Abstract
Selenocysteine (Sec) residues occur in thiol oxidoreductase families, and functionally characterized selenoenzymes typically have a single Sec residue used directly for redox catalysis. However, how new Sec residues evolve and whether non-catalytic Sec residues exist in proteins is not known. Here, we computationally identified several genes with multiple Sec insertion sequence (SECIS) elements, one of which was a methionine-R-sulfoxide reductase (MsrB) homolog from Metridium senile that has four in-frame UGA codons and two nearly identical SECIS elements. One of the UGA codons corresponded to the conserved catalytic Sec or Cys in MsrBs, whereas the three other UGA codons evolved recently and had no homologs with Sec or Cys in these positions. Metabolic 75Se labeling showed that all four in-frame UGA codons supported Sec insertion and that both SECIS elements were functional and collaborated in Sec insertion at each UGA codon. Interestingly, recombinant M. senile MsrB bound iron, and further analyses suggested the possibility of binding an iron-sulfur cluster by the protein. These data show that Sec residues may appear transiently in genes containing SECIS elements and be adapted for non-catalytic functions.
Keywords: Antioxidant, Computation, Evolution, Methionine, Oxidation-Reduction, Selenium, SECIS Element, TGA Codon, Methionine Sulfoxide Reductase, Selenocysteine
Introduction
Selenium is an important biological trace element. It is co-translationally inserted into protein in the form of selenocysteine (Sec),2 the naturally occurring 21st amino acid, in response to an in-frame UGA codon (1–5). In eukaryotes, Sec insertion requires the presence of a stem loop structure, known as selenocysteine insertion sequence (SECIS) element, in the 3′-untranslated region (UTR) and several trans-acting factors, such as SBP2, EFsec, and other components (5–10).
Unique features of SECIS elements and occurrence of in-frame TGA codons have been used for in silico identification of Sec-containing proteins (selenoproteins). The SECIS element is composed of an apical loop, SECIS core, two helices, and an internal loop, forming a thermodynamically stable stem-loop structure. It contains conserved sequences including AA in the apical loop and AUGA at the base of one of the stems (8, 9, 11). By recruiting SBP2, AUGA is involved in the formation of the translational complex for Sec insertion at the UGA codon. This SECIS motif, together with an in-frame UGA, are key features in the design of an algorithm for identification of selenoprotein genes (8, 12, 13). By using this strategy, mammals were found to contain 24–25 selenoprotein genes (14).
Selenoproteomes (full sets of selenoproteins) have been characterized for a variety of organisms (15–23). Interestingly, almost all functionally characterized selenoproteins contain a single Sec and a single SECIS element. Prominent exceptions include SelP (a selenium transport protein in plasma of animals) and SelL that occurs in aquatic organisms (24). It was also reported that the single Sec in selenoproteins has a catalytic function and corresponds to the position of catalytic Cys in thiol oxidoreductases (5, 25–27). The two Sec in SelL were also suggested to participate in redox catalysis through a diselenide bond (24), and even several Sec residues in SelP correspond to Cys in other homologs, including one Sec that corresponds to Cys in the catalytic sites of thiol oxidoreductases. The role of Sec as a catalytic residue is supported by numerous experimental data wherein a change of Sec to Cys reduced enzyme activity 100–1000-fold in glycine reductase (28), methionine sulfoxide reductase B1 (MsrB1) (29), glutathione peroxidase (30, 31), type I deiodinase (32), and several other selenoproteins. Therefore, it is now widely accepted that new Sec residues primarily evolve to substitute Cys residues to carry out redox catalysis. However, recently evolved Sec residues have not been described. The availability of sequence information for hundreds of genomes provides an opportunity to identify transient Sec residues.
MsrB is a thiol oxidoreductase that reduces methionine-R-sulfoxide (Met-R-SO) to Met, and a Sec-containing form of this protein, MsrB1 (described previously as SelR or SelX), has also been described (18, 33, 34). MsrBs are present in most organisms, but their Sec-containing forms were only found in eukaryotes. The change of Sec to Cys led to a decrease in catalytic efficiency by ∼500-fold. This makes MsrB a typical thiol oxidoreductase that has both Sec and Cys forms (29).
In the present study, we identified an MsrB from Metridium senile that contains four in-frame TGA codons and two SECIS elements in the 3′-UTR. We investigated the role of these multiple Sec residues and SECIS elements and report a non-canonical use of Sec in this protein. This study also suggests a model for selenocysteine evolution.
EXPERIMENTAL PROCEDURES
Searches for EST Sequences with Dual SECIS Elements
The EST database “est_others” was downloaded from the NCBI; it contained 51,484,119 sequences (29,098,406,053 bp). It was initially scanned with PatScan to detect SECIS-like structures. Because some SECIS elements (such as human SelM and SelO structures) lack the unpaired AA in the apical loop, the “default” pattern used in this search was modified to include such cases. Only EST sequences with two or more candidate SECIS elements were further analyzed. This subset was then examined with standalone SECISearch (14) using the pattern described above. In the final step, candidate sequences were analyzed using a covariance model, and all candidates with a score lower than 15 were dismissed. The remaining sequences were clustered using UCLUST 2.0.591 and then manually analyzed.
M. senile MsrB Constructs
M. senile MsrB nucleotide sequence, including the 3′-UTR, was synthesized and inserted into the pEGFP-C3 vector at HindIII/BamHI sites. The construct was subjected to mutagenesis using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The following primers were used to change TGA to TGC: 264 TGA to TGC, 5′-GCTTGGTTATAAATGCCAAAACGGATGACCTG-3′; 276 TGA to TGC, 5′-CAAAACGGATGCCCTGCAGACAACAAGTTC-3′; 297 TGA to TGC, 5′-CAGACAACAAGTTCTGCTTTTGAATCTTCTCTGC-3′; 303 TGA to TGC, 5′-CAAGTTCTGATTTTGCATCTTCTCTGCAGCAC-3′; 297 and 303 TGA to TGC, 5′-CAAGTTCTGCTTTTGCATCTTCTCTGCAGCAC-3′. These primers were used to change TG to AC in the SECIS element core: first SECIS element TG to AC, 5′-CTCTTTTAAGTAGTTTAACATGTAATCCATCTCTATCC-3′; second SECIS element TG to AC, 5′-GGAAGCAGTTTAACATGTAATCCATCACTATCC-3′. The following primers were used to change TGC to TCC: 264 TGC to TCC, 5′-GCTTGGTTATAAATCCCAAAACGGATGCCC-3′; 264 and 276 TGC to TCC, 5′-GCTTGGTTATAAATCCCAAAACGGATCCCC-3′; 297 TGC to TCC, 5′-GCAGACAACAAGTTCTCCTTTTGCATCTTCTC-3′; 303 TGC to TCC, 5′-CAAGTTCTGCTTTTCCATCTTCTCTGCAGC-3′; 297 and 303 TGC to TCC, 5′-CAAGTTCTCCTTTTGAATCTTCTCTGCAGC-3′.
Transfection of Recombinant Constructs and Metabolic Labeling of Cells with 75Se
HEK 293 cells were cultured in DMEM (Invitrogen) containing fetal bovine serum and an antibiotic-antimycotic mixture (Invitrogen) of 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B in 6-well plates maintained at 37 °C in the atmosphere of 5% CO2. At 60% confluency, these cells were transfected using LipofectamineTM 2000 reagent (Invitrogen) according to the manufacturer's protocol. After 24 h, 0.01 mCi of freshly neutralized [75Se]selenious acid (specific activity 1,000 Ci/mmol, University of Missouri Research Reactor, Columbia, MO) was added in each well, and cells were incubated at 37 °C for 24 h followed by harvesting the cells. Cell extracts (50–80 μg of total protein) were electrophoresed on 10% Bis-Tris gels and transferred onto PVDF membranes (Invitrogen). The 75Se radioactivity pattern was visualized by using a PhosphorImager (GE Healthcare).
Cloning, Expression, and Purification
M. senile MsrB forms including either TGC for Cys or TCC for Ser in place of the TGA codon were transferred to the pET28a vector (Novagen, Madison, WI) without SECIS elements, and then these recombinant vectors were transformed into Escherichia coli BL21 (DE3) cells for protein expression. Also, recombinant MsrB1-Cys in the pET21b vector was transformed into E. coli BL21 cells (29). These cells were grown in LB medium containing 100 μg/ml ampicillin until A600 reached 0.5, and then isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm. Induction of protein synthesis proceeded at 37 °C for 4 h, and cells were then harvested by centrifugation at 4,000 rpm. Cell pellets were washed with PBS and recentrifuged. Harvested cells were stored at −70 °C until use.
To purify recombinant proteins, cell pellets were mixed with the resuspension buffer (50 mm Tris-HCl, pH 7.5, 15 mm imidazole, 300 mm NaCl), and protease inhibitor mixture (Roche Applied Science, Basel, Switzerland) was added to this mixture. After sonication, the supernatant was collected by centrifugation at 8,000 rpm for 30 min and loaded onto a Cobalt Talon resin (Clontech) pre-equilibrated with the resuspension buffer. Following washing with the same buffer, proteins were eluted with elution buffer (50 mm Tris-HCl, pH 7.5, 300 mm imidazole, 300 mm NaCl). The eluted fractions were analyzed by SDS-PAGE, the fractions containing MsrBs were pooled, and then their buffer was exchanged to PBS by using Amicon Ultra 3K (Millipore, Billerica, MA).
MsrB Activity Assays and Metal Analysis by ICP-MS
MsrB activity was assayed in cell lysates (400 μg) and with purified proteins (50–100 μg). Briefly, the reaction (100 μl) was carried out at 37 °C for 30 min in the presence of DTT (20 mm), and 200 μm dabsyl-methionine-R-sulfoxide was added to the reaction mixture. After the reaction was stopped by adding 200 μl of acetonitrile, it was centrifuged at 4 °C for 15 min at 13,000 rpm, and then supernatant (50 μl) was injected onto a C18 column (ZORBAX Eclipse XDB-C18, Agilent Technologies, Lexington, MA) to quantify dabsyl-methionine in an HPLC procedure.
To analyze for metal content of M. senile MsrB and mouse MsrB1-Cys, elemental analysis was performed using inductively coupled plasma mass spectroscopy (ICP-MS). The samples were digested with 2% v/v of HNO3 and 0.5 m H2O2 with 50 ppb of gallium added as an internal standard. The analysis was performed using an Agilent ICP-MS model 7500ce using 3.5 ml/min of H2 and 1.5 ml/min of helium in the reaction/collision cell. Purified M. senile MsrB forms, which alternatively contained Cys or Ser in place of Sec, were also subjected to spectrophotometric analysis using an HP 845x spectrophotometer (Hewlett-Packard, Palo Alto, CA).
Complementation Assay
Each Cys mutant of M. senile MsrB and yeast MsrB was cloned into the yeast high copy p423GPD vector and overexpressed in triple MSR (MsrAΔ/MsrBΔ/fRMsrΔ) mutant cells. Wild-type cells transformed with an empty vector served as control. Logarithmically growing wild-type and triple mutant cells, cultured in YNB medium, were harvested and washed with distilled water. The cultures were diluted to an A600 of 0.25 and spotted on appropriate YNB plates missing methionine but containing a racemic mixture of methionine sulfoxide (20 mg/liter). Plates were then incubated at 30 °C for 4 days and photographed.
RESULTS
Multiple In-frame TGA Codons and SECIS Elements in M. senile MsrB
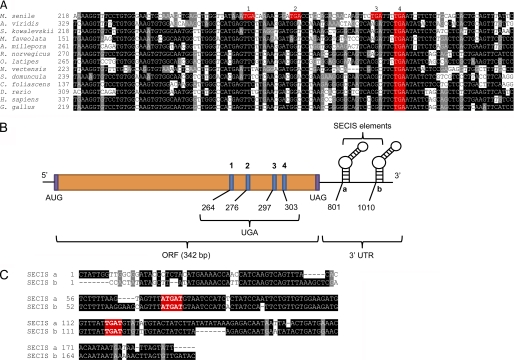
By searching for EST sequences with multiple SECIS elements (Table 1), we identified an MsrB homolog from M. senile (brown sea anemone) that had four in-frame TGA codons and two nearly identical SECIS elements in the 3′-UTR (Fig. 1). This is only the second selenoprotein identified (besides SelP) to have more than one Sec and more than one SECIS element. In addition, two Sec residues that form a catalytic diselenide bond were reported for SelL, a selenoprotein present in aquatic organisms (24). Also, certain selenoprotein genes were found to have two SECIS elements, e.g. SelW1 in Ciona intestinalis (35, 36), but only its second SECIS element was compatible with the strict pattern when analyzed by SECISearch (35). Thus, four in-frame TGA codons and two SECIS elements in M. senile MsrB represent a unique case, and studying the role of multiple Sec and SECIS elements in this protein may provide insights into selenoprotein evolution.
TABLE 1.
Search for EST sequences containing more than one SECIS element
| Number of hits | |
|---|---|
| PatScan search for SECIS-like structures | 8,723,411 |
| Sequences containing two SECIS elements | 800,484 |
| SECISearch hits | 282,870 |
| SECISearch with two SECIS elements | 45,288 |
| 90% identity clusters | 22,591 |
| 50% identity clusters | 13,977 |
| Cove score >15 | 2,282 |
| Final list of candidates | 535 |
FIGURE 1.
Multiple in-frame TGA codons and two nearly identical SECIS elements in M. senile MsrB. A, multiple nucleotide sequence alignment of MsrBs. Conserved nucleotides are highlighted and in-frame TGA codons are colored in red. Accession numbers are: M. senile (FC834691.1), Amaranthus viridis (FK755697.1), Saccoglossus kowalevskii (FF491760.1), Montastraea faveolata (GW275275.1), Acropora millepora (GH999895.1), Rattus norvegicus (FM079831.1), Oryzias latipes (DK011817.1), Nematostella vectensis (DV084978.1), Suberites domuncula (GH567716.1), Carteriospongia foliascens (GO082776.1), Danio rerio (NM_178288.4), Homo sapiens (NM_016332.2), and Gallus gallus (NM_001135558.1). B, mRNA organization of M. senile MsrB. This transcript contains four in-frame TGA codons and two nearly identical SECIS elements. C, alignment of two SECIS elements (FC826394.1 and FC834690.1) of M. senile MsrB. Conserved nucleotides are highlighted, and those essential for SECIS function are colored in red.
Evolution of M. senile MsrB
Sec often evolves to replace catalytic Cys, and thus, Cys homologs of nearly all selenoproteins are known. M. senile MsrB possesses one TGA codon that codes for the conserved catalytic Sec (Fig. 1, A and B, Sec residue 4) but also three non-conserved TGA codons (Fig. 1, A and B, Sec residues 1–3). Analysis of MsrB sequences revealed that the first three TGA codons can be obtained by single nucleotide replacements (or two nucleotides in one case) from other MsrBs (Fig. 1A). Moreover, these TGA codons are only found in M. senile MsrB, i.e. they are absent in all other MsrB sequences available in NCBI databases (supplemental Figs. S1 and S2). These observations suggest that M. senile MsrB only recently evolved these three in-frame TGA codons.
4 Selenocysteines and Low Conservation near the Catalytic Site in M. senile MsrB
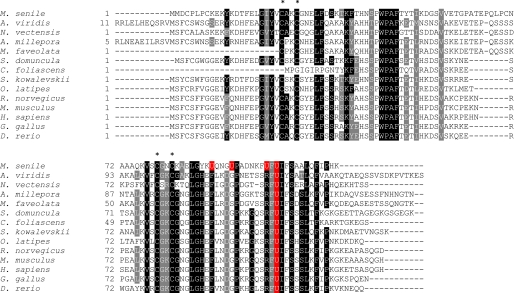
We aligned Sec-containing MsrBs (Fig. 2) and their Cys-containing forms (supplemental Fig. S1). MsrBs have two conserved CXXC motifs (Fig. 2) for zinc binding and a single catalytic Sec (or Cys). Zinc serves a structural role, and this function is preserved in M. senile MsrB. However, this protein has additional changes in conserved amino acids near the catalytic site. For example, Sec-containing MsrBs have well conserved Leu-84, Gly-85, His-86, Glu-87, Phe-88, Asp-91, Gly-92, Pro-93, Ser-98, Arg-99, Phe-100, Sec-101, Ile-102, and Ser-104 residues (Fig. 2). In particular, His-86 and Arg-99 are essential for MsrB activity. However, in M. senile MsrB, His-86 is replaced with Tyr, Glu-87 is replaced with Lys, Asp-91 is replaced with Gly, and Ser-98 is replaced with Phe (Fig. 2). Interestingly, no Cys homolog was found for any of the three newly evolved Sec. The presence of four Sec residues in the M. senile protein indicates a non-catalytic function for at least some of them.
FIGURE 2.
Multiple sequence alignment of MsrB proteins. MsrB homologs were collected from protein and EST databases by using BLASTp and tBLASTn. Conserved amino acids are highlighted. Four conserved Cys in two CXXC motifs are marked with an asterisk. Sec residues are highlighted in red. Accession numbers are: M. senile (FC834691.1), A. viridis (FK755697.1), S. kowalevskii (FF491760.1), M. faveolata (GW275275.1), A. millepora (GH999895.1), R. norvegicus (FM079831.1), O. latipes (DK011817.1), N. vectensis (DV084978.1), S. domuncula (GH567716.1), C. foliascens (GO082776.1), D. rerio (AAH67380.1), H. sapiens (NP_057416.1), G. gallus (NP_001129030.1), and Mus musculus (NP_038787.1).
Characterization of Four Sec in M. senile MsrB and the Role of SECIS Duplication
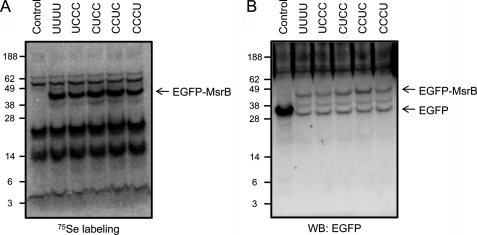
Metabolic labeling of cells with 75Se was used to examine the roles of in-frame TGA codons in inserting Sec residues (Fig. 3). Three of the four TGA codons were changed to TGC codons (to insert Cys in place of Sec), and a single remaining TGA codon was left unchanged. This approach was individually applied to all four TGA codons, the resulting constructs were transfected into HEK 293 cells, and cells were metabolically labeled with 75Se. We found that Sec was inserted at each TGA position, indicating that each TGA codes for Sec (Fig. 3).
FIGURE 3.
Insertion of four Sec residues into M. senile MsrB. A, HEK 293 cells expressing EGFP alone (Control), wild-type EGFP-MsrB with four Sec (UUUU), or single Sec-containing EGF-MsrBs (UCCC, CUCC, CCUC, or CCCU) were labeled with 75Se and analyzed by SDS-PAGE and PhosphorImager detection of 75Se. EGFP-MsrBs (wild-type and Sec forms) were detected at ∼43 kDa. B, expression of EGFP-MsrB confirmed by Western blotting (WB) using anti-GFP antibodies.
As mentioned above, M. senile MsrB has two nearly identical SECIS elements in the 3′-UTR (Fig. 1C), suggesting that they recently evolved by duplication. On the other hand, in Sepp1, two SECIS elements are of different types, e.g. they show no homology. To investigate the functions of duplicated SECIS elements, we mutated conserved nucleotides in the SECIS core to disrupt one or both SECIS elements and examined Sec insertion by these structures by metabolic 75Se labeling (Fig. 4). Single SECIS mutants still supported Sec insertion regardless of the position of the TGA codon, whereas mutation of both SECIS elements completely disrupted Sec insertion. Thus, both SECIS elements in M. senile MsrB were functional and supported Sec insertion at all TGA positions. Interestingly, the data suggested that the two SECIS elements collaborated for Sec insertion. For example, expression of MsrB from the construct with either SECIS element mutated was less efficient than MsrB expression when both SECIS elements were intact (Fig. 4B). Therefore, we conclude that SECIS duplication led to an increase in Sec insertion efficiency in M. senile MsrB. This appears to be the first finding of SECIS elements collaborating in Sec insertion (SECIS elements in SelP show position-specific Sec insertion).
FIGURE 4.
Function of two SECIS elements in M. senile MsrB. A, constructs used in Fig. 3 were mutated wherein conserved TG nucleotides were changed to AC separately in each SECIS element (i.e. SECIS a and SECIS b). Expression of selenoproteins was analyzed by 75Se labeling and Western blotting (WB) with anti-GFP antibodies. Mu, mutant. B, quantification of relative Sec insertion into each Sec site in M. senile MsrB based on 75Se labeling experiments.
Selenoproteins with Dual SECIS Elements
By searching EST databases for sequences with more than one SECIS element, we found that the majority of identified sequences corresponded either to SelP (the only previously known selenoprotein that requires two SECIS elements for incorporation of multiple Sec residues) or to “orphan” sequences (i.e. there was insufficient sequencing data to identify ORFs). We also detected C. intestinalis SelW, which was recently reported to contain two SECIS elements (35). Of interest was also the observation of three unique EST sequences/clusters (GenBankTM accession numbers FC826394.1+FC834690.1; FC842550.1; and FC850510.1 + FC849224.1) in M. senile (supplemental Fig. S3). One of them corresponded to MsrB, whereas the other two were orphan sequences. It appears that M. senile is enriched for duplicated SECIS elements, but further studies are needed to examine this possibility.
Curiously, we identified a C. intestinalis sequence (GenBank accession number FK073247.1) that was predicted to have five SECIS-like elements. Although four of them satisfied all search criteria, most likely they do not represent real SECIS elements because they belong to the sequence that is homologous to a heavy neurofilament protein, and no suitable upstream ORF with in-frame UGA codons could be found. This case highlights the importance of multiple criteria used in selenoprotein identification (37).
Other selenoprotein genes with dual SECIS elements identified in our search (Table 1) included SelP, MsrB, SelW, glutathione peroxidase, deiodinase, Sep15, and membrane selenoprotein (MSP) selenoproteins. In most cases, both SECIS elements were highly similar, suggesting that duplication events took place. However, unlike M. senile MsrB, no newly evolved Sec residues were detected in these proteins. It is possible that these dual SECIS elements increase efficiency of Sec insertion in just one position.
Based on the observations described so far, we propose the following evolutionary model. Initially, the anemone M. senile (or its ancestor) possessed a regular, single-Sec MsrB. Then, at some point, an additional in-frame UGA codon appeared. Because all components of the Sec insertion machinery were readily available, this mutation did not result in early termination of translation. Instead, an additional Sec was inserted. This process was repeated with two additional Sec residues. Subsequently, SECIS duplication occurred to support more efficient incorporation of multiple Sec residues. The extremely high similarity between two SECIS elements indicates that this event took place recently.
Thus, we identified a gene with multiple in-frame UGA codons that likely occurred as a result of random mutations. In the absence of a SECIS element in the 3′-UTR, these mutations would have resulted in early termination, counterselecting this sequence. However, availability of the existing SECIS element and Sec insertion machinery allowed M. senile to overcome this problem, generating a functional protein with four Sec residues. Perhaps the presence of multiple Sec sites required more efficient Sec insertion, and to increase its efficiency, a segment of the 3′-UTR, including the SECIS element, was duplicated.
M. senile MsrB Does Not Have MsrB Activity
M. senile MsrB contains a conserved catalytic Sec, whereas its sequence near the active site is not fully conserved. To examine for MsrB activity of this protein, we assayed its ability to reduce dabsylated Met-R-SO in the presence of DTT. We examined both cells expressing M. senile MsrB and purified Cys-containing M. senile MsrB (in which all Sec residues were replaced with Cys) (Fig. 5, A and B). Cell lysates of HEK 293 cells expressing MsrB with four Sec residues (UUUU) or a single Sec wherein other Sec positions were occupied by Cys or Ser (UCCC, CUCC, CCUC, CCCU, or SSSU) showed no MsrB activity (Fig. 5A). To further characterize MsrB activity of the M. senile protein, we prepared recombinant MsrB forms in which all Sec residues were replaced with Cys. It is known that Cys mutants of selenoproteins show a significant decrease in catalytic activity, but the activity is still detectable. We used mouse MsrB1-Cys (in which Sec was replaced with Cys) as a positive control. Again, no activity was detected in M. senile MsrB preparations, whereas MsrB1-Cys showed activity (∼4200 pmol/min/mg). We also carried out in vivo complementation assays of yeast cells lacking all three Msrs and supplemented with Met-R-SO in place of Met. The yeast strain lacking MsrA, MsrB, and fRMsr was transformed with the construct coding for Cys-containing M. senile MsrB (CCCC) or its other mutant forms (CCSS, SSSS, SSSC, or CCCS) and then tested for growth on Met-R-SO-supplemented, Met-deficient medium. Except for controls, all tested yeast strains failed to grow. Overall, these analyses supported the idea that M. senile MsrB does not have a typical MsrB activity.
FIGURE 5.
Lack of reduction of Met-R-SO by M. senile MsrB. A, lysates of HEK 293 cells expressing control EGFP, wild-type EGFP-MsrB with four Sec (UUUU), or mutant EGFP-MsrBs with single Sec (UCCC, CUCC, CCUC, CCCU, or SSSU) were subjected to activity assays, and specific activities are shown. B, purified Cys-containing MsrB forms in which Cys (C) or Ser (S) replace the four Sec residues (CCCC, SSSS, SSSC, or CCCS forms) and a Cys mutant of mouse MsrB1 (MsrB1-Cys) were subjected to activity assays. Error bars in panels A and B represent S.D. C, triple mutant yeast strain (MsrAΔ/MsrBΔ/fRMsrΔ) was transformed with a control vector or plasmids coding for Cys-containing MsrB forms (CCSS, CCCC, SSSS, SSSC, or CCCS) and subjected to complementation assay by replacing methionine with free Met-R-SO in the medium.
Iron-Sulfur Cluster in M. senile MsrB
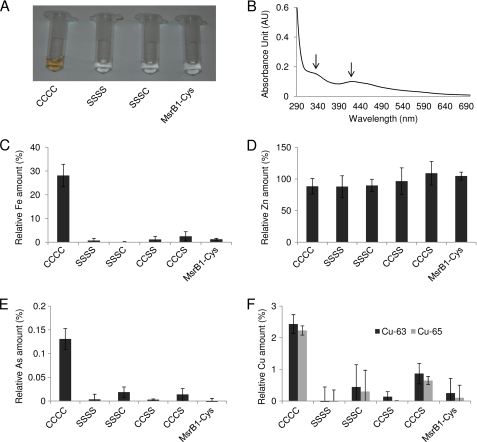
The finding that M. senile MsrB was not active in reducing Met-R-SO offered a challenge to identify the role of the four Sec residues. A clue was provided by an MsrB form containing four Cys residues in place of four Sec. This recombinant protein had brownish color (Fig. 6A) and showed absorbance peaks at 338 and 412 nm (Fig. 6B). These properties were compatible with the occurrence of a cofactor, most likely an iron-sulfur cluster. To test for the presence of iron in this protein, recombinant M. senile MsrB (the CCCC form), its mutants (SSSC, SSSS, CCSS, and CCCS), and mouse MsrB1-Cys were subjected to metal analysis using ICP-MS. Indeed, 0.3 equivalents of iron were found in M. senile MsrB (CCCC), whereas iron was not detected or was present at very low levels in other mutant forms (SSSC, SSSS, CCSS, and CCCS) and in MsrB1-Cys (Fig. 6C). In addition, M. senile MsrB and its mutants (as well as MsrB1-Cys) had approximately one equivalent of zinc (Fig. 6D), indicating preservation of this structural metal ion. We also observed substoichiometric levels of arsenic and copper (Fig. 6, E and F), which were dependent on the same four Cys. Overall, the biochemical data suggested the presence of an iron-sulfur cluster that was coordinated by four Cys residues in the M. senile mutant MsrB.
FIGURE 6.
Metal binding by M. senile MsrB. A, purified Cys-containing MsrB forms (CCCC, SSSS, and SSSC) and mouse MsrB1-Cys. B, spectrum of a purified Cys-containing MsrB form (CCCC). AU, absorbance units. C–F, purified Cys-containing MsrBs (CCCC, SSSS, SSSC, CCSS, and CCCS forms) and mouse MsrB1-Cys were subjected to metal analysis by ICP-MS. C, iron (Fe). D, zinc (Zn). E, arsenic (As). F, copper (Cu). (Data are shown for two isotopes, Cu-63 and Cu-65.) Error bars in panels B–F represent S.D.
DISCUSSION
Cys residues are often engaged in metal binding and disulfide bonds. The presence of selenium in place of sulfur would make these residues even more reactive because Sec has a higher polarizability and lower pKa (∼5.2) than Cys (∼8.3) and therefore could readily react with incoming substrate or other proximal residues. On the other hand, Cys often requires assistance from its microenvironment for deprotonation and nucleophilic attack. Thus, the use of Sec in place of Cys could greatly enhance catalytic efficiency of a thiol oxidoreductase and explain the need for Sec as the 21st proteinogenic amino acid.
M. senile MsrB has one conserved Sec, which is located in place of the catalytic Sec (or Cys) in other MsrBs, but it also has three other Sec that could not be all catalytic as they are located in the same domain and in positions that correspond to neither Sec nor Cys in homologs. Moreover, activity assays and growth complementation data revealed no activity of M. senile MsrB in reducing free Met-R-SO. These data show that some, if not all, Sec residues in this protein function as non-catalytic residues. Consistent with this idea, we found that Cys residues in the recombinant MsrB (which replaced Sec in the wild-type protein) bound 0.3 equivalents of iron, suggesting a possibility that they coordinated an iron-sulfur cluster. Selenium is positioned below sulfur in the periodic table of elements, and so its chemical properties are similar to those of sulfur (38). Therefore, the use of Sec to bind a metal is not unexpected from a physico-chemical point of view. In support of this possibility, Sec-metal complexes were reported in several proteins, e.g. E. coli (39) and Desulfovibrio gigas (40) formate dehydrogenases. Moreover, genetically engineered azurin was shown to possess an Se-Cu bond (41). Molybdenum, tungsten, copper, and nickel are known to be coordinated by Sec residues, and it is also feasible that other transition metals (including iron) could be coordinated by Sec.
Sec often evolves in place of Cys to serve as a catalytic redox residue. One exception has been SelP. It has multiple Sec residues, at least some of which are not used for catalysis. SelP is a selenium transport protein that delivers this trace element from liver to other organs by incorporating it into multiple Sec residues in its C-terminal region. Other functionally characterized selenoproteins contain a single Sec. The idea of interchangeability of catalytic redox Sec and Cys has been used for developing a strategy for identifying catalytic redox Cys residues in proteins (27).
In this regard, our analysis of M. senile MsrB revealed an unusual Sec occurrence. Three Sec residues in this protein were not found in any other MsrB sequences. Moreover, even Cys could not be found in these sites in other homologs. Together with the observation that the three non-conserved Sec could originate by simple nucleotide substitutions, this finding suggested that these Sec residues evolved recently. Previous analyses of Sec evolution focused on the incidence of Sec in catalytic sites, but the identification of three non-conserved Sec suggests that additional Sec may evolve if a selenoprotein sequence already has SECIS element(s). The three Sec could have disappeared immediately; instead, in M. senile MsrB, they appear to have acquired a new function, i.e. metal binding.
The lack of other non-conserved Sec residues in examined protein sequences suggests that new Sec residues appear and rapidly disappear from the sequence universe, probably because Sec decreases the rate of protein synthesis and it is too reactive a residue to be maintained in a protein unless it is functional. In this regard, M. senile MsrB offers a useful case because it represents a rare example when newly evolved Sec residues were preserved. It is likely that further searches will yield additional examples of newly evolved Sec residues in proteins.
M. senile MsrB contains two nearly identical SECIS elements in the 3′-UTR, which is also an extremely rare situation in selenoprotein genes. Except for SelP, very few sequences are known to possess two SECIS elements. Both SECIS structures in M. senile MsrB are functional, and Sec insertion efficiency is maximal when both are present. Their nearly identical sequences strongly suggest that they evolved recently by duplication, further supporting the idea of recent evolution of M. senile MsrB. Interestingly, our search for multiple SECIS elements in ESTs identified 535 candidates. Although further characterization is required to confirm the function of these SECIS elements, this finding showed that multiple SECIS elements do occur in genes.
Overall, our study identified the first example of recently evolved Sec residues. These residues are non-catalytic as: (i) they replace non-conserved residues; (ii) they are present in a thiol oxidoreductase domain, which in all known cases has a single Cys or Sec acting as the catalytic residue; and finally (iii) the enzyme has four Sec, which cannot all be catalytic. Some of these Sec residues must possess a different function or, less likely, they may be non-functional. Regardless of these possibilities, we identified a new situation with respect to Sec distribution and function in proteins.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AG021518 and GM061603 (to V. N. G.) and by the Intramural Research Program at the Center for Cancer Research, NCI, National Institutes of Health (to D. L. H.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Sec
- selenocysteine(s)
- SECIS
- selenocysteine insertion sequence
- SelP
- selenoprotein P
- Met-R-SO
- methionine-R-sulfoxide
- MsrB
- methionine-R-sulfoxide reductase
- EST
- expressed sequence tag
- ICP-MS
- inductively coupled plasma mass spectroscopy
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Cone J. E., Del Río R. M., Davis J. N., Stadtman T. C. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 2659–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zinoni F., Birkmann A., Stadtman T. C., Böck A. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 4650–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stadtman T. C., Davis J. N., Ching W. M., Zinoni F., Böck A. (1991) Biofactors 3, 21–27 [PubMed] [Google Scholar]
- 4. Hatfield D. L., Gladyshev V. N. (2002) Mol. Cell Biol. 22, 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stadtman T. C. (1996) Annu. Rev. Biochem. 65, 83–100 [DOI] [PubMed] [Google Scholar]
- 6. Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., Harney J. W., Larsen P. R. (1991) Nature 353, 273–276 [DOI] [PubMed] [Google Scholar]
- 7. Driscoll D. M., Copeland P. R. (2003) Annu. Rev. Nutr. 23, 17–40 [DOI] [PubMed] [Google Scholar]
- 8. Hubert N., Walczak R., Carbon P., Krol A. (1996) Nucleic Acids Res. 24, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee B. J., Worland P. J., Davis J. N., Stadtman T. C., Hatfield D. L. (1989) J. Biol. Chem. 264, 9724–9727 [PubMed] [Google Scholar]
- 10. Hoffmann P. R., Berry M. J. (2005) Thyroid 15, 769–775 [DOI] [PubMed] [Google Scholar]
- 11. Hubert N., Walczak R., Sturchler C., Myslinski E., Schuster C., Westhof E., Carbon P., Krol A. (1996) Biochimie 78, 590–596 [DOI] [PubMed] [Google Scholar]
- 12. Fletcher J. E., Copeland P. R., Driscoll D. M., Krol A. (2001) RNA 7, 1442–1453 [PMC free article] [PubMed] [Google Scholar]
- 13. Kryukov G. V., Kryukov V. M., Gladyshev V. N. (1999) J. Biol. Chem. 274, 33888–33897 [DOI] [PubMed] [Google Scholar]
- 14. Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigó R., Gladyshev V. N. (2003) Science 300, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 15. Lobanov A. V., Hatfield D. L., Gladyshev V. N. (2009) Biochim. Biophys. Acta. 1790, 1424–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conrad M., Schweizer U. (2010) Antioxid. Redox Signal. 12, 851–865 [DOI] [PubMed] [Google Scholar]
- 17. Dikiy A., Novoselov S. V., Fomenko D. E., Sengupta A., Carlson B. A., Cerny R. L., Ginalski K., Grishin N. V., Hatfield D. L., Gladyshev V. N. (2007) Biochemistry 46, 6871–6882 [DOI] [PubMed] [Google Scholar]
- 18. Fomenko D. E., Novoselov S. V., Natarajan S. K., Lee B. C., Koc A., Carlson B. A., Lee T. H., Kim H. Y., Hatfield D. L., Gladyshev V. N. (2009) J. Biol. Chem. 284, 5986–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lobanov A. V., Delgado C., Rahlfs S., Novoselov S. V., Kryukov G. V., Gromer S., Hatfield D. L., Becker K., Gladyshev V. N. (2006) Nucleic Acids Res. 34, 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lobanov A. V., Fomenko D. E., Zhang Y., Sengupta A., Hatfield D. L., Gladyshev V. N. (2007) Genome Biol. 8, R198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lobanov A. V., Gromer S., Salinas G., Gladyshev V. N. (2006) Nucleic Acids Res. 34, 4012–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taskov K., Chapple C., Kryukov G. V., Castellano S., Lobanov A. V., Korotkov K. V., Guigó R., Gladyshev V. N. (2005) Nucleic Acids Res. 33, 2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Fomenko D. E., Gladyshev V. N. (2005) Genome Biol. 6, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shchedrina V. A., Novoselov S. V., Malinouski M. Y., Gladyshev V. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13919–13924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johansson L., Gafvelin G., Arnér E. S. (2005) Biochim. Biophys. Acta. 1726, 1–13 [DOI] [PubMed] [Google Scholar]
- 26. Lee B. C., Dikiy A., Kim H. Y., Gladyshev V. N. (2009) Biochim. Biophys. Acta. 1790, 1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fomenko D. E., Xing W., Adair B. M., Thomas D. J., Gladyshev V. N. (2007) Science 315, 387–389 [DOI] [PubMed] [Google Scholar]
- 28. Garcia G. E., Stadtman T. C. (1992) J. Bacteriol. 174, 7080–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim H. Y., Gladyshev V. N. (2005) PLoS Biol. 3, e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rocher C., Lalanne J. L., Chaudière J. (1992) Eur. J. Biochem. 205, 955–960 [DOI] [PubMed] [Google Scholar]
- 31. Ursini F., Maiorino M., Brigelius-Flohé R., Aumann K. D., Roveri A., Schomburg D., Flohé L. (1995) Methods Enzymol. 252, 38–53 [DOI] [PubMed] [Google Scholar]
- 32. St Germain D. L., Schwartzman R. A., Croteau W., Kanamori A., Wang Z., Brown D. D., Galton V. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7767–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim H. Y., Gladyshev V. N. (2004) Mol. Biol. Cell. 15, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kryukov G. V., Kumar R. A., Koc A., Sun Z., Gladyshev V. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang L., Liu Q., Ni J. (2010) BMC Genomics 11, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumaraswamy E., Malykh A., Korotkov K. V., Kozyavkin S., Hu Y., Kwon S. Y., Moustafa M. E., Carlson B. A., Berry M. J., Lee B. J., Hatfield D. L., Diamond A. M., Gladyshev V. N. (2000) J. Biol. Chem. 275, 35540–35547 [DOI] [PubMed] [Google Scholar]
- 37. Kryukov G. V., Gladyshev V. N. (2002) Methods Enzymol. 347, 84–100 [DOI] [PubMed] [Google Scholar]
- 38. Wessjohann L. A., Schneider A., Abbas M., Brandt W. (2007) Biol. Chem. 388, 997–1006 [DOI] [PubMed] [Google Scholar]
- 39. Gladyshev V. N., Khangulov S. V., Axley M. J., Stadtman T. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7708–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Almendra M. J., Brondino C. D., Gavel O., Pereira A. S., Tavares P., Bursakov S., Duarte R., Caldeira J., Moura J. J., Moura I. (1999) Biochemistry 38, 16366–16372 [DOI] [PubMed] [Google Scholar]
- 41. Berry S. M., Gieselman M. D., Nilges M. J., van Der Donk W. A., Lu Y. (2002) J. Am. Chem. Soc. 124, 2084–2085 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.