Abstract
N-glycosylation of eukaryotic proteins helps them fold and traverse the cellular secretory pathway and can increase their stability, although the molecular basis for stabilization is poorly understood. Glycosylation of proteins at naïve sites (ones that normally are not glycosylated) could be useful for therapeutic and research applications, but currently results in unpredictable changes to protein stability. We show that placing a Phe residue two or three positions prior to a glycosylated Asn in distinct reverse turns facilitates stabilizing interactions between the aromatic side chain and the first N-acetylglucosamine (GlcNAc) of the glycan. Glycosylating this portable structural module, an “enhanced aromatic sequon”, in three different proteins stabilizes their native states by −0.7 to −2.0 kilocalories per mole and increases cellular glycosylation efficiency.
In eukaryotic cells, N-glycosylation occurs co-translationally as the ribosome inserts proteins into the endoplasmic reticulum (ER). The oligosaccharyl transferase (OST) attaches the Glc3Man9GlcNAc2 (where Glc, Man, and GlcNAc are glucose, mannose, and N-acetylglucosamine, respectively) oligosaccharide to the Asn side chain within some Asn-Xxx-Thr/Ser sequons (where Xxxx denotes any amino acid except Pro) (1). The factors governing glycosylation efficiency are incompletely understood. Asn-linked N-glycans extrinsically affect protein folding efficiency in the ER because the glycan tag can direct the protein into the calnexin/calreticulin (CNX/CRT) folding versus degradation pathway (2, 3). Glycans can also intrinsically stabilize proteins (4–11).
Protein stabilization through N-glycosylation decreases the population of the aggregation-prone, protease-sensitive unfolded state relative to the folded ensemble, increasing serum half-life, improving shelf-life, and shielding immunogenic epitopes (12). Conferring these properties by adding N-glycans to naïve proteins (not normally glycosylated) or to glycoproteins at naïve sites could have useful applications. However, attempts to add N-glycans to proteins by trial and error have unpredictable thermodynamic consequences, and often lead to destabilization (13–15). Thus, we sought an increased understanding of glycan-protein interactions that mediate stabilization in order to identify a portable structural module that could reliably stabilize proteins upon N-glycosylation.
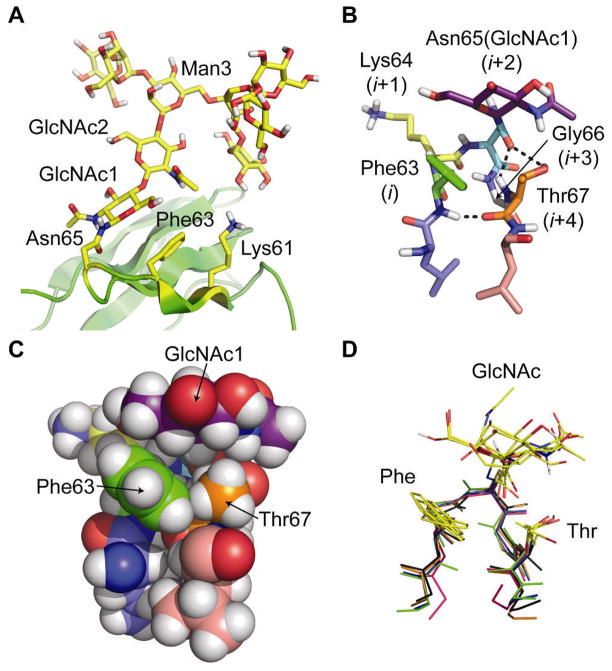
Previous work on the adhesion domain of human CD2 (HsCD2ad, Fig. 1A) led us to believe that it might contain such a module. HsCD2ad is known to be unfolded without glycosylation at Asn65 (16). We previously showed that glycosylated HsCD2ad is −3.1 kcal mol−1 more stable and unfolds 50× more slowly than its non-glycosylated counterpart (11), a large effect that is atypical of most glycoproteins. Published NMR-based structural studies (16) reveal that N-glycosylated Asn65 occupies the i+2 position of a type I β-turn with a G1 β-bulge (Fig. 1B), hereafter called a type I β-bulge turn (also classified as a 3:5 hairpin (17)). This turn spans Phe63 (i) to Thr67 (i+4), with Gly66 occupying the i+3 bulge position (16). These NMR data (16) show that the Phe63 side chain interacts with the hydrophobic α-face of GlcNAc1 on Asn65 and the Thr67 side chain, forming a compact structure (Fig. 1C), and that the Lys61 side chain is close to GlcNAc2.
Figure 1.
(A) Ribbon diagram of the published NMR structure of HsCD2ad (PDB: 1GYA) (16). The N-glycan, Asn65, Phe63, and Lys61 are highlighted in yellow. (B) Stick and (C) space-filling representations of the type I β-bulge turn in HsCD2ad; the (i+2)-position Asn65(GlcNAc1) packs against the i-position Phe63, and the i+4 Thr67. Dashed lines indicate hydrogen bonds. (D) Glycosylated type I β-bulge turns with an i-position Phe from known proteins in the PDB (green: HsCD2ad; blue: 1G82; orange: 1FJR; red: 1ICF; and black: 3DMK). All structures rendered in Pymol (www.pymol.org).
As we began this work, the energetic contributions of these interactions had not been assessed directly, but they were thought to play a key role in stabilizing HsCD2ad (11, 16), and in accelerating its folding (11). General stabilization of HsCD2ad via glycan-induced unfolded-state destabilization had also been hypothesized (11, 16), though recent studies do not support this proposed effect as a universal consequence of N-glycosylation (13–15). If the tripartite interaction between Phe63, GlcNAc1, and Thr67 (in the context of a type I β-bulge turn) contributes to the large stabilizing effect of N-glycosylation on HsCD2ad, it could represent a stabilizing portable structural module that could be engineered into many proteins, since the vast majority of proteins contain reverse turns. A survey of the Protein Data Bank (PDB) supports this possibility; four other known proteins contain type I β-bulge turns with Phe at the i position, an Asn(glycan) at i+2, Gly at i+3, and Thr at i+4 (Fig. 1D, see Fig. S1 for examples where Trp, Tyr, and His occupy the i position) (18).
We probed the energetic consequences of these interactions using the rat ortholog of HsCD2ad (RnCD2ad, Fig. 2A), because it does not require N-glycosylation to fold. Published structural data (19) reveal that residues 63–67 of RnCD2ad have the same type I β-bulge turn geometry found in HsCD2ad (Fig. 2A, inset), but this turn is not glycosylated because it lacks a glycosylation sequon. We installed the HsCD2ad Asn65-Gly66-Thr67 sequon into RnCD2ad by mutating Asp67 to Thr (Asn65 and Gly66 were already present). To ensure that RnCD2ad would be glycosylated only at Asn65, we removed three naturally occurring sequons (by Asn72Gln, Asn82Gln and Asn89Asp mutations) (18). The RnCD2ad sequence with these four mutations is referred to as RnCD2*.
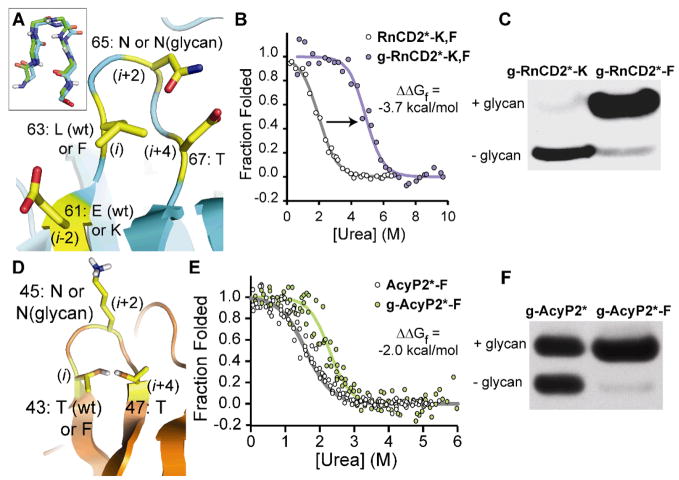
Figure 2.
(A) Ribbon diagram of a type I β-bulge turn in the known crystal structure of RnCD2ad (PDB: 1HNG) (19), rendered in Pymol; inset shows the backbone alignment of this turn with the corresponding reverse turn from HsCD2ad. Overlaid text shows the amino acids to be perturbed at positions 61 (i-2), 63 (i), 65 (i+2), and 67 (i+4), which are highlighted in yellow. wt, wild type (B) Fluorescence-monitored urea denaturation curve for glycoprotein g-RnCD2*-K,F (purple circles) and non-glycosylated RnCD2*-K,F (white circles). Fits of the data (18) appear as purple and gray lines, respectively. (C) Western blot (anti-FLAG tag) of conditioned media from infected Sf9 cells expressing g-RnCD2*-K and g-RnCD2*-F. (D) Ribbon diagram of a reverse turn in the known NMR structure of horse AcyP2 (PDB: 1APS; 95% identical to human, with 100% identity in residues 43 to 47) (22), rendered in Pymol. Overlaid text shows the amino acids to be perturbed at positions 43 (i), 45 (i+2), and 47 (i+4), which are highlighted in yellow. (E) Fluorescence-monitored urea denaturation curve for glycoprotein g-AcyP2*-F (green circles) and non-glycosylated AcyP2*-F (open circles). Fits of the data to a two-state folding model (18) appear as green and gray lines, respectively. (F) Western blot (anti-FLAG tag) of conditioned media from infected Sf9 cells expressing g-AcyP2* and g-AcyP2*-F.
RnCD2* has Glu and Leu at positions 61 and 63, instead of Lys and Phe, as in HsCD2ad (cf. Fig. 1B and Fig. 2A, the latter showing the published structure of non-glycosylated wild-type RnCD2ad (19)). These differences make RnCD2* ideal for studying the energetics of the interactions between the N-glycan, Lys61, and Phe63, using eight RnCD2* variants in a triple mutant cycle (20). Glycosylated variants (denoted by the prefix “g−”) were produced in Sf9 insect cells (see Table S1 for ESI-MS data), whereas non-glycosylated counterparts were produced in E. coli (18). Table 1 summarizes the thermodynamic and kinetic data determined by fluorescence-monitored chaotrope denaturation (Fig. 2B and Fig. S2) and stopped-flow kinetics experiments (Fig. S3) (18).
Table 1.
Sequences (positions 61 to 67 for RnCD2*, 41–47 for ActP2*, and 15–21 for WW: positions where amino acids were perturbed appear in boldface), folding free energies, and folding and unfolding rate constant (k) ratios for RnCD2ad-, AcyP2- and Pin WW-derived glycoproteins and their non-glycosylated counterparts.†
| Protein | Sequence‡ | Δ Gf (kcal/mol) | ΔΔ Gf (kcal/mol) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61 | 67 | ||||||||||||||
| RnCD2* | -- | E | I | L | A | N | G | T | -- | −5.1 ± 0.4 | −0.6 ± 0.6 | 1.2 ± 0.1 | 0.5 ± 0.6|| | ||
| g-RnCD2* | -- | E | I | L | A | N | G | T | -- | −5.7 ± 0.5 | |||||
| RnCD2*-K | -- | K | I | L | A | N | G | T | -- | −4.0 ± 0.5 | −1.5 ± 0.6 | 1.5 ± 0.4 | 0.1 ± 0.1 | ||
| g-RnCD2*-K | -- | K | I | L | A | N | G | T | -- | −5.5 ± 0.4 | |||||
| RnCD2*-F | -- | E | I | F | A | N | G | T | -- | −4.2 ± 0.2 | −1.8 ± 0.4 | 1.0 ± 0.2 | 0.06 ± 0.01 | ||
| g-RnCD2*-F | -- | E | I | F | A | N | G | T | -- | −6.0 ± 0.3 | |||||
| RnCD2*-K,F | -- | K | I | F | A | N | G | T | -- | −2.1 ± 0.4 | −3.7 ± 0.6 | 2.4 ± 0.7 | 0.005 ± 0.004¶ | ||
| g-RnCD2*-K,F | -- | K | I | F | A | N | G | T | -- | −5.8 ± 0.5 | |||||
| 41 | 47 | ||||||||||||||
| AcyP2* | -- | K | N | T | A | N | G | T | -- | −4.1 ± 0.7 | 0.5 ± 0.8 | N.D.§ | N.D. | ||
| g-AcyP2* | -- | K | N | T | A | N | G | T | -- | −3.6 ± 0.4 | |||||
| AcyP2*-F | -- | K | N | F | A | N | G | T | -- | −2.5 ± 0.5 | −2.0 ± 0.7 | N.D. | N.D. | ||
| g-AcyP2*-F | -- | K | N | F | A | N | G | T | -- | −4.5 ± 0.8 | |||||
| 15 | 21 | ||||||||||||||
| WW | -- | M | S | R | S | N | G | R | -- | −0.11 ± 0.05 | 0.26 ± 0.06 | 0.70 ± 0.06 | 1.0 ± 0.1 | ||
| g-WW | -- | M | S | R | S | N | G | R | -- | 0.15 ± 0.04 | |||||
| WW-F | -- | M | F | R | S | N | G | R | -- | 0.29 ± 0.04 | 0.07 ± 0.07 | 0.89 ± 0.09 | 1.0 ± 0.1 | ||
| g-WW-F | -- | M | F | R | S | N | G | R | -- | 0.36 ± 0.05 | |||||
| WW-T | -- | M | S | R | S | N | G | T | -- | 0.19 ± 0.06 | 0.08 ± 0.08 | 0.86 ± 0.07 | 1.0 ± 0.1 | ||
| g-WW-T | -- | M | S | R | S | N | G | T | -- | 0.28 ± 0.05 | |||||
| WW-F,T | -- | M | F | R | S | N | G | T | -- | 0.69 ± 0.05 | −0.70 ± 0.08 | 2.0 ± 0.3 | 0.7 ± 0.1 | ||
| g-WW-F,T | -- | M | F | R | S | N | G | T | -- | −0.01 ± 0.06 | |||||
Tabulated data are given as mean ± standard error at 25 °C for RnCD2* and AcyP2*, and are calculated at 55 °C for WW variants. For tabulated rate constants, see Fig. S3 and Table S3 (18).
N= Asn(glycan). The glycan is mostly Man6–8 oligomannose for RnCD2* variants, is mostly fucosylated paucimannose for AcyP2* variants, and is GlcNAc for WW variants (see Table S1) (18).
N.D., not determined.
This unfolding rate ratio is within error of 1, indicating that the unfolding rates are not significantly different. It is also within error of being negative; this is an artifact of treating this naturally positive quantity as being normally distributed.
Glycosylation does not significantly stabilize g-RnCD2* relative to RnCD2* (difference in Gibbs free energy of folding (ΔΔGf) = −0.6 ± 0.6 kcal mol−1, −2.5 kcal mol−1 less than the glycosylation-induced stabilization of HsCD2ad) (Table 1). In contrast, g-RnCD2*-K and g-RnCD2*-F are −1.5 ± 0.6 and −1.8 ± 0.4 kcal mol−1 more stable than their non-glycosylated counterparts, respectively. These effects are each ~ −1 kcal mol−1 greater than the glycosylation-induced stabilization of g-RnCD2*.
While the Glu61Lys and Leu63Phe mutations are individually destabilizing to RnCD2* (Table 1), triple mutant cycle analysis allows the energetic contributions of the stabilizing interactions between the N-glycan, Lys61, and Phe63 to be clearly quantified. The interaction energies of Lys61 (ΔΔGf [g-RnCD2*-K]–ΔΔGf [g-RnCD2*]) and Phe63 (ΔΔGf [g-RnCD2*-F]–ΔΔGf [g-RnCD2*]) with the N-glycan are comparable: − 0.9 ± 0.8 and −1.2 ± 0.7 kcal mol−1, respectively (Table 1). Glycosylated variants harboring Phe63 unfold 20 to 200× more slowly than their non-glycosylated counterparts (Table 1), suggesting that the Phe-glycan interaction in the type I β-bulge turn of RnCD2* stabilizes the native state of RnCD2* more than the transition state (maximal influence on folding rate is 2×). This result is consistent with reports that corresponding reverse turns in similar Ig domains are not highly structured in the transition state (21). Analogous native-state interactions between the N-glycan, Phe63, and Lys61 are also likely central to the unusually large stabilizing effect of N-glycosylation on HsCD2ad. Thr67 is probably also energetically important, but we did not test this hypothesis in RnCD2* because efficient cellular glycosylation requires Thr.
RnCD2* variants appear to be more efficiently glycosylated when Phe is present, as observed by Western blot of conditioned media from Sf9 baculovirus infections expressing g-RnCD2*-K and g-RnCD2*-F (Fig. 2C and Fig. S4) (18). This observation could have important implications for N-glycoprotein production, as discussed below.
The Phei-Xxx-Asn(glycan)-Gly-Thri+4 sequence within a type I β-bulge turn appears to be a stabilizing structural module, which we call an enhanced aromatic sequon. We next explored whether it could stabilize the native states of other proteins. We engineered the enhanced aromatic sequon into a reverse turn of human muscle acylphosphatase (AcyP2). AcyP2 is cytosolic and thus, glycosylation-naïve. Reverse turn residues 43 to 47 are not well enough defined in the published NMR structure of horse AcyP2 (95% identical to human, with 100% identity in residues 43 to 47) to discern their precise conformation (Fig. 2D shows one conformation of wild-type AcyP2 that is consistent with the NMR data) (22), but previously reported crystallographic data (23) show that homologous residues in human common type acylphosphatase (57% identical, also non-glycosylated) adopt a type I β-bulge turn. The i+4 position in AcyP2 is already Thr (Fig. 2D); thus, Thr43Phe (i) and Lys45Asn (i+2) mutations (Fig. 2D) install the enhanced aromatic sequon. Additional sequons present in wild-type AcyP2 were removed by Ser to Ala mutations to produce a modified version of AcyP2 (called AcyP2*) that is glycosylated only at Asn45 (18).
Four AcyP2* variants, differing at position 43 (Phe or Thr) and 45 (Asn or Asn(N-glycan)) were prepared (Table 1, see Table S1 for ESI-MS data). Their folding free energies were determined by fluorescence-monitored urea denaturation experiments (Fig. 2E and Fig. S5) (18). Glycoprotein g-AcyP2*-F is −2.0 ± 0.7 kcal mol−1 more stable than non-glycosylated AcyP2*-F (Table 1). In contrast, g-AcyP2* is not significantly stabilized by glycosylation (ΔΔGf [g-AcyP2*] = +0.5 ± 0.8 kcal mol−1). Thus, introducing the N-glycan into g-AcyP2*-F is −2.5 ± 1.1 kcal mol−1 more stabilizing than introducing the N-glycan into g-AcyP2* (Table 1). This interaction energy compares favorably with the corresponding interaction energy in RnCD2* (Table 1). We infer from these data that the stabilization is due to an interaction between Phe43 and the Asn45(glycan) (and perhaps Thr47), consistent with the positioning of these side chains that is predicted by the structure of non-glycosylated wild-type AcyP2.
Phe-containing enhanced aromatic sequons in RnCD2* and AcyP2* appear to be much more efficiently glycosylated by OST in baculovirus-infected Sf9 insect cells (Figs. 2C, F and Figs. S4, S6) than sequons lacking Phe (18). A published PDB survey (24) is consistent with this observation, as aromatic amino acids are overrepresented two positions before glycosylated Asn residues. A likely explanation for this observation is that the enhanced aromatic sequon is a better substrate for OST; if this feature of the enhanced aromatic sequon proves to be general, it would be useful for enhancing glycoprotein yields in research and pharmacologic applications. Even in the absence of enzymological data, it is tempting to speculate that OST evolved to favor sequences that are likely to stabilize proteins upon glycosylation.
The data in Fig. 1 and 2 suggest that the stability conferred by glycosylating the enhanced aromatic sequon depends on the type I β-bulge turn conformation, which potentially brings Phe, GlcNAc1, and Thr into contact, enabling carbohydrate-π interactions (25), burial of hydrophobic surface area (Fig. 1C), and, at least in the case of HsCD2ad, formation of three solvent-excluded hydrogen bonds (Fig. 1B) (16). In principle, other reverse turns could facilitate these stabilizing interactions by similarly positioning Phe, GlcNAc1, and Thr.
We tested this hypothesis using the 34-residue WW domain from human Pin 1 (Pin WW), a glycosylation-naïve protein with three anti-parallel β-strands connected by two loops. Previous structural data (26) on this non-glycosylated protein reveals that loop 1 contains a four-residue type II β-turn within a larger six-residue loop (Fig. 3A). The side chain β-carbons of Ser16 (i), Ser19 (i+3), and Arg21 (i+5) are within 5–6 Å of each other (26). Cβ-Cβ distances between i, i+2, and i+4 positions in the type I β-bulge turn of HsCD2ad (16) are similar, suggesting that introducing Phe, Asn(glycan), and Thr at positions 16, 19, and 21 in Pin WW by mutagenesis could enable a stabilizing tripartite interaction. In this reverse turn context, Phe is three residues before the glycosylated Asn.
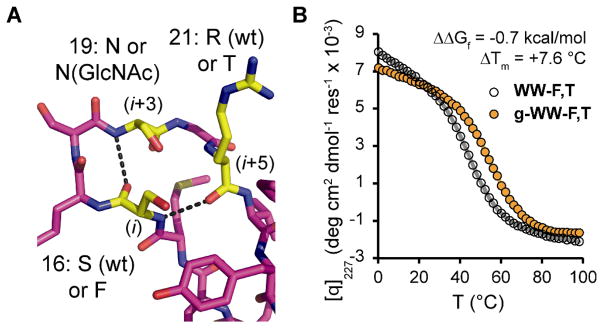
Figure 3.
(A) Stick representation of loop 1 in the known crystal structure of Pin WW (PDB: 1PIN) (26), rendered in Pymol. Overlaid text shows the amino acids to be perturbed at positions 16 (i), 19 (i+3), and 21 (i+5), which are highlighted in yellow. Dashed lines indicate hydrogen bonds in wild-type Pin WW. (B) Variable temperature circular dichroism data (monitored at 227 nm) for glycoprotein g-WW-F,T (orange circles) and non-glycosylated WW-F,T (open circles). Fits of the data to a two-state folding model (18) are shown in orange and gray lines, respectively.
We chemically synthesized Pin WW to examine the interactions of both the Phe and Thr side chains with the N-glycan (18). Glycosylated Pin WW variants contained a single Asn-linked GlcNAc (Asn(GlcNAc)) for synthetic simplicity; previous work (11, 15, 16) demonstrated that a single GlcNAc can still confer substantial stability. We prepared eight Pin WW variants for a triple mutant cycle (20), comprising all combinations of Ser16Phe, Asn19Asn(GlcNAc), and Arg21Thr mutations (Tables 1 and S2). The thermodynamic stability and folding and unfolding rate ratios (Table 1, see Table S3 for folding and unfolding rate constants) were determined by variable temperature circular dichroism spectroscopy (Fig. 3B, Figs. S7–S10) and laser temperature jump fluorescence experiments (Figs. S11–S18), respectively (18).
As was observed for RnCD2*, the Ser16Phe, Asn19Asn(GlcNAc), and Arg21Thr mutations are each individually destabilizing. However, triple mutant cycle analysis allows quantification of the energetic contributions of stabilizing interactions between Phe16, Asn19(GlcNAc), and Thr21. The N-glycan-dependent contribution of Phe16 to Pin WW stability at 55°C increases from −0.19 ± 0.09 kcal mol−1 (ΔΔGf [g-WW-F] – ΔΔGf [g-WW]) to −0.78 ± 0.11 kcal mol−1 (ΔΔGf [g-WW-F,T] – ΔΔGf [g-WW-T]) when Arg21 is changed to Thr (Table 1). Similarly, the N-glycan-dependent contribution of Thr21 at 55°C increases from −0.18 ± 0.10 kcal mol−1 (ΔΔGf [g-WW-T] – ΔΔGf [g- WW]) to −0.77 ± 0.10 kcal mol−1 (ΔΔGf [g-WW-F,T] – ΔΔGf [g-WW-F]) when Ser16 is changed to Phe. These data suggest that both Phe and Thr contribute to the stabilizing effect of glycosylating the enhanced aromatic sequon and that the tripartite interaction between Phe, GlcNAc, and Thr can be successfully applied in reverse turns other than type I β-bulge turns. Glycoprotein g-WW-F,T folds 2× faster and unfolds 1.4× slower than its non-glycosylated counterpart, presumably because the interaction between Phe, GlcNAc, and Thr stabilizes the transition state and native state conformations of loop 1 (this loop is structured in the folding transition state of WW domains). Thr has long been known to be essential for OST-mediated glycosylation (1, 27); these results provide evidence for its energetic importance.
The high frequency of glycosylation in the reverse turns of secreted proteins (24, 28) suggests that this portable stabilizing module should be readily incorporated into proteins at naïve sites, including those within protein drugs, which are often multiply glycosylated (2–4 glycosylation sites per glycoprotein) (29). It is important to note that N-glycosylation and the Phe mutation alone do not consistently improve protein stability as isolated modifications (Table 1) (18). Rather, it is the tripartite interaction between Phe, the N-glycan, and likely Thr in a reverse turn context that stabilizes the native states of RnCD2*, AcyP2* and Pin WW by −0.7 to −2.0 kcal mol−1. GlcNAc1 of the N-glycan interacts with the Phe and Thr side chains to form a stabilizing hydrophobic core that would likely not otherwise form. In addition to native state stabilization, the enhanced aromatic sequon notably increases glycosylation efficiency in RnCD2* and AcyP2*, which facilitates glycoprotein production and purification. Owing to the compact structure of the enhanced aromatic sequon (Fig. 1C) and the central location of the N-glycan (Fig. 1B), the one or two local mutations that install this portable stabilizing module into proteins should be shielded by the glycan, minimizing immunogenicity. These features and the additional beneficial pharmacokinetic attributes of N-glycosylation discussed above highlight the potential for using the enhanced aromatic sequon as a portable stabilizing module for research and therapeutic applications. (30)
Supplementary Material
References and Notes
- 1.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Molinari M. Nat Chem Biol. 2007;3:313. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 3.Helenius A, Aebi M. Science. 2001;291:2364. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 4.Glozman R, et al. J Cell Biol. 2009;184:847. doi: 10.1083/jcb.200808124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wormald MR, et al. Eur J Biochem. 1991;198:131. doi: 10.1111/j.1432-1033.1991.tb15995.x. [DOI] [PubMed] [Google Scholar]
- 6.Andreotti AH, Kahne D. J Am Chem Soc. 1993;115:3352. [Google Scholar]
- 7.Imperiali B, Rickert KW. Proc Natl Acad Sci USA. 1995;92:97. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Eufemi M, Turano C, Giartosio A. Biochemistry. 1996;35:7299. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi H, Uchida M. J Biochem. 1996;120:474. doi: 10.1093/oxfordjournals.jbchem.a021435. [DOI] [PubMed] [Google Scholar]
- 10.Kern G, Schülke N, Schmid FX, Jaenicke R. Protein Sci. 1992;1:120. doi: 10.1002/pro.5560010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson SR, et al. Proc Natl Acad Sci USA. 2009;106:3131. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh G, Jefferis R. Nat Biotechnol. 2006;24:1241. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 13.Hackenberger CPR, Friel CT, Radford SE, Imperiali B. J Am Chem Soc. 2005;127:12882. doi: 10.1021/ja051855k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott S, Chang D, Delorme E, Eris T, Lorenzini T. J Biol Chem. 2004;279:16854. doi: 10.1074/jbc.M311095200. [DOI] [PubMed] [Google Scholar]
- 15.Price JL, et al. J Am Chem Soc. 2010;132:15359. doi: 10.1021/ja106896t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyss DF, et al. Science. 1995;269:1273. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 17.Sibanda BL, Blundell TL, Thornton JM. J Mol Biol. 1989;206:759. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- 18.see supporting online material for details.
- 19.Jones EY, Davis SJ, Williams AF, Harlos K, Stuart DI. Nature. 1992;360:232. doi: 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- 20.Horovitz A, Fersht AR. J Mol Biol. 1992;224:733. doi: 10.1016/0022-2836(92)90557-z. [DOI] [PubMed] [Google Scholar]
- 21.Geierhaas CD, Paci E, Vendruscolo M, Clarke J. J Mol Biol. 2004;343:1111. doi: 10.1016/j.jmb.2004.08.100. [DOI] [PubMed] [Google Scholar]
- 22.Pastore A, Saudek V, Ramponi G, Williams RJP. J Mol Biol. 1992;224:427. doi: 10.1016/0022-2836(92)91005-a. [DOI] [PubMed] [Google Scholar]
- 23.Yeung RCY, Lam SY, Wong KB. Acta Crystallogr F. 2006;62:80. doi: 10.1107/S174430910504145X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrescu AJ, Milac AL, Petrescu S, Dwek RA, Wormald MR. Glycobiology. 2004;14:103. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 25.Laughrey ZR, Kiehna SE, Riemen AJ, Waters ML. J Am Chem Soc. 2008;130:14625. doi: 10.1021/ja803960x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranganathan R, Lu KP, Hunter T, Noel JP. Cell. 1997;89:875. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 27.Imperiali B, Shannon KL, Unno M, Rickert KW. J Am Chem Soc. 1992;114:7944. [Google Scholar]
- 28.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Cell. 2010;141:897. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Apweiler R, Hermjakob H, Sharon N. Biochim Biophys Acta. 1999;1473:4. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 30.We thank R. A. Dwek, I. A. Wilson, J. C. Paulson, S. Deechongkit, and D.L. Powers for presubmission reviews and for helpful prepublication discussions, and SeniorEditor P. D. Szuromi for a useful revision strategy. This work was supported in part by the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation and by NIH grant GM051105 to J.W.K. and E.T.P. J.L.P was supported in part by NIH post-doctoral fellowship F32 GM086039. A.D. and M.G. were supported by National Science Foundation grant MCB 1019958. S.R.H. and C.-H.W. were supported by NIH grant AI072155. The Scripps Research Institute has filed a provisional patent application covering the results described in this paper.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.