Abstract
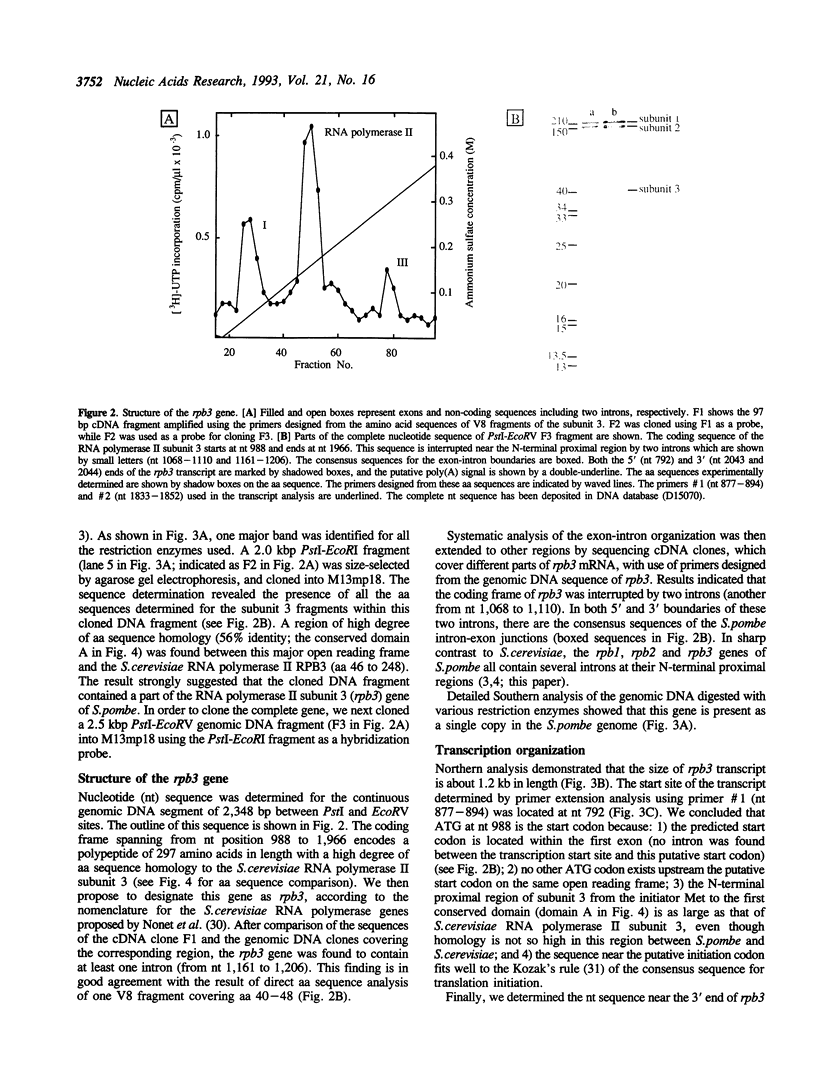
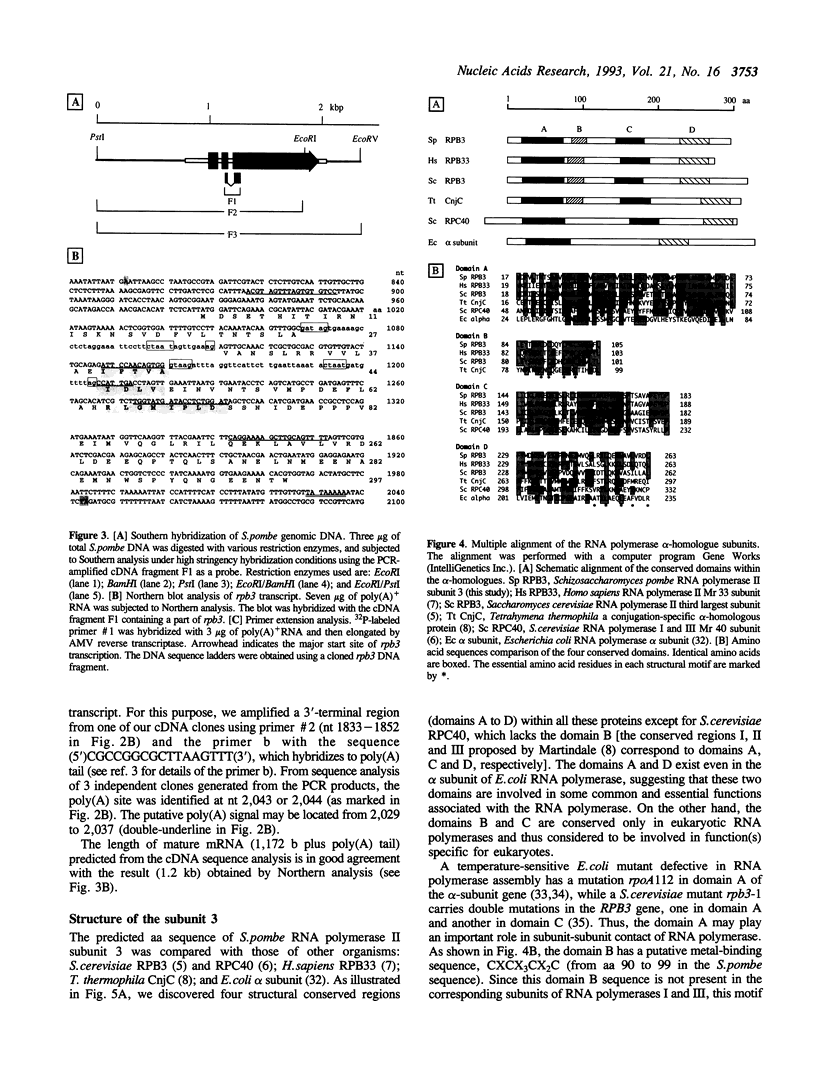
To improve our understanding of the structure and function of eukaryotic RNA polymerase II, we purified the enzyme from the fission yeast Schizosaccharomyces pombe. The highly purified RNA polymerase II contained more than eleven polypeptides. The sizes of the largest the second-, and the third-largest polypeptides as measured by SDS-polyacrylamide gel electrophoresis were about 210, 150, and 40 kilodaltons (kDa), respectively, and are similar to those of RPB1, 2, and 3 subunits of Saccharomyces cerevisiae RNA polymerase II. Using the degenerated primers designed after amino acid micro-sequencing of the 40 kDa third-largest polypeptide (subunit 3), we cloned the subunit 3 gene (rpb3) and determined its DNA sequence. Taken together with the sequence of parts of PCR-amplified cDNA, the predicted coding sequence of rpb3, interrupted by two introns, was found to encode a polypeptide of 297 amino acid residues in length with a molecular weight of 34 kDa. The S. pombe subunit 3 contains four structural domains conserved for the alpha-subunit family of RNA polymerase from both eukaryotes and prokaryotes. A putative leucine zipper motif was found to exist in the C-terminal proximal conserved region (domain D). Possible functions of the conserved domains are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azuma Y., Yamagishi M., Ueshima R., Ishihama A. Cloning and sequence determination of the Schizosaccharomyces pombe rpb1 gene encoding the largest subunit of RNA polymerase II. Nucleic Acids Res. 1991 Feb 11;19(3):461–468. doi: 10.1093/nar/19.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Corden J. L. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987 Feb;7(2):586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Fujita N., Ishihama A. Identification of a subunit assembly domain in the alpha subunit of Escherichia coli RNA polymerase. J Mol Biol. 1991 Mar 5;218(1):1–6. [PubMed] [Google Scholar]
- Igarashi K., Fujita N., Ishihama A. Sequence analysis of two temperature-sensitive mutations in the alpha subunit gene (rpoA) of Escherichia coli RNA polymerase. Nucleic Acids Res. 1990 Oct 25;18(20):5945–5948. doi: 10.1093/nar/18.20.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Hanamura A., Makino K., Aiba H., Aiba H., Mizuno T., Nakata A., Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase: two modes of transcription activation by positive factors. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8958–8962. doi: 10.1073/pnas.88.20.8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Ishihama A. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991 Jun 14;65(6):1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Promoter selectivity of prokaryotic RNA polymerases. Trends Genet. 1988 Oct;4(10):282–286. doi: 10.1016/0168-9525(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Role of the RNA polymerase alpha subunit in transcription activation. Mol Microbiol. 1992 Nov;6(22):3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- Ishihama A., Shimamoto N., Aiba H., Kawakami K., Nashimoto H., Tsugawa A., Uchida H. Temperature-sensitive mutations in the alpha subunit gene of Escherichia coli RNA polymerase. J Mol Biol. 1980 Feb 25;137(2):137–150. doi: 10.1016/0022-2836(80)90321-6. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Transcription signals and factors in Escherichia coli. Adv Biophys. 1986;21:163–173. doi: 10.1016/0065-227x(86)90021-3. [DOI] [PubMed] [Google Scholar]
- Jendrisak J. J., Burgess R. R. A new method for the large-scale purification of wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1975 Oct 21;14(21):4639–4645. doi: 10.1021/bi00692a012. [DOI] [PubMed] [Google Scholar]
- Jokerst R. S., Weeks J. R., Zehring W. A., Greenleaf A. L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989 Jan;215(2):266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- Kawagishi M., Yamagishi M., Ishihama A. Cloning and sequence determination of the Schizosaccharomyces pombe rpb2 gene encoding the subunit 2 of RNA polymerase II. Nucleic Acids Res. 1993 Feb 11;21(3):469–473. doi: 10.1093/nar/21.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P. A., Woychik N., Liao S. M., Young R. A. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol Cell Biol. 1990 May;10(5):1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol Cell Biol. 1991 Sep;11(9):4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer N. F., Simanis V., Nurse P. Fission yeast Schizosaccharomyces pombe correctly excises a mammalian RNA transcript intervening sequence. Nature. 1985 Nov 7;318(6041):78–80. doi: 10.1038/318078a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D. W., Hayward R. S. Nucleotide sequence of the rpoA-rplQ DNA of Escherichia coli: a second regulatory binding site for protein S4? Nucleic Acids Res. 1984 Jul 25;12(14):5813–5821. doi: 10.1093/nar/12.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Pati U. K., Weissman S. M. The amino acid sequence of the human RNA polymerase II 33-kDa subunit hRPB 33 is highly conserved among eukaryotes. J Biol Chem. 1990 May 25;265(15):8400–8403. [PubMed] [Google Scholar]
- Russell P. R. Transcription of the triose-phosphate-isomerase gene of Schizosaccharomyces pombe initiates from a start point different from that in Saccharomyces cerevisiae. Gene. 1985;40(1):125–130. doi: 10.1016/0378-1119(85)90031-9. [DOI] [PubMed] [Google Scholar]
- Schultz L. D., Hall B. D. Transcription in yeast: alpha-amanitin sensitivity and other properties which distinguish between RNA polymerases I and III. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1029–1033. doi: 10.1073/pnas.73.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Toda T., Ohashi M. Purification and identification of intermediate catabolic products in the in vivo degradation of pig liver phosphofructokinase. J Biol Chem. 1986 Sep 25;261(27):12455–12461. [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol. 1989 Jul;9(7):2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]




