Abstract
N-acetyltransferase 2 (NAT2), an important enzyme in clinical pharmacology, metabolizes antibiotics such as isoniazid and sulfamethoxazole, and catalyzes the transformation of aromatic and heterocyclic amines from the environment and diet into carcinogenic intermediates. Polymorphisms in NAT2 account for variability in the acetylator phenotype and the pharmacokinetics of metabolized drugs. Native Americans, settled in rural areas and large cities of Latin America, are under-represented in pharmacogenetics studies; therefore, we sequenced the coding region of NAT2 in 456 chromosomes from 13 populations from the Americas, and two from Siberia, detecting nine substitutions and 11 haplotypes. Variants *4 (37%), *5B (23%) and *7B (24%) showed high frequencies. Average frequencies of fast, intermediate and slow acetylators across Native Americans were 18, 56 and 25%, respectively. NAT2 intra-population genetic diversity for Native Americans is higher than East Asians and similar to the rest of the world, and NAT2 variants are homogeneously distributed across native populations of the continent.
Keywords: acetylator phenotype, Native American, Latin-American populations, drug-metabolizing enzyme, admixture
Introduction
The N-acetyltransferase 2 (NAT2) is an important enzyme in clinical pharmacology. It metabolizes xenobiotic compounds containing aromatic amines by N- or O-acetylation. NAT2 is the metabolizing enzyme of commonly prescribed antibiotics, such as the antituberculosis drug isoniazid (INH)1 and sulfamethoxazole, prescribed for secondary infections in AIDS patients.2 NAT2 also catalyzes the transformation of aromatic and heterocyclic amines present in cigarette smoke and overcooked meat into carcinogenic intermediates.3
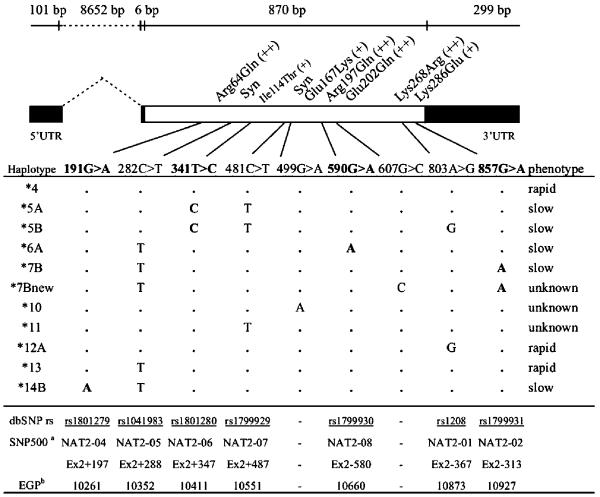
The NAT2 is encoded by NAT2 (OMIM 243400), a gene located at 8p22. NAT2 has two exons, the second of which includes a single open-reading frame of 870 bp (Figure 1, Blum et al.5). Besides the wild-type reference haplotype NAT2*4 (GenBank NM_000015), 35 variants have been described (http://www.louisville.edu/medschool/pharmacology/NAT2.html), most of which have been associated with impaired metabolic activity.6-9 The most common substitutions are 191G>A (Arg64Gln), 341T>C (Ile114Thr), 590G>A (Arg197Gln), 803A>G (Lys268Arg), 857G>A (Lys286Glu), 282C>T and 481C>T (Figure 1), which are unevenly distributed across autochthonous human populations.10
Figure 1.
Representation of human NAT2 locus and definition of haplotypes. The white box represents the coding sequence. Nucleotide changes identified in this study are indicated along with the haplotypes that carry them as in the official nomenclature (www.louisville.edu/medschool/pharmacology/NAT2.html). Mutations that cause a change in the protein activity are in bold. Amino-acid changes are also shown (+ + = conservative, + = moderately conservative; Grantham4). aSNP500Cancer Database (http://snp500cancer.nci.nih.gov/home.cfm) and bNIEHS Environmental Genome Project (http://egp.gs.washington.edu).
NAT2 polymorphism is responsible for variation in the acetylator phenotype. Individuals can be classified as slow or fast acetylators (or metabolizers) on the basis of their NAT2 genotype,11,12 although some authors consider a third intermediate category.13 Slow, intermediate and fast acetylators are defined as carriers of zero, one or two full functioning haplotypes, respectively,7 although exceptions have been reported, in particular in HIV-infected patients.14,15
NAT2 variants are also responsible for variation in pharmacokinetics of INH.16 Tuberculosis is a major health problem in Latin America and other developing countries, and its therapy, based on INH and other first-line medications, is cheap and effective (http://www.who.int). However, adverse drug reactions such as hepatotoxicity can occur,17,18 leading to decreased medication adherence and to drug resistance.19 Whether the incidence of hepatotoxicity differs among slow and fast acetylators is a controversial topic. Whereas recent studies have suggested that slow acetylator status is a risk factor,20-23 some investigations have shown an opposite trend.24,25
In this study, we analyzed NAT2 genetic variation in 12 native and one admixed populations from the Americas and two populations from Siberia, the region that hosted the Pleistocene ancestors of current Native Americans.26,27 Prior sequencing data for NAT2 are not available for these ethnic groups. Previous studies genotyped common single-nucleotide polymorphisms (SNPs) identified in individuals from other continents,28,29 which could lead to ascertainment bias. Therefore, mutations important to define region-specific variation could pass undetected. Because our aim was to identify genetic variants and haplotypes characteristic of the Americas, we sequenced the coding region in each sampled individual. We described the pattern of genetic variation at this locus and tested if haplotype diversity in the American continent fits the population genetics model of isolation-by-distance,30 which explains the geographical pattern of diversity of neutral loci in part of the continent.31 American and Siberian NAT2 haplotype diversities were compared with those of other regions of the world. Finally, we inferred the frequencies of acetylator phenotypes and discussed its pharmacogenetic implications in populations with different demographic history and geographic origin.
Results
Molecular variation, haplotypes and distribution of NAT2 variants
In a sequence analysis of 900 bp that includes the coding region as well as part of the 3′ UTR (Untranslated Region) of NAT2 on 456 chromosomes, nine substitutions were observed (Figure 1), eight of which were reported previously (http://www.louisville.edu/medschool/pharmacology/NAT2.html): the non-synonymous substitutions 191G>A, 341T>C, 499G>A, 590G>A, 803A>G and 857G>A, and the silent substitutions 282C>T and 481C>T. Functional studies have determined that mutations 341T>C and 590G>A reduce the expression or activity of NAT2, and mutations 191G>A, and 857G>A, and in minor part 590G>A, reduce the protein stability.32,9 Despite their phenotypic effect, 341T>C, 590G>A and A857G>A are common substitutions.
We inferred 11 different haplotypes (Figure 1). The non-synonymous 607G>C substitution, detected for the first time in a Coyaima individual, was observed on the background of the slow-function haplotype *7B, and thus the haplotype was called *7Bnew. Haplotype frequencies determined in the studied populations are in Table I of Supplementary Information.
The network in Figure 2 represents the mutational relationships between NAT2 haplotypes and groups of haplotypes (i.e. haplogroups) and shows how each haplotype is distributed across groups of populations. For this analysis, we included data from this study and haplotype frequencies obtained from the literature (see the legend of Figure 2 for references). Although the network encompasses a region of only 900 bp, it contains loops, suggesting that recombination or recurrent mutations have influenced the pattern of diversity in the coding region of NAT2. One NAT2 sequence of chimpanzee was used to root the network. Based on our survey and sequencing data from the SNP500Cancer database and from Patin et al.,33 we identified 13 substitutions between the chimpanzee and the ancestral human haplotype *4, 12 of which fixed and one polymorphic in chimpanzee. This haplotype is modal among East Asians and Native Americans. Haplogroup *5 (defined by 341T>C) shows high frequencies, especially in Europe and in the Hispanic sample, and includes the haplotype *5B, the common and ubiquitous slow-function haplotype with frequency of 23.2% among our sample of the American continent. Haplogroup *7 (defined by 857G>A) is typical of Asian and Native American populations, but is rare in Africa and Europe. Haplogroup *6 (defined by 590G>A) is common in Eurasia and Africa, but rare among Native Americans, and therefore, when observed at moderated frequencies, could be used as indicator of post-Columbian admixture. Altogether, the haplotype distribution shown in Figure 2 is consistent with previous descriptions of allele distribution of NAT2.10,36,37
Figure 2.
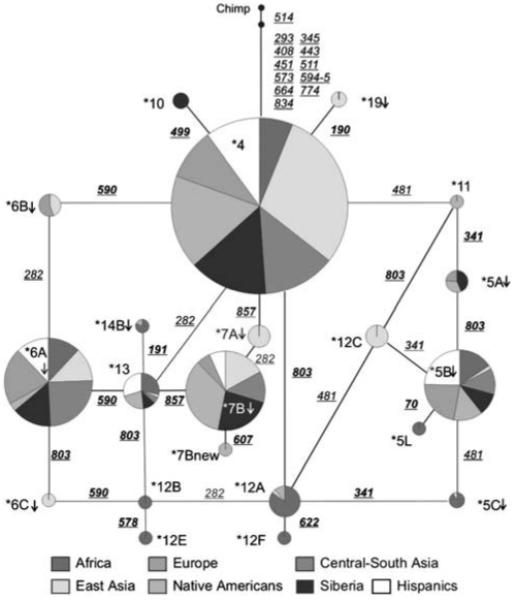
Network of haplotypes. Underlined numbers indicate mutations (non-synonymous in bold) and vertical arrows identify slow function variants. Different colors represent different groups of populations: Africans (African ancestry plus African: n = 24 from SNP500Cancer: http://snp500cancer.nci.nih.gov; Bantu, n = 10;33 Bakola, n = 1033); East Asia (Japan, n = 48;34 Korea, n = 1000;35 Pacific Rim n = 24 from SNP500Cancer); Europe (United States European ancestry, n = 31 from SNP500Cancer; Ashkenazi, n = 10;33 French, n = 10;33 Saami, n = 10;33 Sardinians, n = 633); Central-South Asia (Gujarati, n = 10 and Thai, n = 1433); Native Americans (data from this study); black: Siberians (data from this study) and white: Hispanics (admixed n = 23 from SNP500Cancer).
The frequencies of different haplotypes in each population of this study are represented in Figure 3. The non-marginal frequencies of haplogroup *6 – an indicator of recent admixture from Europe or Africa, among the Coyaima (5/26 chromosomes, 19.2%), Cree (3/28 chromosomes, 10.7%) and Maya (3/32 chromosomes, 9.4%) – suggest the presence of admixture for these Native American populations. In particular, the Coyaima population displays similar frequencies of NAT2*4 and NAT2*6A to the Hispanics from SNP500Cancer, and exhibit the haplotype NAT2*14, which is typical from Africa.38 The observed frequency (9.4%) of the haplogroup *6 in the Maya sample is also consistent with evidence of European admixture in this population, estimated using 377 microsatellites.39
Figure 3.
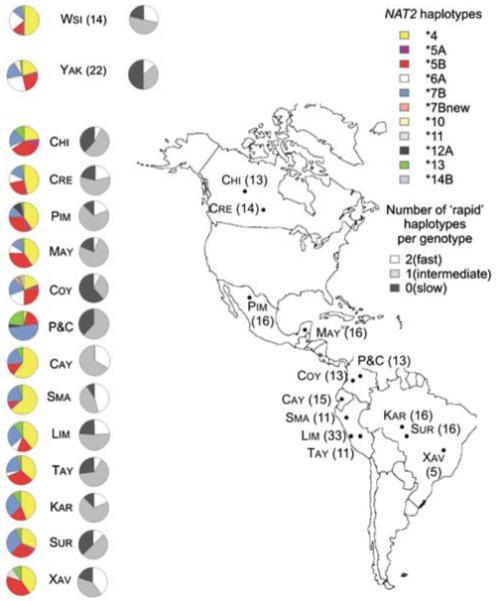
Geographic distribution of samples, haplotype frequencies and number of full function copies of NAT2 haplotypes in Siberians and populations from the American continent. Population codes: CHI: Chypewian; CRE: Cree; PI: Pima; MAY: Maya; COY: Coyaima; P&C: Piapoco and Curripaco; CAY: Cayapa; SMA: San Martín; TAY: Tayacaja; LIM: Las Pampas, Lima; KAR: Karitiana; SUR: Surui and XAV: Xavante. In parentheses number of individuals genotyped per population.
Interestingly, the ‘admixed’ urban sample of tuberculosis patients from the shantytown of Las Pampas (LIM) shows haplotype frequencies typical for Native South-American populations and, in particular, is very similar to that of the native sample of Tayacaja (TAY, Tarazona-Santos et al.40), from the rural area of the Peruvian Central Andes (FST: 0.001; P>0.05).
Population structure
Table 1 shows estimators of intra-population genetic diversity. We also calculated genetic diversity indexes for 13 populations from the literature for which NAT2 coding region was sequenced (Table 1). Although sequencing allows to detect new polymorphisms, most of these screenings only identified the known common mutations 282C>T, 341T>C, 481C>T, 590G>A, 803A>G and 857G>A. The exception were the rare 190C>T in seven individuals from a large Korean sample (n = 2000), and two Bantu (70T>A and 578C>T) and one Bakola (622T>C) from Africa (Figure 2). The comparison across major geographic regions of the world shows that Native Americans have higher NAT2 intra-population genetic diversity than East Asians, and similar values to populations from other continents (Table 1).
Table 1. Intra-population diversity indices in Siberians and Native Americans from this study, and in populations from the literature for which NAT2 coding region sequences were available.
| Population samples |
Diversity indexes
|
||||
|---|---|---|---|---|---|
| S | Number of haplotypes | h | π (× 103) | θs/site (× 103) | |
| West Siberian (n = 14) (66.08°N, 76.63°E) | 6 | 6 | 0.70 | 1.92 | 1.71 |
| Yakut (n = 22) (62–64°N, 129–130°E) | 7 | 7 | 0.82 | 2.72 | 1.79 |
| Siberia | 7 | 7 | 0.79 | 2.42 | 1.6 |
| Chipewyan (n = 13) (59.32°N, 107.18°W) | 6 | 6 | 0.8 | 2.62 | 1.75 |
| Cree (n = 14) (50.38°N, 102.57°W) | 6 | 6 | 0.73 | 2.22 | 1.71 |
| Pima (n = 16) (29°N, 108°W) | 5 | 6 | 0.75 | 2.13 | 1.38 |
| Maya (n = 16) (19°N, 91°W) | 6 | 5 | 0.71 | 2.43 | 1.66 |
| North-Central America (Fst = 0.0) | 6 | 7 | 0.74 | 2.32 | 1.25 |
| Coyaima (n = 13) (3.8°N, 75.2°W) | 8 | 8 | 0.84 | 2.96 | 2.33 |
| Piapoco and Curripaco (n = 13) (3°N, 68°W) | 5 | 5 | 0.68 | 2.16 | 1.46 |
| Cayapa (n = 15) (0, 79°W) | 5 | 4 | 0.6 | 1.61 | 1.40 |
| San Martin (n = 11) (7°S, 77°W) | 5 | 5 | 0.56 | 1.36 | 1.52 |
| Lima (n = 33) (12°S, 77°W) | 6 | 5 | 0.74 | 2.09 | 1.4 |
| Tayacaja (n = 11) (12.33°S, 75.83°W) | 6 | 5 | 0.74 | 2.53 | 1.83 |
| Karitiana (n = 16) (10°S, 63°W) | 5 | 4 | 0.71 | 2.05 | 1.38 |
| Surui (n = 16) (11°S, 62°W) | 5 | 5 | 0.74 | 2.48 | 1.38 |
| South America (Fst = 0.04*) | 8 | 9 | 0.75 | 2.21 | 1.48 |
| America (Fst = 0.03*) | 8 | 9 | 0.75 | 2.27 | 1.37 |
| Hispanic (n = 23) a | 6 | 6 | 0.73 | 2.62 | 1.52 |
| African ancestry (n = 24)a | 5 | 5 | 0.79 | 2.45 | 1.25 |
| Bakola (n = 10)33 | 6 | 6 | 0.77 | 1.88 | 1.88 |
| Bantu (n = 10)33 | 9 | 8 | 0.79 | 2.73 | 2.5 |
| Africa | 9 | 11 | 0.82 | 2.43 | 1.98 |
| European (n = 31)a | 6 | 5 | 0.69 | 2.76 | 1.42 |
| Ashkenazi (n = 10)33 | 5 | 3 | 0.61 | 2.85 | 1.57 |
| French (n = 10)33 | 5 | 4 | 0.72 | 2.61 | 1.57 |
| Saami (n = 10)33 | 6 | 4 | 0.74 | 2.68 | 1.88 |
| Europe | 6 | 6 | 0.69 | 2.73 | 1.22 |
| Gujarati (n = 10)33 | 6 | 4 | 0.73 | 2.68 | 1.88 |
| Thai (n = 14)33 | 6 | 5 | 0.73 | 2.20 | 1.71 |
| Central-South Asia | 6 | 5 | 0.72 | 2.41 | 1.50 |
| Japanese (n = 48)34 | 6 | 7 | 0.66 | 1.47 | 1.30 |
| Korea (n = 1000)35 | 7 | 12 | 0.51 | 1.19 | 0.94 |
| Pacific Rim (n = 24)a | 6 | 4 | 0.70 | 1.92 | 1.50 |
| East Asia | 7 | 12 | 0.53 | 1.23 | 0.94 |
S: number of polymorphic sites; h: haplotype diversity;41 π: nucleotide diversity;42 θs43 (but on base pair basis).
P<0.01.
In parentheses number of individuals and geographic coordinates.
From SNP500Cancer Database (http://snp500cancer.nci.nih.gov/home.cfm).
The FST (i.e. the among-population component of genetic variance) calculated from worldwide populations included in Table 1 is 0.15 (P<0.01), and is similar to the average value estimated across the genome.44,45 For the American continent, the differentiation among populations for NAT2 haplotype frequencies is low (FST = 0.03, P<0.01).
Geographic distribution of NAT2 genetic diversity
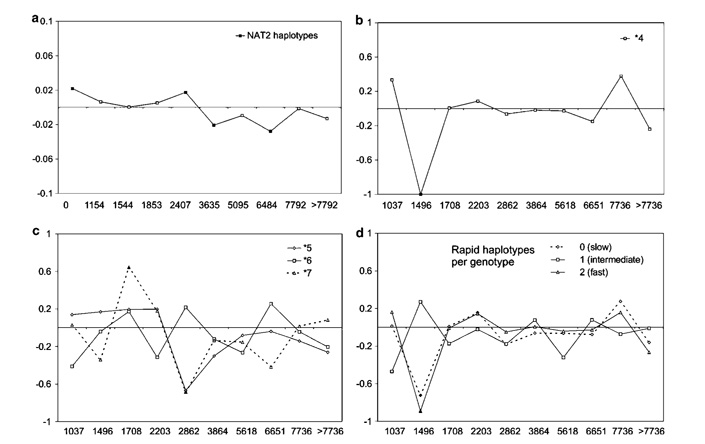
To describe the geographic pattern of genetic diversity at NAT2 in the American continent, we used the spatial autocorrelation analysis on the populations of this study. The spatial autocorrelation coefficients test whether the similarity among haplotypes or samples of haplotypes collected from different locations depends upon geographic distances. The scatterplot of the autocorrelation indexes against classes of geographic distance among populations (i.e. correlogram) can be informative with respect to the evolutionary processes generating it.46 Three analyses were performed: (1) among all observed haplotypes, considering their frequencies and differences in the number of substitutions between them (Figure 4a), using the autocorrelation index for DNA analysis (AIDA) algorithm;47 (2) considering separately the frequencies of each of the common haplotypes *4, *5,*6 and *7 (Figures 4b and c); and (3) considering separately frequencies of inferred phenotypic classes (fast, intermediate and slow acetylators; Figure 4d). Analyses of haplotype and phenotype frequencies (2 and 3) were performed by the classical spatial autocorrelation analysis.48 The observed correlograms were not compatible with a simple isolation-by-distance model,46 whereby an asymptotically decreasing shape is expected. An association between geographic and genetic distances appeared significant only for few classes of distances. In particular, the high frequencies of slow function haplotypes in Colombian populations (COY and P&C) with respect to their low frequencies in CAY and SMA (Figure 3) account for the negative autocorrelation observed on the second distance classes in Figures 4b and c. Taken together, these analyses suggest that geographic distances do not account for the distributions of haplotype or inferred phenotype frequencies in the American continent.
Figure 4.
Spatial autocorrelation analysis in populations from the American continent of this study. X axis: Higher limit of geographic distance classes (in km) between localities. Y axis: Autocorrelation indexes II (a) or I (b–d). (a) Haplotypes defined as in Figure 1. (b) Common fast haplotype: *4. (c) Common slow haplotypes *5A + B; *6A and *7B. (d) Phenotype frequencies: two rapid haplotypes (fast); one rapid and one slow haplotype (intermediate) and two slow haplotypes (slow). Filled symbols indicate significant values. Classes of distance were chosen to contain the same number of comparisons.
Prediction of phenotypes and pharmacogenetic implications
The proportions of inferred acetylator phenotypes based on genotype data obtained in this study are shown in Figure 3 and Table I of Supplementary Information. The highest frequencies of the slow acetylator phenotype are observed in Europe and the Middle East, and the lowest in East Asia.33 On the other hand, the distribution of frequencies of acetylator phenotypes in Africa is heterogeneous.33 A previous study on Native Americans from Panama29 reported frequencies of slow acetylators of 14.7%. Here, based on a larger and more widely distributed sample of Native Americans, we observed a frequency of slow acetylators of ~25%, mainly owing to the common slow-function haplotypes *5B, *6A and *7B. This value is similar to those observed in Central-South Asia, Oceania and in some African populations (see Figure 5 of Patin et al.33).
Discussion
A re-sequencing study of NAT2 coding region in 15 populations of the Americas and Siberia confirmed that mutations common in other continents, known to reduce NAT2 activity (341T>C, 590G>A and 857G>A), are also frequent across the American continent. Notably, we did not observe novel common variants specific for the Americas or Siberia.
The similarity in haplotype structure in an admixed sample from the shantytown of Las Pampas (Lima) to Native Americans population is interesting. To confirm this observation, we sequenced the NAT2 coding region in additional 95 individuals from this population. The resulting haplotype frequencies (see the Extended Lima sample in Table I of Supplementary Information) confirm that this sample resembles Native American ones. Moreover, by using 15 micro satellites, we estimated that the genetic contributions of Native Americans, Europeans and Africans to the Las Pampas (LIM) sample are 82, 12 and 6%, respectively (data not shown), both in the case of tuberculosis patients and healthy individuals. This result confirms that Las Pampas population is predominantly Native American, with a low degree of European admixture evidenced by the presence of haplotype *6A (i.e. a marker of post-Columbian admixture): 7.6% in Lima and 8.9% in the extended Lima sample. The Las Pampas sample was considered admixed or mestizo because of the population history of Lima (Peru), a city that has received contributions from European, Native American and African gene pools. In Latin-American countries, categories such as admixed or mestizos have a strong cultural and socioeconomic basis and do not necessarily reflect the genetic background of individuals or populations. Often, the same individual can be considered Native American or admixed at different ages, depending on cultural and socioeconomic changes. Therefore, these categories should be used with caution in genetic studies on Latin-American populations, as suggested also by other works focused on Brazilian individuals.49,50
Within-population diversity of NAT2 in Native American populations was similar or higher than those characteristics of other geographic regions (Table 1). This contrasts the lower diversity usually seen among Native American populations at different loci.51 Moreover, NAT2 genetic variation appears evenly distributed across populations of the Americas, which do not match the results of recent studies that include part of the samples considered in our survey. On the basis of large sets of microsatellites, Serre and Paabo52 have described the geographical pattern of diversity of the Americas as clinal, and Ramachandran et al.,53 as a product of serial founder effects.
The overall FST is rather low (0.03, P<0.01) compared with other loci,51,54 and consistently, we did not detect any correlation between geographic distances and NAT2 diversity. This low between-population diversity also contrasts with the identification at genomic level of regions where the geographic pattern of diversity drastically changes.55 Our analyses suggest that overall, native populations from the Americas can be considered homogeneous with respect to the distribution of acetylator phenotypes, which may be of significance for pharmacogenetic applications.
In conclusion, we observed a peculiar apportionment of genetic diversity of NAT2 coding region in Native Americans, although a systematic comparison with other loci is not possible owing to the scanty information about genetic variation of these populations.51 Patin et al.33 have recently evidenced that the haplotype structure of NAT2 in Western Eurasia is not compatible with a neutral model of evolution and suggested that positive natural selection have increased the frequency of the slow function haplotype *5B. In principle, their results (i.e. positive values for the neutrality test of Tajima56 for three West-Eurasian populations) are also compatible with a model of balancing natural selection. Similarly, our results show that the nucleotide diversity (π) is consistently higher than θs (based on the number of segregating sites) across the Native American populations (Table 1), so that all the populations but one (i.e. San Martin) show positive, although not significant, Tajima’s D values (data not shown). Based on these results and the low between-population differentiation (Fst = 0.03), also consistent with the signature of balancing selection, it would be tempting to speculate that natural selection could have produced the observed pattern in the Americas. However, we do not have sufficient statistical power to rigorously test this hypothesis on the basis of sequencing analysis of 900 bp, and thus, additional studies based on sequencing or SNPs genotyping across a larger region, such as that performed by Patin et al.,33 are necessary in Native American populations. Moreover, caution to claim balancing selection on Native American population is required, as the founder effect associated with the peopling of the Americas can produce patterns of diversity similar to those generated by balancing selection. On the other hand, the population from the shantytown of Las Pampas in the city of Lima was found to be predominantly Native American. This result stresses the fact that in many Latin-American countries, such as those enveloping the Andes, Native Americans are not a minority group restricted to isolated settlements, but are present in large numbers in urban areas.
Materials and methods
Samples and populations genotyped in this study
We re-sequenced the coding region and part of the 3′ UTR (900 bp) of NAT2 gene from the ATG starting codon of exon 2 (positions 108–1007 GenBank NM_000015), using standard polymerase chain reaction (PCR) and sequencing methods. PCR and sequencing primers and conditions used can be requested from the authors. Our sample includes 456 chromosomes from the following 14 native populations from Siberia and the American continent (codes used in Figure 3 and previous studies on these samples are in parentheses): Western Siberians (WSI), Yakut Siberians (YAK), Chipewyan and Cree from Canada (CHI and CRE), Mexican Pima and Maya (PIM, MAY), Colombian Piapoco and Curripaco (P&C) and Coyaima (COY), Cayapa from Ecuador (CAY),57 San Martín and Tayacaja from Peru (SMA, TAY),31,40,58 and Karitiana (KAR), Surui (SUR), Xavante (XAV) from Brazil. YAK, PIM, MAY, P&C, KAR and SUR individuals are from the HGDP-CEPH Human Genome Diversity Cell Line Panel.59 Figure 3 shows the geographic distributions of the samples and their sizes. Additionally, we have studied 66 chromosomes from tuberculosis patients from the admixed population of Las Pampas, a shantytown in Lima, Peru (LIM). All samples were collected with informed consent of the donors and have been anonymized. We also sequenced the same genomic region in one chimpanzee provided by the European Collection of Cell Cultures (Ecacc, ref. EB176 (JC) 89072704) to identify ancestral states of alleles.
For the admixture analyses of the Las Pampas sample, we used 15 microsatellites included in the AmpFLSTR Identifiler Kit (Applied Biosystems, Foster City, CA, USA).
Statistical analyses
Haplotypes were inferred using the software PHASE version 2.1.60 Phylogenetic relationships between different haplotypes were explored by a network implemented in the software TCS.61
The within-population variability was estimated by haplotype diversity (h),41 and by calculating two estimators of the parameter θ = 4Neμ, (where Ne is the effective population size and μ the mutation rate per generation): (1) nucleotide diversity (π), which is the per-site mean number of pair-wise differences between sequences42 and (2) θs, based on the number of segregating sites (S).43 Xavante from Brazil were excluded from this analysis because of the small sample size. Native Americans were grouped into North-Central America and South America, and standard FST values were calculated to estimate the between-population component of genetic variance for each of these groups, using the software Arlequin 2.0.62
We applied two kinds of spatial autocorrelation analyses. In the first case, we considered the whole set of populations and haplotypes and calculated the similarity index II by means of the AIDA software (Autocorrelation Index for DNA Analysis47 http://web.unife.it/progetti/genetica/Giorgio/giorgio_soft.html). In the second case, a classic spatial autocorrelation analysis was performed on haplotype and phenotype frequencies using the software PASSAGE.63
The admixture was estimated using a sample of 58 tuberculosis patients and 65 healthy individuals from Las Pampas (LIM). To represent the parental populations, we included 42 natives from TAY and SAM, and 31 Europeans and 24 African ancestry individuals from SNP500Cancer. The estimation of admixture was performed using the software Structure,64 assuming that individuals are admixed, and that the allele frequencies of parental populations are correlated.
Supplementary Material
NAT2 haplotype and inferred phenotype frequencies in Siberians and Native Americans from this study, and in populations from the literature for which NAT2 coding region sequences were available.
Acknowledgments
We are grateful to the individuals who, contributing samples, made this study possible; to Cristina Fabbri, Guido Barbujani, Giorgio Bertorelle, Carolina Bonilla and two reviewers for criticisms, to Etienne Patin and Lluis Quintana-Murci for sharing data and to the Brazilian Fundação Nacional do Indio for logistic help. This study was partially funded by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brazil) to ET-S, FMS and SLB; the University of Bologna to DP, DL and ET-S; the Fundaçao de Amparo à Pesquisa do Estado de Rio Grande do Sul to FMS and SLB, and the Wellcome Trust to CAE.
Footnotes
Duality of interest
The authors declare that do not have conflict of interest.
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
References
- 1.American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Treatment of tuberculosis. Am J Respir Crit Care Med. 2003;167:603–662. doi: 10.1164/rccm.167.4.603. [DOI] [PubMed] [Google Scholar]
- 2.Grimwade K, Gilks C. Cotrimoxazole prophylaxis in adults infected with HIV in low-income countries. Curr Opin Infect Dis. 2001;14:507–512. doi: 10.1097/00001432-200110000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 4.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 5.Blum M, Grant DM, McBride W, Heim M, Meyer UA. Human arylamine N-acetyltransferase genes: isolation, chromosomal localization, and functional expression. DNA Cell Biol. 1990;9:193–203. doi: 10.1089/dna.1990.9.193. [DOI] [PubMed] [Google Scholar]
- 6.Cascorbi I, Drakoulis N, Brockmoller J, Maurer A, Sperling K, Roots I. Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am J Hum Genet. 1995;57:581–592. [PMC free article] [PubMed] [Google Scholar]
- 7.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 8.Shishikura K, Hohjoh H, Tokunaga K. Novel allele containing 190C>T nonsynonymous substitution in the N-acetyltransferase (NAT2) gene. Hum Mut. 2000;15:581. doi: 10.1002/1098-1004(200006)15:6<581::AID-HUMU17>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 9.Zang Y, Zhao S, Doll MA, States JC, Hein DW. The T341C (Ile114Thr) polymorphism of N-acetyltransferase 2 yields slow acetylator phenotype by enhanced protein degradation. Pharmacogenetics. 2004;14:717–723. doi: 10.1097/00008571-200411000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Lin HJ, Han CY, Lin BK, Hardy S. Ethnic distribution of slow acetylator mutations in the polymorphic N-acetyltransferase (NAT2) gene. Pharmacogenetics. 1994;4:125–134. doi: 10.1097/00008571-199406000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Cascorbi I, Roots I. Pitfalls in N-acetyltransferase 2 genotyping. Pharmacogenetics. 1999;9:123–127. doi: 10.1097/00008571-199902000-00016. [DOI] [PubMed] [Google Scholar]
- 12.Donald PR, Sirgel FA, Venter A, Parkin DP, Seifart HI, van de Wal BW, et al. The influence of human N-acetyltransferase genotype on the early bactericidal activity of isoniazid. Clin Infect Dis. 2004;39:1425–1430. doi: 10.1086/424999. [DOI] [PubMed] [Google Scholar]
- 13.Parkin DP, Vandenplas S, Botha FJ, Vandenplas ML, Seifart HI, van Helden PD, et al. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am J Respir Crit Care Med. 1997;155:1717–1722. doi: 10.1164/ajrccm.155.5.9154882. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann GR, Wenk M, Taeschner W, Peterli B, Gyr K, Meyer UA, et al. N-acetyltransferase 2 polymorphism in patients infected with human immunodeficiency virus. Clin Pharmacol Ther. 1996;60:62–67. doi: 10.1016/S0009-9236(96)90168-X. [DOI] [PubMed] [Google Scholar]
- 15.O’Neil WM, Drobitch RK, MacArthur RD, Farrough MJ, Doll MA, Fretland AJ, et al. Acetylator phenotype and genotype in patients infected with HIV: discordance between methods for phenotype determination and genotype. Pharmacogenetics. 2000;10:171–182. doi: 10.1097/00008571-200003000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Evans DAP, Manley KA, McKusick VA. Genetic control of isoniazid metabolism in man. BMJ. 1960;2:485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqui MA, Khan IA. Isoniazid-induced lupus erythematosus presenting with cardiac tamponade. Am J Ther. 2002;9:163–165. doi: 10.1097/00045391-200203000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Maddrey WC. Drug-induced hepatotoxicity: 2005. J Clin Gastroenterol. 2005;39(Suppl 2):S83–S89. doi: 10.1097/01.mcg.0000155548.91524.6e. [DOI] [PubMed] [Google Scholar]
- 19.Mitchison DA. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int J Tuberc Lung Dis. 1998;2:10–15. [PubMed] [Google Scholar]
- 20.Pande JN, Singh SP, Khilnani GC, Khilnani S, Tandon RK. Risk factors for hepatotoxicity from antituberculosis drugs: a case–control study. Thorax. 1996;51:132–136. doi: 10.1136/thx.51.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno M, Yamaguchi I, Yamamoto I, Fukuda T, Yokota S, Maekura R, et al. Slow N-acetyltransferase 2 genotype affects the incidence of isoniazid and rifampicin-induced hepatotoxicity. Int J Tuberc Lung Dis. 2000;4:256–261. [PubMed] [Google Scholar]
- 22.Yew WW. Risk factors for hepatotoxicity during anti-tuberculosis chemotherapy in Asian populations. Int J Tuberc Lung Dis. 2001;5:99–100. [PubMed] [Google Scholar]
- 23.Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, Yang SY, et al. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology. 2002;35:883–889. doi: 10.1053/jhep.2002.32102. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell JR, Long MW, Thorgeirsson UP, Jollow DJ. Acetylation rates and monthly liver function tests during one year of isoniazid preventive therapy. Chest. 1975;68:181–190. doi: 10.1378/chest.68.2.181. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto T, Suou T, Hirayama C. Elevated serum aminotransferase induced by isoniazid in relation to isoniazid acetylator phenotype. Hepatology. 1986;6:295–298. doi: 10.1002/hep.1840060223. [DOI] [PubMed] [Google Scholar]
- 26.Santos FR, Pandya A, Tyler-Smith C, Pena SD, Schanfield M, Leonard WR, et al. The central Siberian origin for native American Y chromosomes. Am J Hum Genet. 1999;64:619–628. doi: 10.1086/302242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol Biol Evol. 2004;21:164–175. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]
- 28.Arias TD, Jorge LF, Griese EU, Inaba T, Eichelbaum M. Polymorphic N-acetyltransferase (NAT2) in Amerindian populations of Panama and Colombia: high frequencies of point mutation 857A, as found in allele S3/M3. Pharmacogenetics. 1993;3:328–331. doi: 10.1097/00008571-199312000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Jorge-Nebert LF, Eichelbaum M, Griese EU, Inaba T, Arias TD. Analysis of six SNPs of NAT2 in Ngawbe and Embera Amerindians of Panama and determination of the Embera acetylation phenotype using caffeine. Pharmacogenetics. 2002;12:39–48. doi: 10.1097/00008571-200201000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuselli S, Tarazona-Santos E, Dupanloup I, Soto A, Luiselli D, Pettener D. Mitochondrial DNA diversity in South America and the genetic history of Andean highlanders. Mol Biol Evol. 2003;20:1682–1691. doi: 10.1093/molbev/msg188. [DOI] [PubMed] [Google Scholar]
- 32.Fretland AJ, Leff MA, Doll MA, Hein DW. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 2001;11:207–215. doi: 10.1097/00008571-200104000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Patin E, Barreiro LB, Sabeti PC, Austerlitz F, Luca F, Sajantila A, et al. Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes. Am J Hum Genet. 2006;78:423–436. doi: 10.1086/500614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekine A, Saito S, Iida A, Mitsunobu Y, Higuchi S, Harigae S, et al. Identification of single-nucleotide polymorphisms (SNPs) of human N-acetyltransferase genes NAT1, NAT2, AANAT, ARD1 and L1CAM in the Japanese population. J Hum Genet. 2001;46:314–319. doi: 10.1007/s100380170065. [DOI] [PubMed] [Google Scholar]
- 35.Lee SY, Lee KA, Ki CS, Kwon OJ, Kim HJ, Chung MP, et al. Complete sequencing of a genetic polymorphism in NAT2 in the Korean population. Clin Chem. 2002;48:775–777. [PubMed] [Google Scholar]
- 36.Grant DM, Hughes NC, Janezic SA, Goodfellow GH, Chen HJ, Gaedigk A, et al. Human acetyltransferase polymorphisms. Mutat Res. 1997;376:61–70. doi: 10.1016/s0027-5107(97)00026-2. [DOI] [PubMed] [Google Scholar]
- 37.Meyer UA, Zanger UM. Molecular mechanisms of genetic polymorphisms of drug metabolism. Annu Rev Pharmacol Toxicol. 1997;37:269–296. doi: 10.1146/annurev.pharmtox.37.1.269. [DOI] [PubMed] [Google Scholar]
- 38.Delomenie C, Sica L, Grant DM, Krishnamoorthy R, Dupret J-M. Genotyping of the polymorphic N-acetyltransferase (NAT2*) gene locus in two native African populations. Pharmacogenetics. 1996;6:177–185. doi: 10.1097/00008571-199604000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 40.Tarazona-Santos E, Carvalho-Silva DR, Pettener D, Luiselli D, De Stefano GF, Labarga CM, et al. Genetic differentiation in South Amerindians is related to environmental and cultural diversity: evidence from the Y chromosome. Am J Hum Genet. 2001;68:1485–1496. doi: 10.1086/320601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- 42.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 44.Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, et al. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am J Hum Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P, International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokal R. Ecological parameters inferred from spatial correlograms. In: Patil G, Rozenzweig M, editors. Contemporary Quantitative Ecology and Related Econometrics. International Co-operative Publishing House; Fairland, MD: 1979. pp. 167–196. [Google Scholar]
- 47.Bertorelle G, Barbujani G. Analysis of DNA diversity by spatial autocorrelation. Genetics. 1995;140:811–819. doi: 10.1093/genetics/140.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokal RR, Oden NL. Spatial autocorrelation in biology. 1. Methodology. Biol J Linn Soc. 1978;10:199–228. [Google Scholar]
- 49.Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD. Color and genomic ancestry in Brazilians. Proc Natl Acad Sci USA. 2003;100:177–182. doi: 10.1073/pnas.0126614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suarez-Kurtz G. Pharmacogenomics in admixed populations. Trends Pharmacol Sci. 2005;26:196–201. doi: 10.1016/j.tips.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Mulligan CJ, Hunley K, Cole S, Long JC. Population genetics, history, and health patterns in native Americans. Annu Rev Genomics Hum Genet. 2004;5:295–315. doi: 10.1146/annurev.genom.5.061903.175920. [DOI] [PubMed] [Google Scholar]
- 52.Serre D, Paabo S. Evidence for gradients of human genetic diversity within and among continents. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- 55.Barbujani G, Belle EM. Genomic boundaries between human populations. Hum Hered. 2006;61:15–21. doi: 10.1159/000091832. [DOI] [PubMed] [Google Scholar]
- 56.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rickards O, Martinez-Labarga C, Lum JK, De Stefano GF, Cann RL. mtDNA history of the Cayapa Amerinds of Ecuador: detection of additional founding lineages for the Native American populations. Am J Hum Genet. 1999;65:519–530. doi: 10.1086/302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luiselli D, Simoni L, Tarazona-Santos E, Pastor S, Pettener D. Genetic structure of Quechua-speakers of the Central Andes and geographic patterns of gene frequencies in South Amerindian populations. Am J Phys Anthropol. 2000;113:5–17. doi: 10.1002/1096-8644(200009)113:1<5::AID-AJPA2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 59.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 60.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 62.Schneider S, Roessli D, Excoffier L. Arlequin Ver. 2.0: A Software for Population Genetics Data Analysis. Genetics and Biometry Laboratory, University of Geneva; Switzerland: 2000. [Google Scholar]
- 63.Rosenberg M. Pattern Analysis, Spatial Statistics, and Geographic Exegesis, version 1.1. 1.1 edn. Department of Biology, Arizona State University; Tempe: 2001. [Google Scholar]
- 64.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NAT2 haplotype and inferred phenotype frequencies in Siberians and Native Americans from this study, and in populations from the literature for which NAT2 coding region sequences were available.