Abstract
The cell envelopes of Gram-positive bacteria comprise two major constituents, peptidoglycan and teichoic acids. Wall teichoic acids (WTAs) are anionic glycophosphate polymers that play important roles in bacterial cell growth, division, and pathogenesis. They are synthesized intracellularly and exported by an ABC transporter to the cell surface, where they are covalently attached to peptidoglycan. We address here the substrate specificity of WTA transporters by substituting the Bacillus subtilis homologue, TagGHBs, with the Staphylococcus aureus homologue, TarGHSa. These transporters export structurally different substrates in their indigenous organisms, but we show that TarGHSa can substitute for the B. subtilis transporter. Hence, substrate specificity does not depend on the WTA main chain polymer structure, but may be determined by the conserved diphospholipid-linked disaccharide portion of the WTA precursor. We also show that the complemented B. subtilis strain becomes susceptible to a S. aureus-specific antibiotic, demonstrating that the S. aureus WTA transporter is the sole target of this compound.
Bacteria are surrounded by complex cell envelopes that mediate interactions with the external milieu, act as filters to allow the passage of selected molecules into and out of the cells, and form a protective layer that stabilizes the plasma membrane against high internal osmotic pressure fluctuations (1). The most conserved component of the cell envelope in bacteria is peptidoglycan (PG), a crosslinked mesh of glycan chains connected through peptide bridges. Since it is conserved, essential, and unique to bacteria, PG is a major target for clinically used antibiotics, such as penicillin and vancomycin. However, multi-resistant pathogenic strains such as methicillin-resistant Staphylococcus aureus (MRSA) pose a major problem to the public, making it crucial to explore other possible cell wall targets.
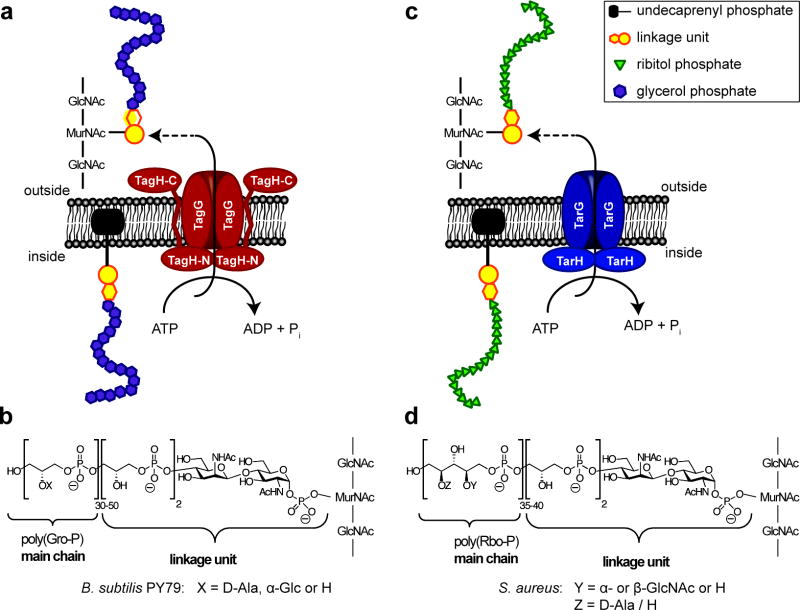
The PG layers of Gram-positive organisms are densely functionalized with anionic polymers called wall teichoic acids (WTAs) (2). These polymers, which comprise as much as 50% of the cell wall mass, are typically composed of linear sugar phosphate repeats, usually glycerol or ribitol phosphates, which are tailored with D-alanyl esters and hexoses (2, 3). WTA precursors are synthesized on an undecaprenyl phosphate carrier lipid (UndP) on the inner surface of the cytoplasmic membrane and then exported through a two-component ATP-binding cassette (ABC) transporter to the cell surface where they are covalently attached to PG (4) (Fig. 1).
Figure 1. Schematic of the WTA exporters of B. subtilis PY79 and S. aureus.
a) Model of WTA precursor export by TagGH from B. subtilis PY79. b) Structure of WTA from B. subtilis PY79 showing the linkage unit, its connection to N-acetyl muramic acid (MurNAc) of peptidoglycan and the poly-glycerol phosphate [poly(GroP)] main chain. c) Schematic view of the WTA exporter TarGH from S. aureus. d) Structure of linkage unit and poly-ribitol phosphate [poly(RboP)] main chain from S. aureus.
WTAs play important roles in determining cell morphology in B. subtilis and are critical for cell division in S. aureus (5–9). They are speculated to scaffold components of the PG biosynthetic machinery as a mechanism for regulating cell envelope biosynthesis (8, 9). WTAs are not essential for survival in vitro (10–12); however, S. aureus strains lacking WTAs are severely impaired in cell division and unable to colonize host tissue and establish infections (8, 12–14). The importance of WTAs in bacterial physiology and host infection make WTA biosynthesis a target for novel antibiotics.
The WTA biosynthetic pathway shares an unusual genetic feature with several other UndP-dependent pathways: many of the downstream genes are essential except in strains having mutations that prevent flux into the pathway (10, 15). The lethality of downstream null mutations may be due to accumulated toxic intermediates and/or inhibition of PG biosynthesis because of sequestration of the UndP carrier (11). It was predicted that small molecules that inhibit these downstream enzymes would have antibacterial activity, and we confirmed this through the discovery of a WTA-active antibiotic in a cell-based high throughput screen that exploited the conditional essentiality of the downstream genes (15). The compound discovered was subsequently optimized for potency to produce a second generation antibiotic named targocil (16).
Targocil has a minimal inhibitory concentration (MIC) below 1 μM against all S. aureus strains examined, including MRSA, and studies in S. aureus have identified its target as TarGH, the essential two-component ABC transporter that exports lipid-linked WTA precursors to the cell surface (Fig. 1)(4, 15). Many Gram-positive organisms make WTAs and contain TarGH orthologs, but targocil is completely specific for S. aureus. It was unknown whether resistance in other organisms was due to intrinsically resistant TarGH transporters, the presence of unidentified, non-homologous transporters, or other mechanisms. We address this question here using a heterologous complementation approach in which the S. aureus transporter is expressed in B. subtilis PY79. This approach provides crucial insights into how WTA transporters select their substrates.
S. aureus makes WTA precursors consisting of a poly-ribitolphosphate chain connected through a linkage unit containing a disaccharide linked to undecapranyl pyrophosphate (17, 18). The ribitolphosphate subunits carry α- or β-O-N-acetyl glucosamine modifications (Fig. 1c, d) (19). The WTA precursors of B. subtilis PY79 consist of a glycerolphosphate polymer tailored with α-O-glucose residues, but the linkage unit is identical to that in S. aureus (Fig. 1a, b). Therefore, the polymeric portions of the native substrates of the WTA transporters TarGHSa and TagGHBs are structurally different.
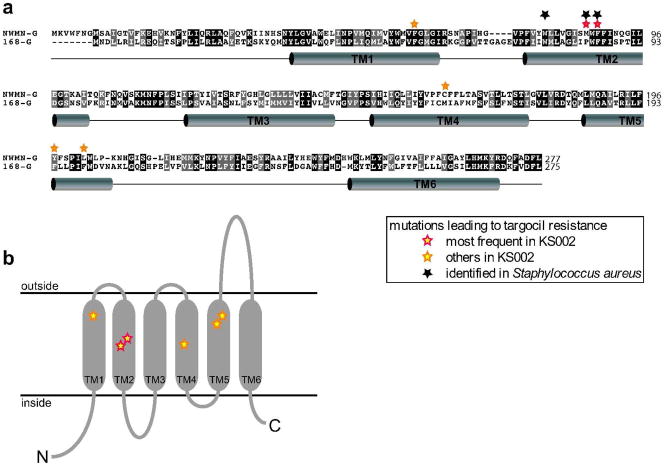
The WTA transporters consist of an ATPase portion (TarHSa or TagHBs) and a transmembrane portion (TarGSa or TagGBs). They belong to a family of polymer-exporting ABC transporters predicted to function as dimers containing two ATPase domains and two transmembrane domains (20, 21). The transmembrane portions are moderately conserved between S. aureus and B. subtilis (36% identity, 57% similarity) (Fig. 2a). Both proteins are predicted to have six transmembrane spanning regions with the N- and C-termini inside (Fig. 2). The ATPase component of the B. subtilis transporter, TarHBs, aligns with high similarity (73%) to the first 264 amino acids of the S. aureus ATPase, but contains an additional 263 residue C-terminal extension that has no significant similarity to any known protein (Supplementary Fig. S1a). A single membrane-spanning region is predicted immediately after the homologous region, which would place a large C-terminal domain outside the cytoplasm (Supplementary Fig. S1b, c). Hence, both the structures of the transporters and the WTA precursors differ significantly between the two organisms.
Figure 2. Alignment of B. subtilis and S. aureus TagG/TarG.
(a) Alignment of S. aureus TarG (NWMN-G) and B. subtilis TagG (168-G). Conserved amino acids are shown with a black background, similar amino acids with a grey background. Barrels indicate predicted transmembrane helices. b) Predicted topology of TarGSa with stars marking the sites of point mutations leading to targocil resistance in B. subtilis KS002.
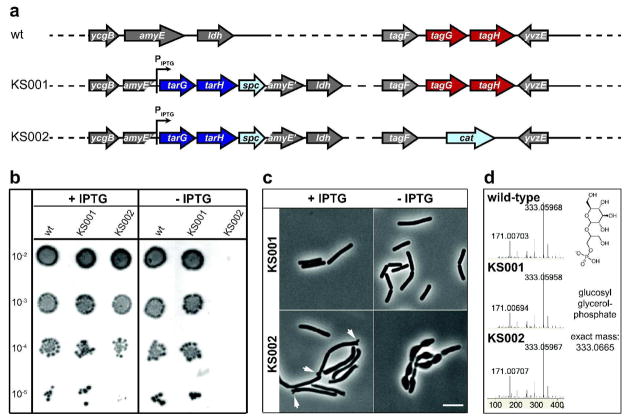
B. subtilis tagGH, like its S. aureus counterpart, is essential for viability (4). Therefore, to determine whether TarGHSa could substitute for TagGHBs, we first constructed a B. subtilis strain with tarGHSa under the control of an IPTG-inducible promoter (strain KS001, Fig. 3a), and we then deleted the endogenous tagGHBs genes (strain KS002, Fig. 3a). The B. subtilis transporter could easily be deleted in the presence of IPTG, showing that TarGHSa can complement the loss of TagGHBs. Many uniform colonies were obtained in several transformations with selection for deletion mutants, indicating that KS002 is unlikely to require additional suppressor mutations for viability. Strain KS002 was dependent on IPTG for growth (Fig. 3b, Supplementary Fig. S2), demonstrating that expression of TarGHSa is essential when TagGHBs is deleted. WTAs extracted from the wild-type, KS001 and KS002 strains were indistinguishable as judged by PAGE analysis of the polymers and LC/MS analysis of chemically degraded subunits (Fig. 3d, Supplementary Fig. S3). Thus, the S. aureus transporter is capable of transporting the polyglycerolphosphate WTA precursors produced by B. subtilis. Since the native substrates for the S. aureus transporter have poly-ribitolphosphate main chains, we conclude that substrate recognition by the ABC transporter is not dependent on the chemical structure of the main chain polymer.
Figure 3. B. subtilis is viable with TarGHSa as the only WTA exporter.
a) Scheme of the genetic background of wild-type B. subtilis PY79 (wt) and strains KS001 and KS002. Indigo arrows indicate S. aureus genes, red arrows B. subtilis genes, light blue arrows antibiotic resistance genes, and grey arrows the flanking genes. b) Dilution series of bacterial cultures spotted on plates with (left) and without IPTG (right). c) Phase contrast microscopic images of strains KS001 and KS002 grown in LB in the presence and absence of 1mM IPTG as indicated. Scale bar 5 μm. d) Mass spectrometry results for WTA monomers of B. subtilis PY79 wild-type, KS001 and KS002.
Examination of KS002 by light microscopy showed that depletion of TarGHSa leads to swelling and rounding of the cells (Fig. 3c), a phenotype consistent with the loss of WTAs (10). Compared to the wild-type, KS002 cells grown in the presence of IPTG had minor defects. They were slightly curved and the cell poles were wider than usual, appearing almost forked (Fig. 3c, arrows). These defects suggest imperfect cell division. KS001 cells grown in the presence of IPTG were identical to wild-type cells (Fig. 3c), indicating that the defects in KS002 are not due to over-expression of TarGHSa. Since IPTG-induced expression of the B. subtilis WTA transporter TagGHBs from the same locus (strain KS005) resulted in full restoration of both growth rate and morphology (Supplementary Fig. S4), the minor defects in KS002 expressing only the S. aureus transporter are due either to less efficient WTA export by the exogenous transporter or possibly the absence of the C-terminal domain of the B. subtilis transporter.
To test whether the lack of this C-terminal domain was responsible for the phenotype of strain KS002, a strain with a truncated version of TagHBs in the native locus was constructed (KS003), leaving only the N-terminal part of the protein from amino acid 1 to 275. Strain KS003 was indistinguishable from the wild-type in cell morphology and growth rate (Supplementary Fig. S5), indicating that the extracellular domain of TagH is not required for function of the ABC transporter, consistent with previous results based on transposon mutagenesis (4). Therefore, the absence of this domain does not lead to the phenotypic changes seen when TarGHSa is the sole transporter.
Having established that the S. aureus transporter complemented the loss of B. subtilis TagGH, albeit imperfectly, we examined whether targocil, a S. aureus-specific antibiotic with a triazoloquinazoline core (Fig. 4a) would have antibiotic activity against KS002. Wild-type B. subtilis PY79 is resistant to targocil at concentrations exceeding 200 μM (Table S2; 15), and expression of TarGHSa in a wild-type background (strain KS001) had no effect on resistance (Fig. 4b, c; Supplementary Table S2). However, the minimal inhibitory concentration (MIC) of targocil against KS002 was 1.25 μM, which is comparable to its MICs against S. aureus. No other compound tested (kanamycin, penicillin G, tunicamycin, carbenicillin, erythromycin) showed a changed MIC in KS002 compared with the wild-type strain, demonstrating that targocil’s activity against this strain was due solely to inhibition of the complementing S. aureus transporter (Supplementary Table S2). Consistent with this interpretation, targocil-treated KS002 cells were morphologically similar to KS002 cells following depletion of TarGHSa. Upon treatment with targocil, KS002 cells progressively rounded up along their cylindrical axis (Fig. 4c). In contrast, the wild-type and KS001 strains were unaffected by growth in the presence of the inhibitor (Fig. 4c).
Figure 4. TarGHSa as the only WTA exporter renders B. subtilis sensitive to the TarG-inhibitor targocil.
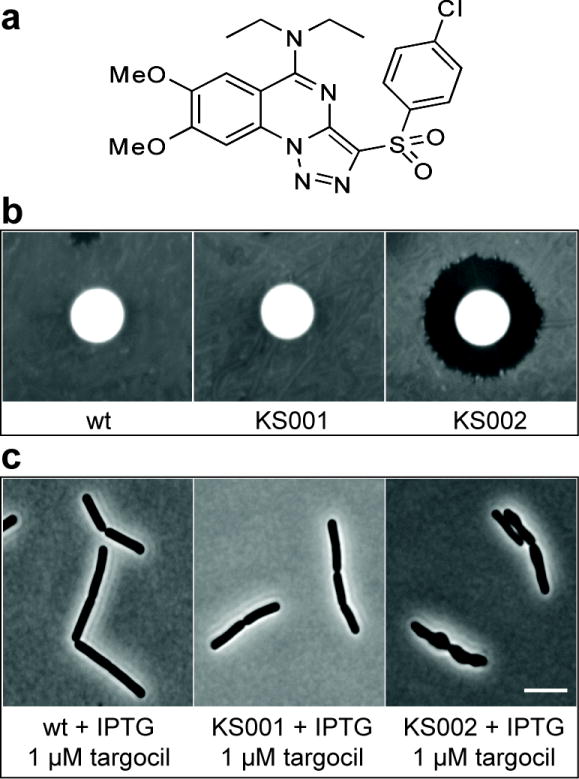
a) Structure of the TarG inhibitor targocil. b) Plate diffusion assay showing degree of sensitivity of wild-type (wt), KS001, and KS002 against targocil. c) Phase contrast microscopy of wild-type, KS001 and KS002 grown in LB containing 1 mM IPTG and treated with 1 μM targocil for 2 hours. Scale bar 5 μm.
Targocil resistant mutants of strain KS002 were selected on plates containing 5 μM targocil and 1 mM IPTG, required for TarGHSa expression. TarG was sequenced from 18 colonies with stable mutations that conferred resistance to targocil. In all cases, tarGSa was found to contain a point mutation at one of several sites (Fig. 2, yellow stars), implying that mutation of the target is the major mechanism of resistance in B. subtilis. In S. aureus, in addition to tarG point mutations, null mutations in tarO or tarA that result in avirulent strains are found. The different resistant populations in the two organisms probably reflect the more severe growth defects following deletion of the WTA pathway in the rod-shaped B. subtilis. The most frequent TarG mutations were M87T (6x) and F89L (6x) and these mutations were also common when targocil resistance was selected in S. aureus (Fig. 2, black stars) (15). The mutated residues are predicted to be located in the middle of the second transmembrane domain of TarG (TM2), and from these and other, less common point mutations conferring targocil resistance (Fig. 2b) we speculate that the binding site for the inhibitor might be located in a tertiary structure formed by predicted transmembrane helices TM1, 2, 4 and 5.
Conclusions
The export of large, highly polar bacterial polymers by ABC transporters is a remarkable but poorly understood process. Almost nothing is known about the mechanism of polymer transport and little is understood about substrate specificity (20). Here we have shown that the WTA transporters from B. subtilis and S. aureus can be exchanged without loss of function, even though they normally transport different substrates, i.e., poly-glycerol and poly-ribitol WTA precursors tailored with different sugars. Therefore, the structure of the polymer itself is not critical for substrate recognition. Since both WTA precursors contain the same disaccharide linkage unit bound to UndP, we speculate that this diphospholipid-anchored disaccharide unit is recognized by the transport machinery. If so, WTA transporters may function as linkage unit “flippases” with main chain export occurring as a result. Alternatively, initiation of transport from the non-reducing end could occur if general chemical characteristics (e.g., repeating phosphates) rather than specific structural features of the main chain polymer are involved in recognition. It has been previously suggested that polymer synthesis and transport are coupled through protein-protein interactions between the biosynthetic enzymes and the transporter, but successful heterologous complementation of B. subtilis with the S. aureus transporter suggests that such interactions are not essential for WTA export.
Since the presence of TarGHSa as the only WTA transporter confers susceptibility against targocil to B. subtilis, this organism’s intrinsic resistance is not due to general differences in uptake, mechanism of action, or the presence of an unknown transporter, but solely to a resistant TagGH transporter. Since the MICs of targocil against S. aureus and KS002 are comparable, the affinity and the mechanism of action of the inhibitor are likely the same in the two organisms. Since targocil blocks the S. aureus transporter in B. subtilis but does not inhibit the B. subtilis transporter, we conclude that the interaction of targocil with S. aureus TarG is highly specific, explaining its narrow spectrum.
Nonetheless, there is potential to develop WTA transport inhibitors with an expanded spectrum since our experiments show that blocking WTA transport is lethal not only in S. aureus, but also in B. subtilis. The transmembrane component for WTA transport is promiscuous with respect to substrate structure and tolerant of many changes in amino acid sequence since different mutations confer resistance without affecting transport. However, it may be possible to identify inhibitors with an expanded spectrum by targeting other regions of the transporter that are involved in more conserved recognition events. These regions include the ATP binding site of the ATPase domain and possibly the WTA linkage unit recognition region (22). We are working towards the reconstitution of a WTA transporter to enable the identification of the structural features that determine how WTA substrates are selected for export, and to lay the groundwork for studies to elucidate how these machines export such large polymers and yet can be inhibited with exquisite specificity by a small molecule.
METHODS
Strain construction
Lists of strains, plasmids and oligonucleotides are given in Table S1.
Since in S. aureus tarG and tarH are oriented in opposite directions and are each transcribed from their own promoter in the intergenic region, the genes were amplified separately and an optimal ribosome binding site was added upstream of each. tarG and tarH were amplified from S. aureus Newman using primer pairs 1+2 and 3+4 respectively. The two PCR products were digested with XbaI, ligated, and the ligation product was used as a template for a PCR using primers 1+4. This product was cloned into pDR111 using SalI and SphI. The plasmid insert was validated by sequencing and then transformed into B. subtilis, where it integrated into the amyE locus, giving strain KS001.
Strain KS002 was constructed by deleting tagGHBs from the chromosome of KS001. For this, approximately 1000 bp up- and down-stream of tarGH were amplified using primers 5+6 and 7+8 respectively, and both PCR products were ligated to the cat resistance cassette from pKM074 using BamHI and EagI. The ligation product was used as a template for a PCR using primers 5+8 and the product was directly transformed into B. subtilis selecting for chloramphenicol resistance. Deletion of tagGHBs was confirmed by PCR.
Transformation of B. subtilis was done according to the method of Anagnostopoulos and Spizizen (23) as modified by Jenkinson (24).
Culture Conditions
Bacterial cultures were grown at 37°C under shaking conditions in LB medium or on Nutrient Agar (NA) plates. If required, antibiotics were added: chloramphenicol 5 μg m−1, spectinomycin 100 μg ml−1, erythromycin 1 μg ml−1. 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) was added to induce expression from the Phyperspank promoter as indicated.
Raising of targocil resistant mutants
Dilutions of exponentially growing cultures of strain KS002 were plated on NA plates containing 5 μM targocil and 1 mM IPTG. After incubation for 24 h at 37°C colonies were picked and streaked on NA plates containing IPTG. Single colonies were then streaked on NA with 5 μM targocil and 1 mM IPTG. From all strains chromosomal DNA was isolated and tarG was amplified and sequenced.
Microscopy
For microscopy, overnight cultures were diluted into fresh medium with or without 1 mM inducer as indicated. Samples were taken at the time points indicated and placed on cover slips with agarose pads. Images were acquired with a Hamamatsu digital camera model ORCA-ER connected to a Nikon Eclipse TE2000-U microscope with X-cite 120 illumination system. Image manipulation was limited to changing brightness and contrast using ImageJ (25).
WTA extraction and analysis
WTAs were extracted, degraded and analyzed by LC/MS as described previously (17).
Determination of Antibiotic Sensitivity
Antibiotic sensitivity was tested by diluting an overnight culture into fresh medium containing IPTG to an OD600 of 0.001. Antibiotics were added in dilution steps 1:2 and the cultures were incubated overnight whilst shaking at 30°C. Lack of visible growth was interpreted as sensitivity against the respective antibiotic concentration.
For the plate diffusion assay overnight cultures were plated on NA containing IPTG, then filter disks soaked with antibiotic were placed on top. The plates were incubated over night at 37°C and zones of clearing around the filter disks were interpreted as sensitivity against the antibiotic.
Bioinformatic predictions
Sequences for TarGSa and TagGBs were obtained from the Genolist webserver (http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList)(26). Alignments were done using the ClustalW webserver (27) and the appearance was modified using Boxshade (Institute Pasteur, France; http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList). The topology model is based on predictions by the TMHMM and the SOSUI webservers.
Supplementary Material
Acknowledgments
We would like to thank D. Rudner for the plasmid pDR111, E. Doud and S. Brown for help with the mass spectrometry analysis and S. Ringgaard for critical reading of the manuscript. This work was funded by the National Institute of Health (1PO1AI083214 and 5R01M078477 to S.W.), and the Taplin Funds for Discovery Program (for the Q-TOF spectrophotometer to S.W).
Footnotes
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swoboda JG, Campbell J, Meredith TC, Walker S. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem. 2010;11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 5.Schirner K, Marles-Wright J, Lewis RJ, Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28:830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee BL, Sekimizu K. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol. 2009;191:141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gründling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell J, Singh AK, Santa Maria JP, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson B, Walker S. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2010 doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2010;107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Elia MA, Millar KE, Beveridge TJ, Brown ED. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol. 2006;188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol. 2006;188:4183–4189. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 13.Weidenmaier C, Peschel A, Xiong YQ, Kristian SA, Dietz K, Yeaman MR, Bayer AS. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis. 2005;191:1771–1777. doi: 10.1086/429692. [DOI] [PubMed] [Google Scholar]
- 14.Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, Götz F, Peschel A. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol. 2008;298:505–513. doi: 10.1016/j.ijmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem Biol. 2009;4:875–883. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, Campbell J, Swoboda JG, Cuny GD, Walker S. Development of improved inhibitors of wall teichoic acid biosynthesis with potent activity against Staphylococcus aureus. Bioorg Med Chem Lett. 2010;20:1767–1770. doi: 10.1016/j.bmcl.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown S, Meredith T, Swoboda J, Walker S. Staphylococcus aureus and Bacillus subtilis W23 make polyribitol wall teichoic acids using different enzymatic pathways. Chem Biol. 2010;17:1101–1110. doi: 10.1016/j.chembiol.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown S, Zhang YH, Walker S. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem Biol. 2008;15:12–21. doi: 10.1016/j.chembiol.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia G, Maier L, Sanchez–Carballo P, Li M, Otto M, Holst O, Peschel A. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem. 2010;285:13405–13415. doi: 10.1074/jbc.M109.096172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuthbertson L, Kos V, Whitfield C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev. 2010;74:341–362. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gronenberg LS, Kahne D. Development of an activity assay for discovery of inhibitors of lipopolysaccharide transport. J Am Chem Soc. 2010;132:2518–2519. doi: 10.1021/ja910361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkinson HF. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J Gen Microbiol. 1983;129:1945–1958. doi: 10.1099/00221287-129-6-1945. [DOI] [PubMed] [Google Scholar]
- 25.Rasband WS. ImageJ. U.S. National Institutes of Health; Bethesda, Maryland, USA: 1997–2009. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 26.Lechat P, Hummel L, Rousseau S, Moszer I. GenoList: an integrated environment for comparative analysis of microbial genomes. Nucleic Acids Res. 2008;36:D469–474. doi: 10.1093/nar/gkm1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.