Summary
We report the immediate effects of estrogen signaling on the transcriptome of breast cancer cells using Global Run-On and sequencing (GRO-seq). The data were analyzed using a new bioinformatic approach that allowed us to identify transcripts directly from the GRO-seq data. We found that estrogen signaling directly regulates a strikingly large fraction of the transcriptome in a rapid, robust, and unexpectedly transient manner. In addition to protein coding genes, estrogen regulates the distribution and activity of all three RNA polymerases, and virtually every class of non-coding RNA that has been described to date. We also identified a large number of previously undetected estrogen-regulated intergenic transcripts, many of which are found proximal to estrogen receptor binding sites. Collectively, our results provide the most comprehensive measurement of the primary and immediate estrogen effects to date and a resource for understanding rapid signal-dependent transcription in other systems.
Keywords: Estrogen, Transcriptome, GRO-seq, Gene annotation, Transcript, Signal-regulated transcription
Introduction
The steroid hormone estrogen, acting through estrogen receptors (ERs), plays key roles in a variety of fundamental developmental and physiological processes, as well as many disease states (Deroo and Korach, 2006). Mammals express two ER isoforms, ERα and ERβ, which exhibit distinct tissue-specific expression patterns and biological roles (Deroo and Korach, 2006; Warner et al., 1999). ERs function primarily as nuclear transcription factors, which dimerize upon binding of the natural ligand, 17β-estradiol (E2), and act as potent regulators of gene expression. ERα binds to >10,000 sites across the genome and acts to (1) promote the recruitment of coregulators that mediate post-translational modification of histones or other transcription factors and (2) regulate the binding or activity of the RNA polymerase II (Pol II) transcriptional machinery, ultimately altering the transcriptome in estrogen-responsive cells (Acevedo and Kraus, 2004; Cheung and Kraus, 2010; Ruhl and Kraus, 2009).
Previous studies analyzing steady-state gene expression patterns in the presence and absence of E2 have failed to reveal a consistent view of the estrogen-regulated gene set. In particular, the use of expression microarrays has produced discrepancies in the numbers of estrogen-regulated genes in the widely used ERα-positive MCF-7 human breast cancer cell line, ranging from 100 to 1,500 (Cheung and Kraus, 2010; Kininis and Kraus, 2008). In addition, genomic ChIP analyses of ERα and Pol II have not produced a clear picture of the estrogen-regulated gene set either. This is due, in part, to the difficulty in assigning ERα binding events to specific gene regulatory outcomes (Carroll et al., 2006; Welboren et al., 2009). Another limitation of these analyses is that they have focused on the effects of estrogen signaling on Pol II transcription, without considering potential effects on Pol I and Pol III.
A fundamental weakness inherent in monitoring estrogen-dependent gene expression by assessing changes in mature mRNA is that longer treatments are required to allow time for mRNA accumulation (~3 to 24 hours). This time allows the accumulation of transcripts from primary ERα target genes, but also leads to a host of secondary transcriptional effects that are not directly mediated by ERα. To address these concerns, preliminary attempts to define the immediate transcriptional effects of estrogen signaling using the translation inhibitor cycloheximide indicated that only 20 to 30% of the genes showing changes in expression are primary targets (Lin et al., 2004). Using cycloheximide to infer primary estrogen target genes is problematic, however, because (1) cycloheximide does not inhibit the effects of non-coding regulatory RNAs on gene expression, which is becoming widely recognized as an important mechanism underlying the regulation of many genes (Krol et al., 2010) and (2) the levels of steady-state mRNA depends not only on transcriptional regulation by E2, but also on the rates of elongation, pre-mRNA processing, and mRNA degradation (Widelitz et al., 1987). Due to these factors, it is clear that a new approach is required to conclusively identify primary estrogen target genes.
Here, we used Global nuclear Run-On and Sequencing (GRO-seq) (Core et al., 2008) to identify the immediate effects of estrogen signaling on the entire transcriptome in MCF-7 cells. GRO-seq is a direct sequencing method that provides a “map” of the position and orientation of all engaged RNA polymerases across the genome at extremely high resolution, providing a direct measure of transcription. Using GRO-seq in combination with a novel bioinformatic approach based on Hidden Markov Models (HMMs), we determined all (i.e., both annotated and unannotated) genomic regions in MCF-7 cells that are transcribed by Pols I, II, and III. In addition, we identified the primary transcriptional targets of E2 signaling by focusing on short treatments (i.e., 0, 10, and 40 min.), prior to the activation of secondary targets. Our unique approach has revealed many unexpected features of E2-regulation, providing the most comprehensive measurement of the primary and immediate effects of E2 signaling to date. Our results provide a model and resource for understanding rapid signal-dependent transcription in other systems.
Results and Discussion
Generation of GRO-seq libraries from estrogen-treated MCF-7 cells
To investigate the immediate effects of estrogen on the transcriptome of human cells, we treated estrogen-deprived ERα-positive MCF-7 human breast cancer cells with a short time course of 17β-estradiol (E2) (0, 10, 40, and 160 min) (Fig. 1A). The estrogen-deprived MCF-7 cells continued to grow actively (Fig. S1A) and the population of cells showed a normal distribution through the cell cycle (Fig. S1B). Nuclei were isolated from two biological replicates of the E2-treated MCF-7 cells and subjected to the GRO-seq procedure to generate ~100 bp libraries representing nascent RNAs, which were sequenced using an Illumina Genome Analyzer (Fig. 1A). Short-reads were aligned to the human reference genome (hg18, NCBI36), including autosomes, the X-chromosome, and one complete copy of an rDNA repeat (GenBank ID: U13369.1). Approximately 13 to 17 million reads were uniquely mapped to the genome for each treatment condition and the biological replicates for each time point were highly correlated (average correlation coefficient = 0.98) (Fig. S1C). GRO-seq returns data from all three RNA Polymerases (Pols I, II, and III). To validate if the reads mapping to the supposed loci transcribed by Pols I, II and III were correlated with the activities of each individual RNA polymerase, we carried out filter binding assays with combinations of polymerase inhibitors to isolate each polymerase. As expected, the activities detected by the filter binding assays were comparable to GRO-seq product fraction, with a slight under-representation of the apparent fraction of Pol I transcripts by GRO-seq due to an enrichment of positions that are not mappable in the repetitive rDNA sequences (Fig. S1D).
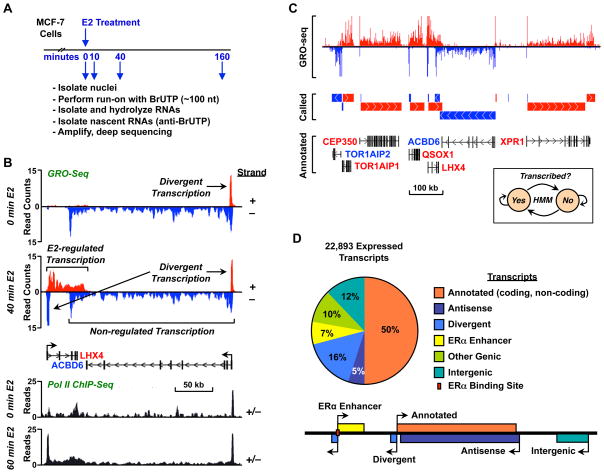
Figure 1. GRO-seq provides a detailed view of the E2-regulated transcriptome in MCF-7 cells.
(A) Overview of the experimental set up for GRO-seq analysis using MCF-7 cells.
(B) Genome browser view for a specific locus showing GRO-seq (top) and Pol II ChIP-seq (bottom) data illustrating the features of transcription and the effects of estrogen treatment.
(C) De novo detection of transcripts using GRO-seq data (top) and an HMM (inset). Called transcripts (middle) match well to RefSeq annotations (bottom).
(D) Classification of transcripts based on the annotation filter (Fig. S1, E and F).
See also Figure S1.
Fig. 1B (top) shows a representative histogram of read counts versus genomic position for a locus containing the LHX4 and ACBD6 genes. Key features of the data set are illustrated in this representation, including strand-specific transcription, divergent transcription near transcription start sites (TSSs), and robust E2-dependent induction for some genes (e.g., LHX4). These features are not readily apparent in ChIP-seq data from the same region (Fig. 1B, bottom).
Unbiased assignment of GRO-seq reads to specific transcripts
To determine the effects of E2 on the entire transcriptome (i.e., annotated and unannotated; coding and non-coding), we developed an unbiased approach for calling transcripts using a two-state hidden Markov model (HMM). The model takes as input information about read counts across the genome and subsequently divides the genome into two states representing “transcribed” and “non-transcribed” regions (Fig. 1C, inset; see Supplemental Data for additional details). An example of the input and output of this algorithm for a gene-rich region of the genome is shown in Fig. 1C. The top panel shows the raw sequence read counts for the GRO-seq data, the middle panel shows the predicted transcripts, and the bottom panel shows the RefSeq annotations.
To evaluate the robustness of our approach, we compared our predicted transcript calls to existing annotations when these were available (see Supplemental Data for details). First, we determined whether our predictions reflect entire transcripts, as opposed to breaking each gene up into a series of smaller units. Then, we determined whether our approach can accurately identify non-transcribed intervals between neighboring, but distinct, gene annotations. We found that 90% of transcribed annotated genes overlap with exactly one transcript, and that 82% of called transcripts overlapping an annotated gene do so with exactly one annotation. Together, these results suggest that our HMM-based transcript calls largely recapitulate public annotations. In many cases our transcript calls provided new or more refined information about TSSs, 5′ exons, and transcription termination sites than was available in existing databases. Using our algorithm, we assigned the genomic reads into 22,893 transcripts at one or more points during the E2 treatment time course, covering ~27% of the MCF-7 genome.
Transcripts called by the HMM were divided using a heuristic approach into six distinct, non-overlapping classes, which describe the best classification of each transcript given currently available annotations and other information (Fig. S1, E and F; see Supplemental Experimental Procedure for additional details). The six classes of transcripts that we defined are illustrated in Fig. 1D and include: (1) annotated genic and non-coding RNA transcripts, (2) antisense (genic) transcripts, (3) divergent transcripts, (4) ERα enhancer transcripts, which likely correspond to the recently described enhancer RNAs (Kim et al., 2010), (5) other transcripts falling into annotated regions, but poorly matching the annotation, and (6) completely unannotated, intergenic transcription. Although each transcript is assigned to only one of these six classes, within each class multiple annotations could be applied, allowing the accurate annotation of miRNA genes that fall inside the introns of protein coding genes. We found that 50.1% of the called transcripts map to previously annotated genes or non-coding RNAs, 5.2% map to antisense transcripts, 16.4% map to divergent transcripts, 6.8% map to ERα binding enhancers, and 12.1% are entirely unannotated intergenic transcripts (Fig. 1D).
Extensive estrogen-dependent changes in the MCF-7 transcriptome
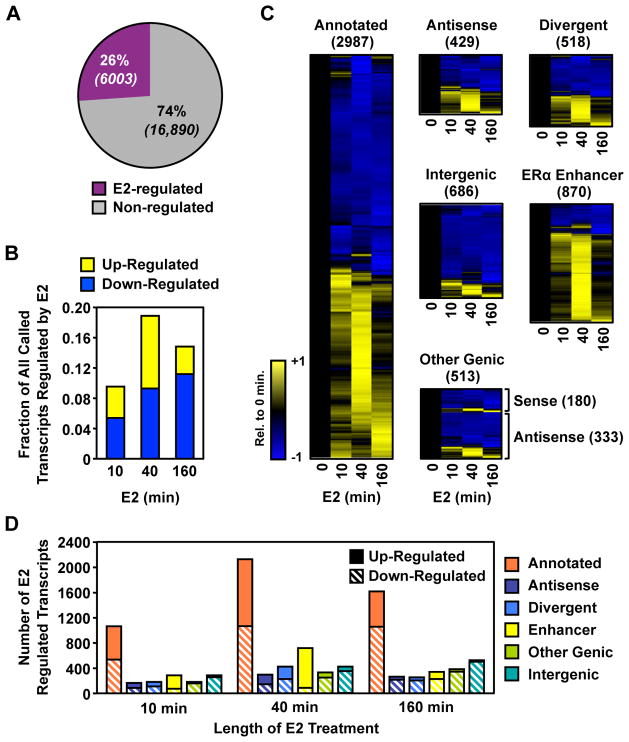
We determined which of the 22,893 transcripts change in response to E2 using a recently described model-based approach (Robinson et al., 2010) that detects changes beyond the global level of variation (Fig. S2A; see Experimental Procedure for details). We focused our analysis on a 12 kb window at the 5′ end of each transcript, as we expect to observe changes during the first 10 minutes in this window that will not yet have spread to the 3′ end of longer transcripts. Surprisingly, we found that transcription of an unexpectedly large fraction (~26%) of the MCF-7 transcriptome is altered (up- or down-regulated relative to the control/untreated condition) upon E2 treatment for at least one point in the time-course (Fig. 2A; comparisons are relative to the untreated condition). Large fractions of the genome are regulated even for the short treatments used in our experiments, strongly suggesting that these are direct actions of ERα. For example, at 10 minutes of E2 treatment, almost 10% of the MCF-7 cell transcriptome was significantly regulated at a false discovery rate of 0.1% (Fig. 2B). Another surprising finding concerns the dynamics of regulation for up and down-regulated transcripts. Through 40 minutes of E2 treatment, the time point at which the largest number of transcripts were regulated in our analyses, roughly equal numbers were up-regulated and down-regulated, but by 160 minutes ~75% of the transcripts were down-regulated (Fig. 2B). Those transcripts showing regulation at 10 or 40 minutes represent the most comprehensive and accurate definition of the immediate transcriptional targets of the estrogen signaling pathway described to date.
Figure 2. A large fraction of MCF-7 transcriptome is regulated by estrogen.
(A) The fraction of all transcripts that are regulated by E2 at any time point.
(B) The fraction of all transcripts that are up- or down-regulated by E2 at the time point shown.
(C) Heatmap representations of time-dependent regulation by E2 for each transcripts class. Values are centered and scaled to the 0 minute time point.
(D) The fraction of each class of transcripts that are up- or down-regulated by E2 at each time point.
See also Figure S2.
Next, we examined the regulation of the different classes of transcripts in more detail. Annotated protein coding and functional RNA transcripts as a group, as well as those unannotated transcripts with possible roles in gene regulation (e.g., divergent and antisense), had approximately equal numbers of up-regulated and down-regulated transcripts at 40 minutes (Figs. 2C and 2D). In contrast, the ERα enhancer transcripts were predominately up-regulated, while the intergenic transcripts were predominantly down-regulated. Together, these results suggest a coordinated transcriptional response in which E2 signaling directs the transcriptional machinery from intergenic regions to those more critical to the estrogen response. In addition, they give a fundamentally different view of estrogen-regulated gene expression than has been obtained using expression microarrays, especially with respect to the timing, magnitude, and extent of regulation.
Regulation of unannotated non-coding transcripts by estrogen: divergent, antisense, and intergenic transcripts
Our GRO-seq data revealed extensive estrogen regulation of a large set of unannotated non-coding transcripts, including divergent, antisense, and intergenic transcripts. Although the functions of these transcripts are largely unknown, their regulation by E2 suggests a role in estrogen-dependent transcriptional responses. The production and accumulation of divergent transcripts was first documented in recent studies using high-throughput genome-wide sequencing approaches with human fibroblasts (Core et al., 2008) and mouse embryonic stem cells (Seila et al., 2008). Divergent transcripts are transcribed in the opposite direction from primary transcripts at the promoters of transcribed genes, and are also produced at enhancers (e.g., eRNAs; (Kim et al., 2010)) and other unannotated regions that are transcribed. The function of divergent transcripts is unknown, but their production has been suggested to promote an open chromatin architecture at promoters through the generation of a nucleosome-free region or negative superhelical tension (Core et al., 2008; Seila et al., 2008; Seila et al., 2009). We identified 518 divergent transcripts associated with the promoters of protein coding genes, enhancers, and other unannotated transcribed regions that are regulated by E2 for at least one time point (FDR q-value < 0.001). Using these annotations, we tested whether production of a given E2-regulated divergent transcript correlates with the synthesis of the corresponding primary transcript. To do so, we tested 844 primary/divergent transcript pairs for which either the divergent, primary, or both transcripts, were regulated by E2 for at least one time point. As shown in Fig. S2B (left), E2-dependent changes in divergent transcription were strongly correlated with E2-dependent changes in the corresponding primary transcripts (Pearson correlation: 0.744; p < 2.2 × 10−16). This result is consistent with a role for divergent transcription in facilitating E2-dependent transcription of the corresponding primary transcript.
Although not well characterized, antisense transcription has been shown to have roles in the degradation of corresponding sense transcripts (Katayama et al., 2005; Werner et al., 2009), as well as gene silencing at the chromatin level (Liu et al., 2010; Morris et al., 2008). Of 1,197 transcripts annotated as antisense to a protein coding transcript, we identified 429 that are regulated by E2 (FDR q-value < 0.001) (Fig. S2C). As with the divergent transcripts, we determined if production of a given E2-regulated antisense transcript correlates with the synthesis of the corresponding primary transcript. Based on 582 sense/antisense transcript pairs, we found a remarkably high correlation between genes and their antisense transcripts (Pearson correlation: 0.654; p < 2.2 × 10−16) (Fig. S2B, right). This is particularly surprising given that, unlike divergent transcripts, antisense transcripts do not share a proximal promoter with the sense transcript, although promoter-promoter contact through genomic looping might allow for coordinated transcriptional responses. If antisense transcripts play a role in the degradation of the sense transcript, as has been suggested previously, then their E2-dependent production may provide a “built in” means of attenuating the steady-state levels of a select set of estrogen-regulated transcripts.
We also identified 2,761 transcripts that have no specific relation to previous genome annotations. Of these, 686 were regulated by E2 for at least one time point. Interestingly, the vast majority of these E2-regulated intergenic transcripts are down regulated by E2 treatment (Fig. 2D). The function of these transcripts is unknown. Some may represent currently unannotated protein coding transcripts or functional RNAs. Ascribing a function to these RNAs and determining their relative stability in the steady-state cellular RNA pool will require additional studies. Their down-regulation by E2, however, suggests a link to the estrogen signaling program. Perhaps they act to antagonize E2-dependent transcriptional responses and must be shut down to achieve a full estrogen response. Alternatively, their antagonism by E2 may be a passive effect of RNA polymerases being diverted to bona fide transcriptional targets of the estrogen signaling pathway, as suggested previously (Carroll et al., 2006).
Rapid, extensive, and transient regulation of protein coding transcripts by estrogen
Numerous studies have examined the steady-state regulation of protein coding transcripts by E2 using expression microarrays (Cheung and Kraus, 2010; Kininis and Kraus, 2008). Given the sensitivity of our approach for detecting immediate transcriptional changes in response to short E2 treatments, we extracted and examined the protein coding transcripts in our GRO-seq data for comparison. We focused on annotations in the RefSeq database, because this set is among the most comprehensive collection of transcripts, and has extensive and well-documented overlap with expression microarrays. As noted above, we used read counts in a 12 kb window at the 5′ end of each annotation and determined regulation by E2 using the edgeR package, filtering for a false discovery rate of 0.1%.
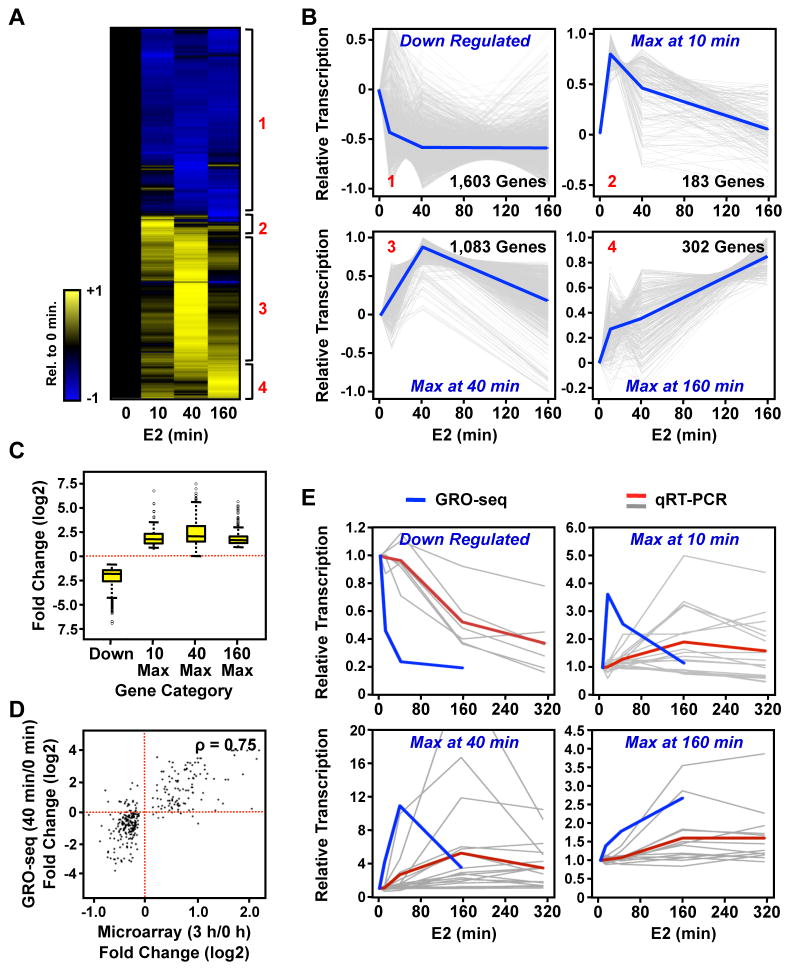
Using this approach, we detected a total of 3,098 protein coding transcripts whose levels changed relative to the control (untreated) condition at one or more of the points in the E2 treatment time course. In total, these transcripts represent ~15% of all genes annotated in RefSeq (~33% of 9,337 expressed genes) that are responsive to E2. This is a considerably larger number of genes than was detected previously at 1 or 3 hours of E2 treatment using expression microarrays ((Cheung and Kraus, 2010; Kininis and Kraus, 2008); Fig. S3A). Surprisingly, we found ~1000 genes total to be up- or down-regulated after only 10 minutes of E2 treatment. We used hierarchical clustering to define four classes of genes sharing similar patterns of regulation, including a class of rapidly down-regulated genes and three classes of genes with maximal transcription at the three E2 treatment time points (10, 40, or 160 minutes) (Fig. 3, A and B). The down-regulated class was the largest, comprising ~50% of the E2-regulated protein coding transcripts. The majority of genes in this class was rapidly down-regulated (by 10 minutes, on average) and tended (with a few exceptions) to stay down regulated throughout the time course. Up-regulated genes with maximal transcription at 40 minutes were the second largest class, comprising ~34% of the E2-regulated protein coding transcripts. Although the time course of induction or repression varied among the four classes, the magnitude of response did not differ between the classes (Fig. 3C). Interestingly, the genes in the “10 minute max” and “40 minute max” classes returned, on average, to the basal levels of transcription by the end of the E2 treatment time course (Fig. 3B), highlighting the rapid and transient nature of the transcriptional response for the majority of the up-regulated genes.
Figure 3. GRO-seq identifies four distinct classes of E2-regulated RefSeq genes.
(A) Heatmap of the time course of E2-dependent regulation of RefSeq genes. Red numbers indicate the four different classes of regulation.
(B) Centered-scaled traces showing the regulation of the four distinct classes of E2 regulation. Grey lines represent GRO-seq data for individual genes and blue lines represent the mean of the individual traces.
(C) Box and whiskers plot showing the E2-dependent fold change for genes in each of the four classes.
(D) Correlation between fold changes measured by GRO-seq and expression microarrays for genes that show a change in the microarray analyses.
(E) Comparison of GRO-seq data to mRNA expression measured by RT-qPCR. Blue lines represent the mean of the GRO-seq data for the genes analyzed. Grey lines represent RT-qPCR data for individual genes and red lines indicate the mean.
Biologically relevant changes in transcription should be accompanied, in most cases, by similar changes in the steady-state level of the corresponding mRNA. We tested this expectation using both genomic and gene-specific comparisons. First, we compared fold-changes in primary transcription detected using our GRO-seq data to fold-changes at the level of steady-state mRNA (3 or 12 hours of E2 treatment) from published expression microarray data for MCF-7 cells. For the subset of genes that we observed to be regulated by GRO-seq, we found that the strongest correlations were between either the 40 or 160 minute GRO-seq time points with the 3 hour microarray time point (Fig. S3, B and C). Note, however, that there are many more genes detected as E2-regulated by GRO-seq than by expression microarray analyses (Fig. S3A). If we limited the analysis to only genes that change in the microarray analysis (FDR corrected q-value < 0.05), we see an even higher correlation between GRO-seq and microarray data (Fig. 3D; Spearman’s correlation: 0.75). This analysis suggests that the early actions of E2 are almost all mediated at the level of transcription, and that E2 does not affect RNA stability or degradation rate directly. These results provide a first indication that transcription, as determined by GRO-seq, is propagated to changes in the steady-state levels of the corresponding mRNAs.
Next, we randomly selected a set of 10 to 20 genes for each of the four classes (54 genes total) and measured the relative steady-state levels of mRNA from each gene over a 6 hour time course of E2 treatment using RT-qPCR. In general, the changes in transcription measured by GRO-seq were reflected in corresponding changes in the steady-state mRNA levels measured by RT-qPCR (Fig. 3E; Fig. S4). In almost all cases, we observed a delay of approximately 1 to 3 hours between the peak fold changes measured by GRO-seq and RT-qPCR. This delay reflects the time necessary for changes in Pol II (measured at the 5′ end in GRO-seq) to reach the 3′ end of the gene and for mRNA to accumulate (or degrade) by a detectable level. As with the comparisons to the microarray expression data, these results indicate that changes in transcription are efficiently translated into changes in the steady-state levels of the corresponding mRNAs. The correspondence was strongest for the down-regulated and the 40 minute max GRO-seq classes (> 80% of genes assayed showed corresponding changes) and weaker for the 10 minute max and 160 minute max classes (~50% of genes assayed showed corresponding changes). The discrepancies between transcription and steady-state mRNA levels may be due to inherent instability of certain nascent transcripts, which prevents them from generating mature transcripts. Alternatively, it may reflect active post-transcriptional regulation of specific transcripts (e.g., by miRNAs; see below). Interestingly, we identified a number of cases for each GRO-seq time point where E2-dependent changes in transcription were accompanied by corresponding changes in the levels of the cognate protein, including the 10 minute max group (e.g., KRT19, MYC, VDR; Fig. S3, D and E).
Gene ontology (GO) analyses of the four classes of genes revealed a similar pattern of enrichment in gene ontological categories for the down-regulated and 40 minute max classes (Table S1, A and C), which differ from or add to those derived previously from microarray expression analyses (Carroll et al., 2006; Frasor et al., 2003). Specifically, there was a significant enrichment in GO terms related to transcription, nucleic acid metabolism, cell surface receptor and G protein-coupled signaling. The fact that the same GO terms, but different genes, are regulated in both the major up- and down-regulated classes suggests a switch from one cellular signaling program (e.g., serum response) to another (i.e., estrogen signaling); each pathway may require the same functional categories of genes, but use a distinct set of genes within each category. Interestingly, the 160 minute max class was significantly enriched in GO terms related to ribosome biogenesis, translation, and protein synthesis (discussed and elaborated below) (Table S1D), whereas a very modest enrichment of GO terms was observed for the 10 minute max class (Table S1B).
Together, our results show that the transcriptional response to estrogen signaling for protein coding genes (and other classes of transcripts, as well; see below) is rapid, extensive, and transient. This represents a different view of the estrogen response than has been provided by microarray expression studies, which have suggested a continually increasing set of regulated genes in response to E2 treatment, many of which are likely to be secondary or tertiary effects (Fig. S3A).
Pol II dynamics in response to E2
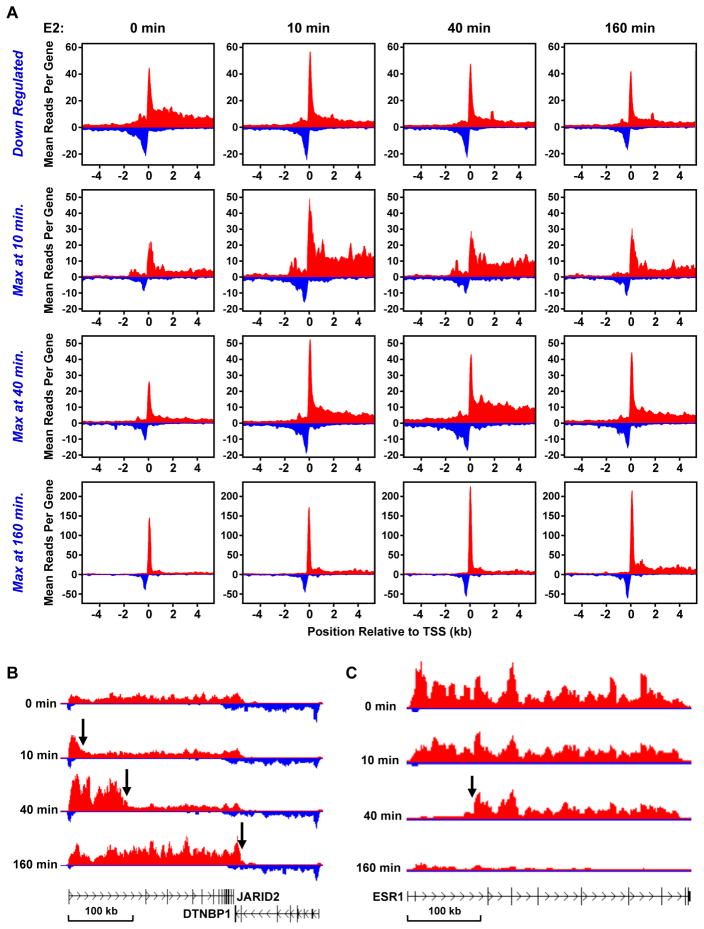
Since the transcriptional response for protein coding genes to estrogen signaling was rapid and transient, we explored the dynamics of Pol II at the promoters of the four classes defined in hierarchical clustering analysis. We performed metagene analyses across the promoter regions of each class from −4 kb to +4 kb for each treatment time point (Fig. 4A). The peak of reads in the immediate vicinity of TSS indicates the presence, on average, of engaged Pol II before and after E2 treatment. The decrease (or increase) of reads in the downstream region indicates the down-regulation (or up-regulation) of transcription in response to E2. This presentation of the GRO-seq results highlights the following: on average (1) loading of Pol II at the TSSs of up-regulated genes increases in response to E2 treatment, (2) divergent transcription of the up-regulated genes increases in response to E2 treatment, (3) down-regulation affects primarily Pol II in the gene bodies, and (4) loading of Pol II at the TSSs and divergent transcription largely follow the Pol II response in the body of the gene.
Figure 4. GRO-seq reveals the dynamics of E2-dependent transcription.
(A) Metagene representations showing the average profile of GRO-seq sequence reads near and at the TSSs of RefSeq genes in each of the four classes during the E2 treatment time course.
(B and C) Gene-specific views of the leading (B) and lagging (C) edges of a Pol II “wave” shown for the up-regulated gene JARID2 (B) and the down-regulated gene ESR1 (C), respectively, during the E2 treatment time course.
See also Figure S4.
The increase in Pol II loading at the TSS in response to E2 suggests that Pol II loads more rapidly than it escapes into the body of the gene for these classes of E2-regulated genes. This is especially evident between the 10 and 40 minute time points for the 40 minute max genes and between the 40 and 160 minutes time points for the 160 minute max genes, where we see increased Pol II loading at the earlier time point followed by an appreciable increase in Pol II in the body of the gene at the later time point. This “delayed” pattern of loading and escape is perhaps unexpected for the 160 minute max genes, since the pausing of Pol II in the promoter proximal region is thought to allow rapid activation of transcription in response to cellular signaling (Lis, 1998). Alternatively, such a response fits well with a recent suggestion that pausing of Pol II in the promoter proximal region allows synchronous gene activation (Boettiger and Levine, 2009).
The dynamics of Pol II can also be clearly observed in examples from specific up- and down-regulated genes (Fig. 4, B and C; Fig. S4). With E2 up-regulated genes, the leading edge of a Pol II wave was observed traveling into the gene body upon E2 treatment (Fig. 4B). In contrast, with E2 down-regulated genes, the lagging edge of a Pol II wave was observed as the polymerases were cleared from the TSS (Fig. 4C). The results from our GRO-seq analysis have provided an unprecedented view of the Pol II dynamics in response to a sustained signal.
Regulation of miRNA gene transcription by estrogen: parallels to the regulation of protein coding genes
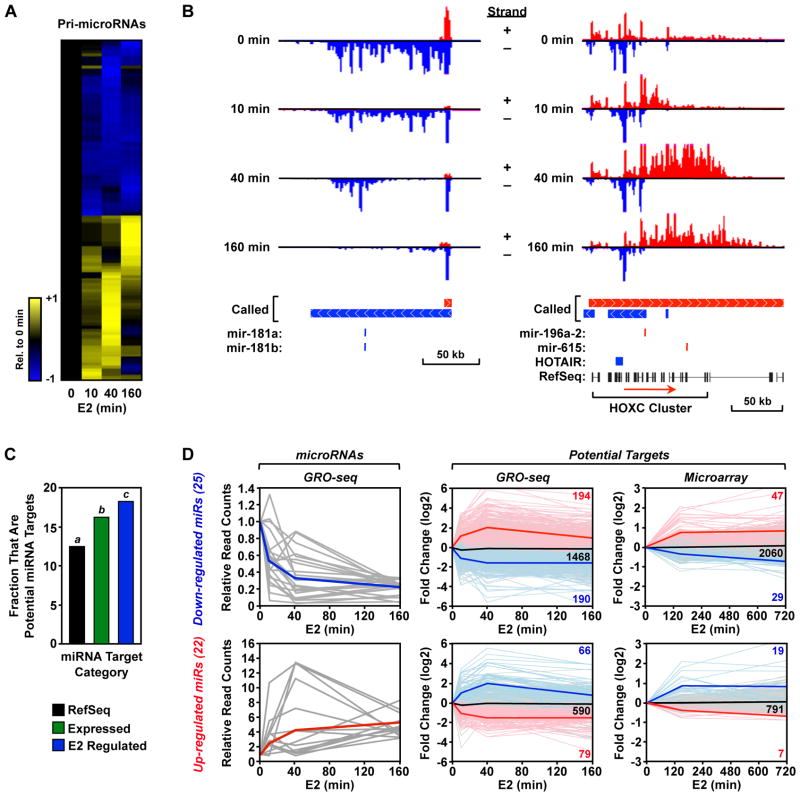
Our GRO-seq approach also provides considerable information regarding the transcriptional regulation of primary microRNA transcripts. Micro RNAs (miRNAs) are ~22 nt non-coding regulatory RNAs that mediate post-transcriptional regulation of gene expression by inhibiting the translation or promoting the degradation of target mRNAs. miRNA precursor transcripts (pri-miRNAs) are generated by Pol II, or in some cases Pol III, either as part of a “host” gene in which they are embedded or from an intergenic region using their own promoter (Krol et al., 2010). Using our GRO-seq data set, we explored the regulation of pri-miRNA gene transcription by E2. We unambiguously identified 322 expressed miRNA-containing transcripts in our data set based on miRBase ver. 14. Of these, 119 (~37%) were regulated by E2 during at least one time point (FDR q-value < 0.001). Regulated pri-miRNAs included some previously published estrogen-regulated miRNAs, including mir-181a, mir-181b and mir-21. Overall, the pattern of regulation depicted in the heatmap shown in Fig. 5A mirrors that observed for the protein-coding transcripts (i.e., approximately half up-regulated and half down-regulated), which is consistent with a large fraction being processed from protein-coding transcripts. Examples of the transcriptional response of specific pri-miRNAs are shown in Fig. 5B. The primary transcript of both examples is considerably larger than the processed miRNA. Therefore, as with the protein-coding genes, the leading (or lagging) edge of the polymerase wave can be seen during the transcriptional response of the up-regulated (or down-regulated) genes. Together, these results suggest that the transcription of pri-miRNA genes are regulated by E2 in a similar pattern and with similar kinetics as protein-coding genes.
Figure 5. E2 regulates the transcription of primary miRNA genes.
(A) Heatmap of the time course of E2-dependent regulation of primary miRNA transcripts.
(B) Gene-specific examples of down-regulated (left) and up-regulated (right) primary miRNA genes. Called transcripts and annotations are shown. In the right panel, the “+ strand” called transcript (red) is actually number of smaller called transcripts that, at the resolution used to represent this region, appear as one transcript.
(C) Fraction of the specified subset of annotated genes that are predicted to be targets of an E2-regulated miRNA based on TargetScan. Bars with different superscripts are significantly different by Fisher’s exact test (p = 3.7 × 10−14 for a/b; p = 0.03 for b/c; p = 1.8 × 10−13 for a/c).
(D) Right panels, GRO-seq data for pri-miRNA transcripts that are up-regulated (top) or down-regulated) ≥ 3-fold by E2. Grey lines = data for individual genes. Blue lines = average for all genes. Middle and left panels, GRO-seq (middle) and expression microarray (right) data for all of the potential targets of miRNAs encoded by the pri-miRNA transcripts shown in the left panels. Faded red, black, and blue lines = data for individual up-regulated, unregulated, and down-regulated genes, respectively (the counts for each type are listed). Bold red, black, and blue lines = averages for all up-regulated, unregulated, and down-regulated genes, respectively.
See also Figure S5.
Next, we determined whether estrogen stimulation involves a coordinated response between pri-miRNA transcripts and the protein coding genes that they ultimately regulate. For this analysis, we reasoned that the subset pri-miRNAs undergoing long-lasting and relatively large regulatory changes are the most likely to be reflected as changes in processed, mature miRNA. Therefore, we focused on 47 of the 119 (~40%) regulated pri-miRNA transcripts that show more than 3-fold up- or down-regulation. These 47 robustly E2-regulated pri-miRNAs potentially target ~2,700 mRNAs according to the TargetScan database (Grimson et al., 2007; Lewis et al., 2005), or ~12.8% of RefSeq annotated mRNAs.
Interestingly, as shown in Fig. 5C, MCF-7 cells express a larger fraction of the ~2,700 target mRNAs than expected, such that 16.6% of expressed genes are targets of these miRNAs (p = 3.7 × 10−14; Fisher’s exact test). This enrichment is consistent with an integrated regulatory program between the miRNAs expressed in a cell and the corresponding mRNA targets, consistent with previous suggestions (Farh et al., 2005). Importantly, the subset of genes regulated by E2 is enriched even further over those genes that are expressed by the cell, such that 18.6% of E2-regulated mRNAs are targets of E2-regulated pri-miRNAs (p = 0.03) (Fig. 5C). Moreover, this pattern of enrichment is robust to selecting a smaller set of miRNAs that are > 5-fold regulated by E2 (p = 0.02), or taking all miRNA transcripts regardless of their fold-change (p = 0.003)]. We found no evidence that E2 specifically coordinates the transcriptional regulation of pri-miRNAs with the direction (i.e. up or down) of regulation of their potential target mRNAs, either by GRO-seq (Fig. 5D, middle panel) or by expression microarrays (Fig. 5D, right panel). In fact, we found evidence for both coordinated and compensatory regulation (Fig. 5D; see Fig. S5 for a detailed explanation). Together, these results suggest an integrated regulatory program for E2-regulated transcription of pri-miRNA transcripts and the mRNAs targeted by the mature miRNAs.
Dramatic up regulation of the protein biosynthetic machinery by estrogen signaling
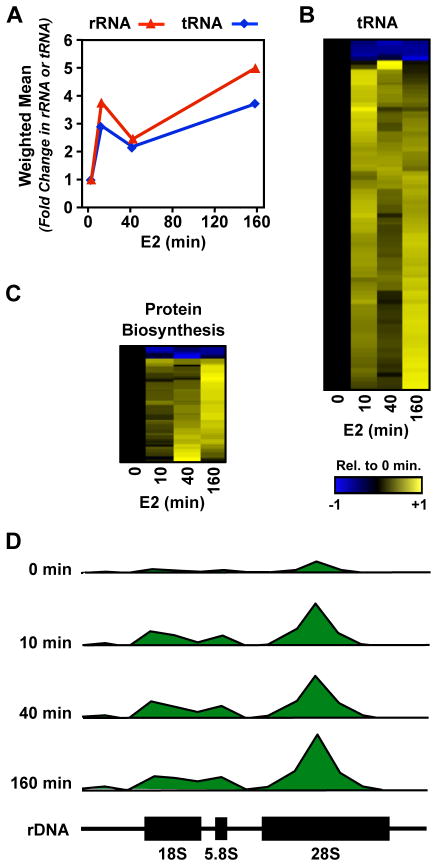
Because our GO analyses showed enrichment in genes with a primary biological function in protein biosynthesis, we asked whether E2 signaling has a broader effect on the protein biosynthetic machinery. GRO-seq provides a measure of all three eukaryotic polymerases; we therefore extracted and analyzed the data for changes in the 45S rRNA (RNA Pol I) and tRNAs (Pol III) annotated in the rnaGene track in the UCSC genome browser. Our analysis revealed that the transcription of Pol I and Pol III transcripts show a similar pattern of regulation by E2: (1) an initial burst at 10 minutes, (2) a slight decrease at 40 minutes, and (3) a maximal increase at 160 minutes (Fig. 6, A and D). These rapid effects are indicative of a primary, rather than secondary, transcriptional response to estrogen signaling.
Figure 6. E2 regulates transcription by Pol I and Pol III.
(A) E2-dependent fold-change in the transcription of the 45S rDNA (Pol I) and tRNA (Pol III) genes.
(B and C) Heatmap of the time course of E2-dependent regulation of tRNA transcripts (B) or protein coding transcripts encoding genes with a biological function or cellular compartment related to the synthesis, metabolism, or function of mature tRNAs or rRNAs (C).
(D) GRO-seq reads mapped to the human rDNA gene (GenBank U13369.1) shown in 1 kb bins relative to the genome location during the time course of E2 treatment.
See also Figure S6.
For individual tRNA genes, changes were strongly biased toward up-regulation, with the transcription of >90% of the tRNA genes showing up-regulation (Fig. 6B). Furthermore, this regulation unambiguously affects 158 of the 486 functional annotated tRNA genes (32%) in at least one of the time points. If the cell is indeed regulating tRNA genes in order to facilitate an increase in translation, one may expect that all 20 amino acids will be up-regulated. Indeed, we found that of the 158 up-regulated tRNA genes, at least one tRNA gene coding for each of the 20 amino acids is represented (p = 0.0012; Fisher’s exact test) (Fig. S6A). In addition to the 20 primary amino acids, we also found the tRNA coding for the amino acid variant selenocysteine, which is thought to play a role in anti-oxidant activity and hormone biosynthesis (Stadtman, 1996), to be regulated by E2. Because each three letter combination of codons is represented multiple times in the 486 annotated tRNA genes, we also asked whether E2 regulates a larger fraction of the 64 possible codon combinations than expected by chance. Indeed, we find 64% of the 64 codon combinations are unambiguously regulated by E2, which is more than expected based on our ability to call 32% of tRNA genes as regulated (p = 0.0027; Fisher’s exact test). These results demonstrate that the observed changes in the protein biosynthetic machinery are applied in a robust and coordinated manner across amino acid and codon variations.
We also conducted a more focused analysis of protein coding genes with functions or cellular localization suggesting a role in protein biosynthesis (e.g. ribosome biogenesis, tRNA aminoacetylation, etc.; see Fig. S6B for all GO terms used). As we observed for tRNA genes, protein coding genes represented in these groups are strongly biased toward up-regulation (Fig. 6C). As suggested by the GO analysis above, these genes are strongly enriched in the 160 minute max class (p = 6.7 × 10−13; Fisher’s exact test), suggesting that these are sustained effects that translate the widespread changes observed in the cellular transcriptome to the proteome.
Taken together, these results demonstrate a potent effect of estrogen signaling on the protein biosynthetic machinery, which fits well with the known mitogenic effects of E2 on MCF-7 cells. In addition, they highlight the fact that estrogen signaling has strong, immediate, and likely direct effects on transcription by all three RNA polymerases, not just Pol II. Up-regulation of the protein biosynthetic machinery is likely a means by which the estrogen signaling pathway prepares the cell for translation of the protein coding transcripts that are newly synthesized in response to estrogen signaling.
Relationship of ERα binding sites to primary estrogen target genes
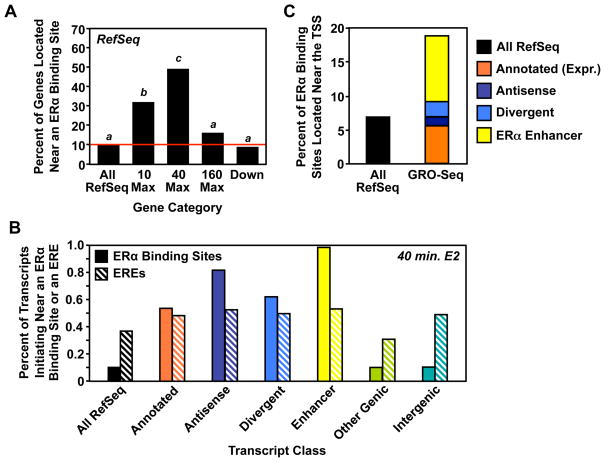
Although most ERα binding sites are located distal to the promoters of protein coding genes, a small but highly significant, enrichment of ERα binding sites has been observed in the proximal promoters of up-regulated genes (Carroll et al., 2005; Carroll et al., 2006), consistent with a direct role of ERα in mediating their regulation. Because our GRO-seq data reflects the direct transcriptional output of the cell, and because our shorter treatment times make it unlikely that we will detect secondary changes in transcription, we reasoned that we should observe a larger fraction of the genes that are regulated by GRO-seq are near ERα binding sites. To test this hypothesis, we used existing ERα ChIP-seq data (Welboren et al., 2009) to determine the fraction of E2-regulated RefSeq genes with a proximal ERα binding site (<10 kb to the transcription start site). Indeed, we found that 46% of genes up-regulated by E2 at shorter time points (i.e., 10 and 40 minutes) contain an ERα binding site within 10 kb of the transcription start site.
Interestingly, when we analyzed the four classes of RefSeq genes (i.e., 10, 40, 160 minute max, and down-regulated) separately, we found striking differences in binding site enrichment between these classes (Fig. 7A). In particular, almost half of the genes in the 40 minute max class are located within 10 kb of an ERα binding site, a striking enrichment over the ~10% found for RefSeq genes in general (p < 2.2 × 10−16; Fisher’s exact test). Genes in the 10 minute max 21 class are also substantially enriched for proximal ERα binding sites (33%; p = 1.2 × 10−12). Up-regulated genes that peak after 160 minutes have a lower level of enrichment that is not statistically significant (12%. p = 0.24), suggesting that a substantial fraction of this subset of genes reflects secondary effects. Conversely, down-regulated genes were slightly less likely than average to be located within 10 kb of an ERα binding site (8%, p = 0.01). This observation strongly suggests that E2 mediates up- and down-regulation by different mechanisms, and that immediate up-regulated genes tend to be the direct, genomic targets of ERα. Those E2-regulated genes that do not have a proximal ERα binding site may be regulated by (1) other promoter-proximally bound transcription factors acting as endpoints of membrane-initiated E2 signaling pathways or (2) looping from distal ERα enhancers to the promoters.
Figure 7. ERα binding sites are enriched in the promoters of primary E2 target genes.
(A) The fraction of the specified subset of RefSeq genes with an ERα binding site found within 10 kb of the TSS. Bars with different superscripts are significantly different by Fisher’s exact test (p < 1.2 × 10−12).
(B) The fraction of the specified transcript class defined at 40 min. E2 treatment that initiates near an ERα binding site or an ERE.
(C) The fraction ERα binding sites found within 1 kb of either all well-annotated RefSeq genes or the specified subset of de novo transcript annotations determined by GRO-seq analysis.
Looking more broadly across the transcript classes, we found that the sets defined at 40 minutes of E2 treatment show a greater enrichment of both ERα binding sites and EREs than the sets defined at the other time points. Interestingly, while the percent of transcripts initiating near a bioinformatically defined estrogen response element (ERE) is not greatly enriched compared to all RefSeq transcripts and is relatively constant across the transcript classes (i.e., ~30 to 50%), the percent of transcripts initiating near an experimentally defined ERα binding site vary considerably (Fig. 7B). We observed the greatest enrichment of ERα binding sites, compared to all RefSeq, near the initiation sites for annotated, antisense, divergent, and enhancer transcripts, suggesting similar modes of E2-dependent regulation as was observed for the protein-coding transcripts (Fig. 7B).
We next determined the fraction of all ERα binding sites that map within the proximal promoter (<1 kb) for each class of transcript defined in our GRO-seq analysis (i.e., looking from an ERα binding site-centric view, as opposed to the transcript-centric view above). We found that ~18% of all ERα binding sites fall near transcripts detected using our HMM in MCF-7 cells (Fig. 7C). This includes ~5 to 6% of ERα binding sites near transcripts matching annotated genes that were specifically found to be expressed in MCF-7 cells using our approach (Fig. 7C, orange bar), as well as an additional ~12% of ERα binding sites found in the proximal promoters of genes producing transcripts that are not currently annotated in public databases (i.e., antisense, divergent, and enhancer transcripts). While this finding still suggests that long-range enhancer-promoter interactions play a pivotal role in actions of ERα, as suggested previously (Fullwood et al., 2009; Pan et al., 2008; Theodorou and Carroll, 2010), it demonstrates a 3- to 4-fold increase in the fraction of ERα binding sites that are located near TSSs.
Collectively, our results provide a new view of signal-dependent transcription events that will suggest new questions and new ways of thinking about specific aspects of the transcriptional response.
Experimental Procedures
Additional details about the experimental procedures can be found in the Supplemental Data.
Cell culture
MCF-7 cells were maintained and propagated as described previously (Kininis et al., 2009).
Generation and analysis of GRO-seq libraries
GRO-seq was performed as described previously (Core et al., 2008), with limited modifications. The data are available from the NCBI’s Gene Expression Omnibus (accession no. GSE27463) and the scripts are available upon request from the corresponding author.
Generation of GRO-Seq libraries
Libraries were generated from two biological replicates of MCF-7 cells grown in estrogen-free medium and treated with 100 nM E2 as indicated. The libraries were sequenced using an Illumina Genome Analyzer.
Transcript calling and annotation
Short-reads were aligned to the human reference genome (hg18, NCBI36), including autosomes, X-chromosome, and one complete copy of an rDNA repeat (GenBank ID: U13369.1) using SOAP2 (Li et al., 2009). A two-state hidden Markov model (HMM) (Durbin, 1998) was used to call transcripts, which were then divided into six distinct, non-overlapping classes, which are intended to describe the function of each transcript. Annotations were made using the decision tree outlined in (Fig. S1E) and based on a set of definitions (Fig. S1F).
Determining estrogen regulation of called transcripts
E2-dependent changes in gene expression were detected using the edgeR package (v.1.4.1) (Robinson et al., 2010). For each GRO-seq time point, reads were counted in a window at the 5′ end of each transcript (+1 to +13 kb). Transcripts that change between the vehicle control and either the 10, 40, or 160 minute time points were collected for analysis if they met a false discovery rate (FDR) corrected q-value threshold (q< 0.001), corresponding to a ~0.1% false discovery rate under the edgeR modeling assumptions.
Clustering, time course, and classification of temporal profiles
We selected all genes with an FDR corrected q-value of 0.001 at any point during the time course for inclusion in the temporal analysis. Computations were performed in R, using the same pipeline we described previously (Danko and Pertsov, 2009).
Additional genomic analyses
In addition to the analyses described above, we performed a set of more focused analyses, as described below. Unless otherwise noted, all computations were performed in R.
Gene Ontology analyses
Gene ontology analyses were performed using GoStat (http://gostat.wehi.edu.au/; (Beissbarth and Speed, 2004)). All expressed genes were used as a background set to analyze GO terms for each class (p < 0.05).
Protein biosynthesis-associated protein coding genes
Protein coding genes with a primary biological function or cellular compartment associated with the ribosome were identified using the Gene Ontology (GO) website (http://www.geneontology.org/) (Fig. S6B).
Comparing E2-induced changes in transcripts called by GRO-seq to changes observed by expression microarrays and Pol II ChIP-seq
Raw CEL files from existing microarray datasets collected using the Affymetrix U133 platform were analyzed together using a previously described pipeline (Danko and Pertsov, 2009). Normalized microarray data were compared to read counts mapping to the +1 to +13 kb window of genes regulated by E2 during at least one point in the GRO-seq time course.
MicroRNA analyses
We identified E2-regulated primary transcripts from our HMM transcript prediction algorithm that contain known miRNAs as described above. Each of these E2-regulated pri-miRNAs was associated with its regulatory targets using the TargetScan database (Lewis et al., 2005). Additional analyses were performed as described in the Supplemental Data.
Comparing the GRO-seq results to known ERα binding sites
For the 10,205 ERα binding sites defined by Welboren et al. (Welboren et al., 2009), we calculated: (1) the fraction of genes in a particular class that are found within 10 kb of an ER-alpha binding site (Fig. 7A), or (2) the fraction of ER-alpha binding sites mapping to within either 1 kb, 5 kb or 10 kb from the 5′ end of the nearest transcript identified de novo using the HMM described above, or in a public database (Fig. 7B).
Correlations between primary transcripts and antisense/divergent transcripts
Transcripts corresponding to sense/antisense or sense/divergent pairs were collected, and the reads were counted and analyzed using R.
Metagene analyses
We used metagene representations to illustrate the distribution of reads near a “typical” transcription start site. Mathematically, we defined a metagene as specified in the Supplemental Data.
RT-qPCR Gene Expression Analyses
Changes in the steady-state levels of the E2-regulated genes were analyzed by RT-qPCR, as previously described (Kininis et al., 2009). The fold expression changes were normalized to GAPDH as an internal standard.
Supplementary Material
Acknowledgments
We thank Andre Martins for helpful insights and suggestions, and Xin Luo and Shrikanth Gadad for critical comments on this manuscript. This work was supported by an NIH training award (T32HD052471) and a postdoctoral fellowship from the PhRMA Foundation to C.G.D., grants from the NIH (GM25232 and HG04845) to J.T.L., and a grant from the NIH/NIDDK (DK058110) to W.L.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acevedo ML, Kraus WL. Transcriptional activation by nuclear receptors. Essays Biochem. 2004;40:73–88. doi: 10.1042/bse0400073. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cheung E, Kraus WL. Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol. 2010;72:191–218. doi: 10.1146/annurev-physiol-021909-135840. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Pertsov AM. Identification of gene co-regulatory modules and associated cis-elements involved in degenerative heart disease. BMC Med Genomics. 2009;2:31. doi: 10.1186/1755-8794-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press; 1998. [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Kraus WL. A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and DNA sequence analysis. Nucl Recept Signal. 2008;6:e005. doi: 10.1621/nrs.06005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Lin CY, Strom A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283:32977–32988. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD, Kraus WL. Chapter 5 biochemical analyses of nuclear receptor-dependent transcription with chromatin templates. Prog Mol Biol Transl Sci. 2009;87:137–192. doi: 10.1016/S1877-1173(09)87005-1. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Theodorou V, Carroll JS. Estrogen receptor action in three dimensions - looping the loop. Breast Cancer Res. 2010;12:303. doi: 10.1186/bcr2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M, Nilsson S, Gustafsson JA. The estrogen receptor family. Curr Opin Obstet Gynecol. 1999;11:249–254. doi: 10.1097/00001703-199906000-00003. [DOI] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- Widelitz RB, Duffy JJ, Gerner EW. Accumulation of heat shock protein 70 RNA and its relationship to protein synthesis after heat shock in mammalian cells. Exp Cell Res. 1987;168:539–545. doi: 10.1016/0014-4827(87)90026-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.