1074/jbc.M505057200. [DOI] [PubMed] [Google Scholar]
- 63.Feig LA, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem.Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 64.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev.Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 65.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG, Der CJ. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat.Cell Biol. 2002;4:621–625. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 66.Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J.Biol.Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 67.Voice JK, Klemke RL, Le A, Jackson JH. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J.Biol.Chem. 1999;274:17164–17170. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 68.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J.Biol.Chem. 2001;276:15609–15615. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- 69.Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem.J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henis YI, Hancock JF, Prior IA. Ras acylation, compartmentalization and signaling nanoclusters. Mol Membr.Biol. 2009;26:80–92. doi: 10.1080/09687680802649582. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Philips MR. Compartmentalized signalling of Ras. Biochem.Soc.Trans. 2005;33:657–661. doi: 10.1042/BST0330657. [DOI] [PubMed] [Google Scholar]
- 72.Neubauer A, Maharry K, Mrozek K, Thiede C, Marcucci G, Paschka P, Mayer RJ, Larson RA, Liu ET, Bloomfield CD. Patients with acute myeloid leukemia and RAS mutations benefit most from postremission high-dose cytarabine: a Cancer and Leukemia Group B study. J.Clin.Oncol. 2008;26:4603–4609. doi: 10.1200/JCO.2007.14.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka T, Slamon DJ, Shimada H, Shimoda H, Fujisawa T, Ida N, Seeger RC. A significant association of Ha-ras p21 in neuroblastoma cells with patient prognosis. A retrospective study of 103 cases. Cancer. 1991;68:1296–1302. doi: 10.1002/1097-0142(19910915)68:6<1296::aid-cncr2820680619>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 74.Van Cruchten S, Van Den BW. Morphological and biochemical aspects of apoptosis, oncosis and necrosis. Anat.Histol.Embryol. 2002;31:214–223. doi: 10.1046/j.1439-0264.2002.00398.x. [DOI] [PubMed] [Google Scholar]
- 75.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat.Rev.Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 76.Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat.Rev.Mol Cell Biol. 2008;9:378–390. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- 77.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 78.Mayo MW, Wang CY, Cogswell PC, Rogers-Graham KS, Lowe SW, Der CJ, Baldwin AS., Jr. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 79.Gulbins E, Brenner B, Schlottmann K, Welsch J, Heinle H, Koppenhoefer U, Linderkamp O, Coggeshall KM, Lang F. Fas-induced programmed cell death is mediated by a Ras-regulated O2- synthesis. Immunology. 1996;89:205–212. doi: 10.1046/j.1365-2567.1996.d01-743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gomez J, Martinez A, Fernandez B, Garcia A, Rebollo A. Critical role of Ras in the proliferation and prevention of apoptosis mediated by IL-2. J.Immunol. 1996;157:2272–2281. [PubMed] [Google Scholar]
- 81.Gomez J, Martinez C, Fernandez B, Garcia A, Rebollo A. Ras activation leads to cell proliferation or apoptotic cell death upon interleukin-2 stimulation or lymphokine deprivation, respectively. Eur.J.Immunol. 1997;27:1610–1618. doi: 10.1002/eji.1830270704. [DOI] [PubMed] [Google Scholar]
- 82.Woessmann W, Chen X, Borkhardt A. Ras-mediated activation of ERK by cisplatin induces cell death independently of p53 in osteosarcoma and neuroblastoma cell lines. Cancer Chemother.Pharmacol. 2002;50:397–404. doi: 10.1007/s00280-002-0502-y. [DOI] [PubMed] [Google Scholar]
- 83.Yang R, Piperdi S, Gorlick R. Activation of the RAF/mitogen-activated protein/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase pathway mediates apoptosis induced by chelerythrine in osteosarcoma. Clin.Cancer Res. 2008;14:6396–6404. doi: 10.1158/1078-0432.CCR-07-5113. [DOI] [PubMed] [Google Scholar]
- 84.Schwieger A, Bauer L, Hanusch J, Sers C, Schafer R, Bauer G. Ras oncogene expression determines sensitivity for intercellular induction of apoptosis. Carcinogenesis. 2001;22:1385–1392. doi: 10.1093/carcin/22.9.1385. [DOI] [PubMed] [Google Scholar]
- 85.Choi JA, Park MT, Kang CM, Um HD, Bae S, Lee KH, Kim TH, Kim JH, Cho CK, Lee YS, Chung HY, Lee SJ. Opposite effects of Ha-Ras and Ki-Ras on radiation-induced apoptosis via differential activation of PI3K/Akt and Rac/p38 mitogen-activated protein kinase signaling pathways. Oncogene. 2004;23:9–20. doi: 10.1038/sj.onc.1206982. [DOI] [PubMed] [Google Scholar]
- 86.Ninomiya Y, Kato K, Takahashi A, Ueoka Y, Kamikihara T, Arima T, Matsuda T, Kato H, Nishida J, Wake N. K-Ras and H-Ras activation promote distinct consequences on endometrial cell survivala. Cancer Res. 2004;64:2759–2765. doi: 10.1158/0008-5472.can-3487-2. [DOI] [PubMed] [Google Scholar]
- 87.Geryk-Hall M, Yang Y, Hughes DP. Driven to death: inhibition of farnesylation increases Ras activity in osteosarcoma and promotes growth arrest and cell death. Mol Cancer Ther. 2010;9:1111–1119. doi: 10.1158/1535-7163.MCT-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez d. C., I, Li C, Thompson CB, Cox AD, Philips MR. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 89.Miranda EI, Santana C, Rojas E, Hernandez S, Ostrosky-Wegman P, Garcia-Carranca A. Induced mitotic death of HeLa cells by abnormal expression of c-H-ras. Mutat.Res. 1996;349:173–182. doi: 10.1016/0027-5107(95)00164-6. [DOI] [PubMed] [Google Scholar]
- 90.Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kurokawa M, Kornbluth S. Stalling in mitosis and releasing the apoptotic brake. EMBO J. 2010;29:2255–2257. doi: 10.1038/emboj.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shao J, Sheng H, DuBois RN, Beauchamp RD. Oncogenic Ras-mediated cell growth arrest and apoptosis are associated with increased ubiquitin-dependent cyclin D1 degradation. J.Biol.Chem. 2000;275:22916–22924. doi: 10.1074/jbc.M002235200. [DOI] [PubMed] [Google Scholar]
- 93.Cheng G, Lewis AE, Meinkoth JL. Ras stimulates aberrant cell cycle progression and apoptosis in rat thyroid cells. Mol.Endocrinol. 2003;17:450–459. doi: 10.1210/me.2002-0344. [DOI] [PubMed] [Google Scholar]
- 94.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J.Biol.Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 95.Liu JJ, Chao JR, Jiang MC, Ng SY, Yen JJ, Yang-Yen HF. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Winston JT, Coats SR, Wang YZ, Pledger WJ. Regulation of the cell cycle machinery by oncogenic ras. Oncogene. 1996;12:127–134. [PubMed] [Google Scholar]
- 97.Arber N, Sutter T, Miyake M, Kahn SM, Venkatraj VS, Sobrino A, Warburton D, Holt PR, Weinstein IB. Increased expression of cyclin D1 and the Rb tumor suppressor gene in c-K-ras transformed rat enterocytes. Oncogene. 1996;12:1903–1908. [PubMed] [Google Scholar]
- 98.Fikaris AJ, Lewis AE, Abulaiti A, Tsygankova OM, Meinkoth JL. Ras triggers ataxia-telangiectasia-mutated and Rad-3-related activation and apoptosis through sustained mitogenic signaling. J.Biol.Chem. 2006;281:34759–34767. doi: 10.1074/jbc.M606737200. [DOI] [PubMed] [Google Scholar]
- 99.Kolch W, Heidecker G, Lloyd P, Rapp UR. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 100.Moodie SA, Wolfman A. The 3Rs of life: Ras, Raf and growth regulation. Trends Genet. 1994;10:44–48. doi: 10.1016/0168-9525(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 101.Cowley S, Paterson H, Kemp P, Marshall CJ. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 102.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 103.Castellone MD, Cirafici AM, De Vita G, De F, V, Malorni L, Tallini G, Fagin JA, Fusco A, Melillo RM, Santoro M. Ras-mediated apoptosis of PC CL 3 rat thyroid cells induced by RET/PTC oncogenes. Oncogene. 2003;22:246–255. doi: 10.1038/sj.onc.1206112. [DOI] [PubMed] [Google Scholar]
- 104.Zhuang S, Schnellmann RG. A death-promoting role for extracellular signal-regulated kinase. J.Pharmacol.Exp.Ther. 2006;319:991–997. doi: 10.1124/jpet.106.107367. [DOI] [PubMed] [Google Scholar]
- 105.Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death-apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 106.Hill JM, Vaidyanathan H, Ramos JW, Ginsberg MH, Werner MH. Recognition of ERK MAP kinase by PEA-15 reveals a common docking site within the death domain and death effector domain. EMBO J. 2002;21:6494–6504. doi: 10.1093/emboj/cdf641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Renganathan H, Vaidyanathan H, Knapinska A, Ramos JW. Phosphorylation of PEA-15 switches its binding specificity from ERK/MAPK to FADD. Biochem.J. 2005;390:729–735. doi: 10.1042/BJ20050378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nowak G. Protein kinase C-alpha and ERK1/2 mediate mitochondrial dysfunction, decreases in active Na+ transport, and cisplatin-induced apoptosis in renal cells. J.Biol.Chem. 2002;277:43377–43388. doi: 10.1074/jbc.M206373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nowak G, Clifton GL, Godwin ML, Bakajsova D. Activation of ERK1/2 pathway mediates oxidant-induced decreases in mitochondrial function in renal cells. Am.J.Physiol. Renal Physiol. 2006;291:F840–F855. doi: 10.1152/ajprenal.00219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Drosopoulos KG, Roberts ML, Cermak L, Sasazuki T, Shirasawa S, Andera L, Pintzas A. Transformation by oncogenic RAS sensitizes human colon cells to TRAIL-induced apoptosis by up-regulating death receptor 4 and death receptor 5 through a MEK-dependent pathway. J.Biol.Chem. 2005;280:22856–22867. doi: 10.1074/jbc.M412483200. [DOI] [PubMed] [Google Scholar]
- 111.DeHaan RD, Yazlovitskaya EM, Persons DL. Regulation of p53 target gene expression by cisplatin-induced extracellular signal-regulated kinase. Cancer Chemother.Pharmacol. 2001;48:383–388. doi: 10.1007/s002800100318. [DOI] [PubMed] [Google Scholar]
- 112.Li DW, Liu JP, Mao YW, Xiang H, Wang J, Ma WY, Dong Z, Pike HM, Brown RE, Reed JC. Calcium-activated RAF/MEK/ERK signaling pathway mediates p53-dependent apoptosis and is abrogated by alpha B-crystallin through inhibition of RAS activation. Mol Biol.Cell. 2005;16:4437–4453. doi: 10.1091/mbc.E05-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brown L, Benchimol S. The involvement of MAPK signaling pathways in determining the cellular response to p53 activation: cell cycle arrest or apoptosis. J.Biol.Chem. 2006;281:3832–3840. doi: 10.1074/jbc.M507951200. [DOI] [PubMed] [Google Scholar]
- 114.Feig LA, Buchsbaum RJ. Cell signaling: life or death decisions of ras proteins. Curr.Biol. 2002;12:R259–R261. doi: 10.1016/s0960-9822(02)00787-x. [DOI] [PubMed] [Google Scholar]
- 115.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, Collard JG. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 116.Chou CK, Liang KH, Tzeng CC, Huang GC, Chuang JI, Chang TY, Liu HS. Dominant-negative Rac1 suppresses Ras-induced apoptosis possibly through activation of NFkappaB in Ha-ras oncogene-transformed NIH/3T3 cells. Life Sci. 2006;78:1823–1829. doi: 10.1016/j.lfs.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 117.Joneson T, Bar-Sagi D. Suppression of Ras-induced apoptosis by the Rac GTPase. Mol.Cell Biol. 1999;19:5892–5901. doi: 10.1128/mcb.19.9.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 119.Kaltschmidt B, Kaltschmidt C, Hofmann TG, Hehner SP, Droge W, Schmitz ML. The pro- or anti-apoptotic function of NF-kappaB is determined by the nature of the apoptotic stimulus. Eur.J.Biochem. 2000;267:3828–3835. doi: 10.1046/j.1432-1327.2000.01421.x. [DOI] [PubMed] [Google Scholar]
- 120.Abo A, Pick E, Hall A, Totty N, Teahan CG, Segal AW. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- 121.Knaus UG, Heyworth PG, Evans T, Curnutte JT, Bokoch GM. Regulation of phagocyte oxygen radical production by the GTP-binding protein Rac 2. Science. 1991;254:1512–1515. doi: 10.1126/science.1660188. [DOI] [PubMed] [Google Scholar]
- 122.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, Goldschmidt-Clermont PJ, Finkel T. Regulation of reactive-oxygen-species generation in fibroblasts by Rac1. Biochem.J. 1996;318(Pt 2):379–382. doi: 10.1042/bj3180379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Joneson T, Bar-Sagi D. A Rac1 effector site controlling mitogenesis through superoxide production. J.Biol.Chem. 1998;273:17991–17994. doi: 10.1074/jbc.273.29.17991. [DOI] [PubMed] [Google Scholar]
- 124.Kim KS, Takeda K, Sethi R, Pracyk JB, Tanaka K, Zhou YF, Yu ZX, Ferrans VJ, Bruder JT, Kovesdi I, Irani K, Goldschmidt-Clermont P, Finkel T. Protection from reoxygenation injury by inhibition of rac1. J.Clin.Invest. 1998;101:1821–1826. doi: 10.1172/JCI1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ozaki M, Deshpande SS, Angkeow P, Suzuki S, Irani K. Rac1 regulates stress-induced, redox-dependent heat shock factor activation. J.Biol.Chem. 2000;275:35377–35383. doi: 10.1074/jbc.M005287200. [DOI] [PubMed] [Google Scholar]
- 126.Johnson TM, Yu ZX, Ferrans VJ, Lowenstein RA, Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc.Natl.Acad.Sci.U.S.A. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Simon HU, Haj-Yehia A, Levi-Schaffer F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis. 2000;5:415–418. doi: 10.1023/a:1009616228304. [DOI] [PubMed] [Google Scholar]
- 128.Holland PM, Suzanne M, Campbell JS, Noselli S, Cooper JA. MKK7 is a stress-activated mitogen-activated protein kinase kinase functionally related to hemipterous. J.Biol.Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 129.Ho DT, Bardwell AJ, Grewal S, Iverson C, Bardwell L. Interacting JNK-docking sites in MKK7 promote binding and activation of JNK mitogen-activated protein kinases. J.Biol.Chem. 2006;281:13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 131.Brenner B, Koppenhoefer U, Weinstock C, Linderkamp O, Lang F, Gulbins E. Fas- or ceramide-induced apoptosis is mediated by a Rac1-regulated activation of Jun N-terminal kinase/p38 kinases and GADD153. J.Biol.Chem. 1997;272:22173–22181. doi: 10.1074/jbc.272.35.22173. [DOI] [PubMed] [Google Scholar]
- 132.Embade N, Valeron PF, Aznar S, Lopez-Collazo E, Lacal JC. Apoptosis induced by Rac GTPase correlates with induction of FasL and ceramides production. Mol Biol.Cell. 2000;11:4347–4358. doi: 10.1091/mbc.11.12.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jin S, Ray RM, Johnson LR. Rac1 mediates intestinal epithelial cell apoptosis via JNK. Am.J.Physiol Gastrointest.Liver Physiol. 2006;291:G1137–G1147. doi: 10.1152/ajpgi.00031.2006. [DOI] [PubMed] [Google Scholar]
- 134.Jin S, Ray RM, Johnson LR. TNF-alpha/cycloheximide-induced apoptosis in intestinal epithelial cells requires Rac1-regulated reactive oxygen species. Am.J.Physiol. Gastrointest.Liver Physiol. 2008;294:G928–G937. doi: 10.1152/ajpgi.00219.2007. [DOI] [PubMed] [Google Scholar]
- 135.Byun JY, Yoon CH, An S, Park IC, Kang CM, Kim MJ, Lee SJ. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis. 2009;30:1880–1888. doi: 10.1093/carcin/bgp235. [DOI] [PubMed] [Google Scholar]
- 136.Korsmeyer SJ. BCL-2 gene family and the regulation of programmed cell death. Cancer Res. 1999;59:1693s–1700s. [PubMed] [Google Scholar]
- 137.Reed JC. Mechanisms of apoptosis. Am.J.Pathol. 2000;157:1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Danial NN. BCL-2 family proteins: critical checkpoints of apoptotic cell death. Clin.Cancer Res. 2007;13:7254–7263. doi: 10.1158/1078-0432.CCR-07-1598. [DOI] [PubMed] [Google Scholar]
- 139.Reed JC, Jurgensmeier JM, Matsuyama S. Bcl-2 family proteins and mitochondria. Biochim.Biophys.Acta. 1998;1366:127–137. doi: 10.1016/s0005-2728(98)00108-x. [DOI] [PubMed] [Google Scholar]
- 140.Leibowitz B, Yu J. Mitochondrial signaling in cell death via the Bcl-2 family. Cancer Biol.Ther. 2010;9:417–422. doi: 10.4161/cbt.9.6.11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ruiz-Vela A, Opferman JT, Cheng EH, Korsmeyer SJ. Proapoptotic BAX and BAK control multiple initiator caspases. EMBO Rep. 2005;6:379–385. doi: 10.1038/sj.embor.7400375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 143.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 144.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat.Rev.Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 145.Chen CY, Faller DV. Direction of p21ras-generated signals towards cell growth or apoptosis is determined by protein kinase C and Bcl-2. Oncogene. 1995;11:1487–1498. [PubMed] [Google Scholar]
- 146.Chen CY, Faller DV. Phosphorylation of Bcl-2 protein and association with p21Ras in Ras-induced apoptosis. J.Biol.Chem. 1996;271:2376–2379. doi: 10.1074/jbc.271.5.2376. [DOI] [PubMed] [Google Scholar]
- 147.Denis GV, Yu Q, Ma P, Deeds L, Faller DV, Chen CY. Bcl-2, via its BH4 domain, blocks apoptotic signaling mediated by mitochondrial Ras. J.Biol.Chem. 2003;278:5775–5785. doi: 10.1074/jbc.M210202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.An HJ, Maeng O, Kang KH, Lee JO, Kim YS, Paik SG, Lee H. Activation of Ras up-regulates pro-apoptotic BNIP3 in nitric oxide-induced cell death. J.Biol.Chem. 2006;281:33939–33948. doi: 10.1074/jbc.M605819200. [DOI] [PubMed] [Google Scholar]
- 149.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim.Biophys.Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 150.Wohlgemuth S, Kiel C, Kramer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J.Mol.Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 151.Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J.Biol.Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 152.Vos MD, Ellis CA, Elam C, Ulku AS, Taylor BJ, Clark GJ. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J.Biol.Chem. 2003;278:28045–28051. doi: 10.1074/jbc.M300554200. [DOI] [PubMed] [Google Scholar]
- 153.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 154.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J.Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 155.Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 156.Ke H, Pei J, Ni Z, Xia H, Qi H, Woods T, Kelekar A, Tao W. Putative tumor suppressor Lats2 induces apoptosis through downregulation of Bcl-2 and Bcl-x(L) Exp.Cell Res. 2004;298:329–338. doi: 10.1016/j.yexcr.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 157.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat.Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 158.Kitagawa D, Kajiho H, Negishi T, Ura S, Watanabe T, Wada T, Ichijo H, Katada T, Nishina H. Release of RASSF1C from the nucleus by Daxx degradation links DNA damage and SAPK/JNK activation. EMBO J. 2006;25:3286–3297. doi: 10.1038/sj.emboj.7601212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, Avruch J. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381–1390. doi: 10.1038/sj.onc.1205192. [DOI] [PubMed] [Google Scholar]
- 160.Cooper WN, Dickinson RE, Dallol A, Grigorieva EV, Pavlova TV, Hesson LB, Bieche I, Broggini M, Maher ER, Zabarovsky ER, Clark GJ, Latif F. Epigenetic regulation of the ras effector/tumour suppressor RASSF2 in breast and lung cancer. Oncogene. 2008;27:1805–1811. doi: 10.1038/sj.onc.1210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Donninger H, Hesson L, Vos M, Beebe K, Gordon L, Sidransky D, Liu JW, Schlegel T, Payne S, Hartmann A, Latif F, Clark GJ. The Ras effector RASSF2 controls the PAR-4 tumor suppressor. Mol.Cell Biol. 2010;30:2608–2620. doi: 10.1128/MCB.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chakraborty M, Qiu SG, Vasudevan KM, Rangnekar VM. Par-4 drives trafficking and activation of Fas and Fasl to induce prostate cancer cell apoptosis and tumor regression. Cancer Res. 2001;61:7255–7263. [PubMed] [Google Scholar]
- 163.El Guendy N, Zhao Y, Gurumurthy S, Burikhanov R, Rangnekar VM. Identification of a unique core domain of par-4 sufficient for selective apoptosis induction in cancer cells. Mol.Cell Biol. 2003;23:5516–5525. doi: 10.1128/MCB.23.16.5516-5525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tommasi S, Dammann R, Jin SG, Zhang XF, Avruch J, Pfeifer GP. RASSF3 and NORE1: identification and cloning of two human homologues of the putative tumor suppressor gene RASSF1. Oncogene. 2002;21:2713–2720. doi: 10.1038/sj.onc.1205365. [DOI] [PubMed] [Google Scholar]
- 165.Eckfeld K, Hesson L, Vos MD, Bieche I, Latif F, Clark GJ. RASSF4/AD037 is a potential ras effector/tumor suppressor of the RASSF family. Cancer Res. 2004;64:8688–8693. doi: 10.1158/0008-5472.CAN-04-2065. [DOI] [PubMed] [Google Scholar]
- 166.Allen NP, Donninger H, Vos MD, Eckfeld K, Hesson L, Gordon L, Birrer MJ, Latif F, Clark GJ. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene. 2007;26:6203–6211. doi: 10.1038/sj.onc.1210440. [DOI] [PubMed] [Google Scholar]
- 167.Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem.J. 2010;425:303–311. doi: 10.1042/BJ20091318. [DOI] [PubMed] [Google Scholar]
- 168.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors ehey eegulate. Mol.Cell.Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lockshin RA, Zakeri Z. Apoptosis, autophagy, and more. Int.J Biochem.Cell Biol. 2004;36:2405–2419. doi: 10.1016/j.biocel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 171.Schworer CM, Mortimore, E. G. Glucagon-induced autophagy and proteolysis in rat liver;mediation by selective deprivation of intracellular amino acids. Proc.Natl.Acad.Sci.U.S.A. 1979;76:3169–3173. doi: 10.1073/pnas.76.7.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J.Biol.Chem. 1981;256:7652–7658. [PubMed] [Google Scholar]
- 173.Dunn WA., Jr. Studies on the mechanisms of autophagy: Maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gordon P, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem. Biophys. Res. Commun. 1988;151:40–47. doi: 10.1016/0006-291x(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 175.Lawrence B, Brown W. Autophagic vacuoles rapidly fuse with pre-existing lysosomes in cultured hepatocytes. J. Cell Sci. 1992;102:515–526. doi: 10.1242/jcs.102.3.515. [DOI] [PubMed] [Google Scholar]
- 176.Dunn WA., Jr. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 177.Zakeri Z, Bursch W, Tenniswood M, Lockshin RA. Cell death:programmed, apoptosis,necrosis or other? Cell Death Differentiat. 1995;2:87–96. [PubMed] [Google Scholar]
- 178.Bursch W, Ellinger A, Gerner C, Frohwein U, Schulte-Hermann R. Programmed cell death (PCD). Apoptosis, autophagic PCD, or others? Ann.N.Y.Acad.Sci. 2000;926:1–12. doi: 10.1111/j.1749-6632.2000.tb05594.x. [DOI] [PubMed] [Google Scholar]
- 179.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 180.Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J.Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol.Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- 182.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat.Rev.Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ogier-Denis E, Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochim.Biophys.Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- 184.Hait WN, Jin S, Yang JM. A matter of life or death (or both): understanding autophagy in cancer. Clin.Cancer Res. 2006;12:1961–1965. doi: 10.1158/1078-0432.CCR-06-0011. [DOI] [PubMed] [Google Scholar]
- 185.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat.Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bursch W, Ellinger E, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the anti-estrogens tamoxifin and ICI 164 384 in human mammary carcioma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 187.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, Domingo D, Yahalom J. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–444. [PubMed] [Google Scholar]
- 188.Kanzawa T, Kondo Y, Ito H, Kondo S, Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003;63:2103–2108. [PubMed] [Google Scholar]
- 189.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death.Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 190.Yao KC, Komata T, Kondo Y, Kanzawa T, Kondo S, Germano IM. Molecular response of human glioblastoma multiforme cells to ionizing radiation: cell cycle arrest, modulation of the expression of cyclin-dependent kinase inhibitors, and autophagy. J.Neurosurg. 2003;98:378–384. doi: 10.3171/jns.2003.98.2.0378. [DOI] [PubMed] [Google Scholar]
- 191.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 192.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin.Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat.Rev.Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat.Rev.Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sperandio S, de B, I, Bredesen DE. An alternative, nonapoptotic form of programmed cell death. Proc.Natl.Acad.Sci U.S.A. 2000;97:14376–14381. doi: 10.1073/pnas.97.26.14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, Ma D. An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J Cell Sci. 2004;117:1525–1532. doi: 10.1242/jcs.00994. [DOI] [PubMed] [Google Scholar]
- 197.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am.J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 198.Trump BF, Berezesky IK, Chang SH, Phelps PC. The pathways of cell death: oncosis, apoptosis, and necrosis. Toxicol.Pathol. 1997;25:82–88. doi: 10.1177/019262339702500116. [DOI] [PubMed] [Google Scholar]
- 199.Suarez Y, Gonzalez L, Cuadrado A, Berciano M, Lafarga M, Munoz A. Kahalalide F, a new marine-derived compound, induces oncosis in human prostate and breast cancer cells. Mol. Cancer Ther. 2003;2:863–872. [PubMed] [Google Scholar]
- 200.Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat.Chem.Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 201.Han W, Li L, Qiu S, Lu Q, Pan Q, Gu Y, Luo J, Hu X. Shikonin circumvents cancer drug resistance by induction of a necroptotic death. Mol. Cancer Ther. 2007;6:1641–1649. doi: 10.1158/1535-7163.MCT-06-0511. [DOI] [PubMed] [Google Scholar]
- 202.Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 203.Syntichaki P, Tavernarakis N. Death by necrosis. Uncontrollable catastrophe, or is there order behind the chaos? EMBO Rep. 2002;3:604–609. doi: 10.1093/embo-reports/kvf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr.Opin.Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 205.Overmeyer JH, Kaul A, Johnson EE, Maltese WA. Active ras triggers death in glioblastoma cells through hyperstimulation of macropinocytosis. Mol.Cancer Res. 2008;6:965–977. doi: 10.1158/1541-7786.MCR-07-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bhanot H, Young AM, Overmeyer JH, Maltese WA. Induction of nonapoptotic cell death by activated Ras requires inverse regulation of Rac1 and Arf6. Mol. Cancer Res. 2010;8 doi: 10.1158/1541-7786.MCR-10-0090. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chi S, Kitanaka C, Noguchi K, Mochizuki T, Nagashima Y, Shirouzu M, Fujita H, Yoshida M, Chen W, Asai A, Himeno M, Yokoyama S, Kuchino Y. Oncogenic Ras triggers cell suicide through the activation of a caspase-independent cell death program in human cancer cells. Oncogene. 1999;18:2281–2290. doi: 10.1038/sj.onc.1202538. [DOI] [PubMed] [Google Scholar]
- 208.Kitanaka C, Kuchino Y. Caspase-independent programmed cell death with necrotic morphology. Cell Death.Differ. 1999;6:508–515. doi: 10.1038/sj.cdd.4400526. [DOI] [PubMed] [Google Scholar]
- 209.Moley JF, Brother MB, Wells SA, Spengler BA, Biedler JL, Brodeur GM. Low frequency of ras gene mutations in neuroblastomas, pheochromocytomas, and medullary thyroid cancers. Cancer Res. 1991;51:1596–1599. [PubMed] [Google Scholar]
- 210.Tanaka T, Sugimoto T, Sawada T. Prognostic discrimination among neuroblastomas according to Ha-ras/trk A gene expression: a comparison of the profiles of neuroblastomas detected clinically and those detected through mass screening. Cancer. 1998;83:1626–1633. doi: 10.1002/(sici)1097-0142(19981015)83:8<1626::aid-cncr19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 211.Kitanaka C, Kato K, Ijiri R, Sakurada K, Tomiyama A, Noguchi K, Nagashima Y, Nakagawara A, Momoi T, Toyoda Y, Kigasawa H, Nishi T, Shirouzu M, Yokoyama S, Tanaka Y, Kuchino Y. Increased Ras expression and caspase-independent neuroblastoma cell death: possible mechanism of spontaneous neuroblastoma regression. J.Natl.Cancer Inst. 2002;94:358–368. doi: 10.1093/jnci/94.5.358. [DOI] [PubMed] [Google Scholar]
- 212.Koizumi H, Hamano S, Doi M, Tatsunami S, Nakada K, Shinagawa T, Tadokoro M. Increased occurrence of caspase-dependent apoptosis in unfavorable neuroblastomas. Am.J.Surg.Pathol. 2006;30:249–257. doi: 10.1097/01.pas.0000184805.60908.37. [DOI] [PubMed] [Google Scholar]
- 213.Ogier-Denis E, Pattingre S, El Benna J, Codogno P. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J.Biol.Chem. 2000;275:39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- 214.Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 215.Byun JY, Kim MJ, Yoon CH, Cha H, Yoon G, Lee SJ. Oncogenic Ras signals through activation of both phosphoinositide 3-kinase and Rac1 to induce c-Jun NH2-terminal kinase-mediated, caspase-independent cell death. Mol. Cancer Res. 2009;7:1534–1542. doi: 10.1158/1541-7786.MCR-08-0542. [DOI] [PubMed] [Google Scholar]
- 216.Kaul A, Overmeyer JH, Maltese WA. Activated Ras induces cytoplasmic vacuolation and non-apoptotic death in glioblastoma cells via novel effector pathways. Cell Signal. 2007;19:1034–1043. doi: 10.1016/j.cellsig.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 218.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 219.Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J.Biol.Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 220.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat.Rev.Mol.Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat.Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 222.Sakata K, Kato S, Fox JC, Shigemori M, Morimatsu M. Autocrine signaling through Ras regulates cell survival activity in human glioma cells: potential cross-talk between Ras and the phosphatidylinositol 3-kinase-Akt pathway. J.Neuropathol.Exp.Neurol. 2002;61:975–983. doi: 10.1093/jnen/61.11.975. [DOI] [PubMed] [Google Scholar]
- 223.Holmen SL, Williams BO. Essential role for Ras signaling in glioblastoma maintenance. Cancer Res. 2005;65:8250–8255. doi: 10.1158/0008-5472.CAN-05-1173. [DOI] [PubMed] [Google Scholar]
- 224.Ishii N, Maier D, Merlo A, Tada M, Sawamura Y, Diserens A-C, Van Meir EG. Frequnt co-alterations of TP53, p16/CDKN2A, p14arf, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]