Abstract
Two forms of the phototaxis receptor sensory rhodopsin I distinguished by differences in its photoactive site have been shown to be directly correlated with attractant and repellent signaling by the dual-signaling protein. In prior studies, differences in the photoactive site defined the two forms, namely the direction of light-induced proton transfer from the chromophore and the pKa of an Asp counterion to the protonated chromophore. Here, we show by both in vivo and in vitro measurements that the two forms are distinct protein conformers with structural similarities to two conformers seen in the light-driven proton transport cycle of the related protein bacteriorhodopsin. Measurements of spontaneous cell motility reversal frequencies, an in vivo measure of histidine kinase activity in the phototaxis system, indicate that the two forms are a photointerconvertible pair, with one conformer activating and the other inhibiting the kinase. Protein conformational changes in these photoconversions monitored by site-directed spin labeling show that opposite structural changes in helix F, distant from the photoactive site, correspond to the opposite phototaxis signals. The results provide the first direct evidence that displacements of helix F are directly correlated with signaling and impact our understanding of the sensory rhodopsin I signaling mechanism and the evolution of diverse functionality in this protein family.
Keywords: Membrane Proteins, Photoreceptors, Phototransduction, Receptors, Rhodopsin, Bacteriorhodopsin, Microbial Rhodopsin, Phototaxis, Retinal, Sensory Rhodopsin
Introduction
Microbial rhodopsins, photoactive retinal-binding membrane proteins found in both prokaryotes and unicellular eukaryotes, carry out diverse functions, including active light-driven ion transport, light-gated passive cation channel formation, photosensory activation of interacting protein transducers, and likely photoregulated transcription (1–5). These proteins contain seven transmembrane helices (helices A–G) enclosing an all-trans-retinylidene chromophore covalently bound to a lysyl residue on the seventh helix through a protonated Schiff base. Retinal photoisomerization to the 13-cis form initiates a photocycle consisting of consecutive transitions through several intermediate protein states ending with a return to the initial state. The microbial rhodopsin family is providing insights into how functional versatility is produced from modifications of a common structural scaffold, a central question in protein science.
The first discovered and best characterized are four functionally distinct microbial rhodopsins found in the archaeon Halobacterium salinarum, bacteriorhodopsin (BR),2 halorhodopsin (HR), and sensory rhodopsins I and II (SRI and SRII). BR and HR function as light-driven proton and chloride pumps (6, 7), respectively, whereas SRI and SRII form complexes with their transducers HtrI and HtrII, respectively, and mediate phototaxis responses (8, 9). SRI absorbs orange-red light and, in complex with HtrI, mediates attractant responses to orange-red light as well as repellent responses to near-UV light, which is absorbed by the photoproduct produced by the first orange-red photon. SRII, on the other hand, absorbs blue-green light and in complex with HtrII mediates repellent responses.
The notion that the sensory rhodopsin signaling mechanism evolved from that of the proton pump was first raised by the demonstration of proton pumping activity in SRI in the absence of the normally tightly bound HtrI (10). SRII was also found to pump protons in the absence of HtrII (11, 12). The remarkably small evolutionary change required to convert a proton pump into a sensor was demonstrated by conversion of BR into an effective repellent-signaling phototaxis receptor by introducing just three mutations in BR enabling hydrogen bonding to HtrII and creating a steric trigger known to be critical in SRII (13, 14). This result suggested common conformational changes occur during proton pumping and during conformational coupling to HtrII to elicit repellent responses for phototaxis. This commonality in protein structural changes was further supported by the finding that the Schiff base proton accessibility (also called “connectivity”) switches from the outward facing side of the protein to the inward facing side during proton pumping by BR, and a similar “Schiff base connectivity switch” plays a crucial role in signaling by SRI (15, 16).
Considering the analogy in the photochemical reactions between BR and SRII (17–19), conformational changes relevant for repellent phototaxis signaling by SRII and the signaling triple mutant of BR are likely to be the outward tilting of helix F in the late M or N state of BR, as has been demonstrated with various techniques (20–25), which is analogous to the conformational change observed with bovine rhodopsin when the active state for G protein activation, metarhodopsin II, is formed (26).
Apparently, the outward helix F displacement (accompanied by counterclockwise rotation (23)) is perceived by HtrII as a repellent signal, i.e. results in activation of CheA kinase bound at the distal end of the cytoplasmic domain eliciting the reversal of flagella motor rotation through CheY phosphorylation. Does the opposite response, i.e. attraction to orange light produced by SRI, result from the same receptor conformational change causing the opposite effect on HtrI? Alternatively, does the attractant response result from the opposite conformational change of the receptor, i.e. inward helix F displacement and clockwise rotation? An observation in favor of the latter possibility is the presence of two forms of the photoreceptor detected by differences in the photoactive site. Light was shown to induce oppositely directed photocurrents caused by the deprotonation of the retinylidene-protonated Schiff base upon photoactivation. This observation suggested two conformational states differing in “Schiff base connectivity” (defined by proton release direction) either inward to the cytoplasmic or outward to the extracellular side of the protein (15). The finding that the attractant-signaling SRI-HtrI is predominantly inwardly connected and the mutant SRI-HtrI exhibiting repellent-signaling (inverted mutants) and SRII-HtrII are outwardly connected (15, 27) argues that the initial conformations of the attractant and the repellent receptors are different. Furthermore, different dark conformations of the complex in wild-type SRI-HtrI and inverted signaling SRI-HtrI mutants were indicated by altered environments of a reporter group on helix F (28). These results and the observation that the connectivity is switched from the outward to the inward side upon activation of wild-type SRI-HtrI (16) prompted us to postulate a two-conformer model in which the initial states of the attractant- and the repellent-signaling conformers are CheA kinase-activating and -inhibiting states, respectively, and that the two conformers are photointerconvertible (16).
The model with two photointerconvertible conformers is based only on the properties of the retinylidene chromophore in SRI, namely the two opposite directions of Schiff base proton release from the chromophore (15) and differences in the pKa of Asp-76, which is a counterion to the protonated Schiff base (16). The model (Fig. 1) makes two specific predictions not demonstrable from measurements of the chromophore properties as follows: (i) the attractant and repellent signaling forms of the SRI-HtrI complex should have opposite effects on CheA kinase activity in the dark, and (ii) the SRI protein structure at sites distant from the chromophore should differ in the two conformers. Conformational differences are predicted especially in the relative positions of helix F, implicated in various studies in signaling by SR-Htr complexes (1–5).
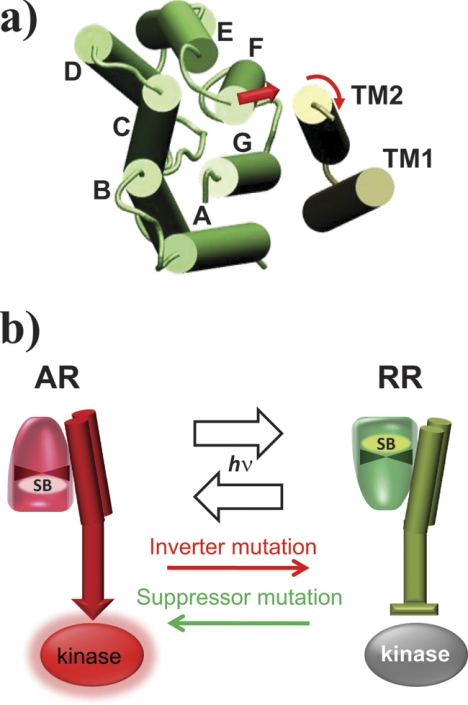
FIGURE 1.
Model for SRI-HtrI signaling. a, modeled helix positions in SRI-HtrI based on the crystal structure of the SRII-HtrII complex (Protein Data Bank code 1H2S (39)) viewed from the cytoplasmic side with the photoreceptor and the transmembrane domain (TM1 and TM2) of the transducer shown in green and gray, respectively. Note that helix F of the photoreceptor is in direct contact with TM2. b, in current models of SR-Htr interaction, a generally accepted feature is that the photoreceptor conformational change couples directly with the transmembrane domain of the transducer (3, 43, 48), which regulates the activity of the kinase bound at the distal end of the transducer. In the model for SRI-HtrI, AR (“Attractant Receptor”) and RR (“Repellent Receptor”) conformers of the photoreceptor, which are distinguished by the direction of chromophore Schiff base (SB) connectivity (see text and Refs. 15, 16), are kinase-activating and kinase-inhibiting conformers, respectively. The wild-type SRI-HtrI complex is dominated by the AR conformer and light converts the AR conformer to the RR conformer causing an attractant response (kinase inhibition). Signal inverting mutations shift the equilibrium toward the RR conformer, which mediates repellent responses to light (kinase activation), and suppressor mutations suppress the inverted phenotype by shifting the equilibrium back to the AR.
To test these predictions, here we report assessment of the level of CheA activation by the receptor-transducer complex in vivo by comparing frequencies of swimming reversals of H. salinarum cells expressing attractant- and repellent-signaling photoreceptor-transducer complexes. The results show that the attractant-signaling SRI-HtrI wild-type is in a CheA-activating state in the dark, whereas the repellent-signaling SRI-HtrI in inverted mutants and SRII-HtrII are in a CheA-inhibiting state. Photostimulation converts each of the CheA-modulating states into the other, validating the model with two photointerconvertible conformers. We then assessed the structural changes of the receptor molecules occurring during their photointerconversions by monitoring their helix F movement with site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy. We found that photolyzed HtrII-free SRII undergoes outward helix F displacement comparable with that of BR, but complexation with HtrII restricts the movement. SRI in complex with HtrI showed inwardly directed helix F movement, which directly confirms the prediction from the model in terms of protein structure. Moreover, the mutation E56Q in HtrI, which inverts the SRI-HtrI complex into a repellent signaling complex like SRII-HtrII, inhibited the light-induced F helix displacement, mimicking the behavior in SRII-HtrII.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
Wild-type and mutants of HsSRII in complex with HsHtrII were expressed under the fdx promoter in the halobacterial plasmid vector pPR5 (29). SRI and HtrI protein fusion constructs are joined with a flexible peptide linker (ASASNGASA) between the C-terminal receptor and N-terminal transducer residues and were expressed under the bop promoter in the halobacterial plasmid vector (30). These plasmid vectors were transformed into H. salinarum strain Pho81Wr− (BR− HR− SRI− HtrI− SRII− HtrII− and carotenoid and restriction-deficient).
Transducer-free NpSRII joined through the peptide linker to NpHtrII truncated at residue 157 (SRII-HtrII157) was cloned into an Escherichia coli expression vector pET21d (Novagen, Merck KgaA, Darmstadt, Germany) between NcoI and HindIII sites under the control of the T7 promoter (31). They were transformed into BL21(DE3) as described. SRI joined through the peptide linker to HtrI truncated at position 147 (SRI-HtrI147) was cloned into Pho81 Wr−. Expression of the genes in BL21(DE3) was induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside and 5 μm all-trans-retinal. Single residue mutations were introduced by the QuickChange® site-directed mutagenesis kits with Pfu Turbo polymerase (Stratagene, La Jolla, CA) or PCR using Phusion polymerase (Finnzymes, Espoo, Finland).
Cell Motility Measurements
Transformed H. salinarum cells were grown in complex medium containing 1 mg ml−1 mevinolin as described previously (29). Cultures at the end of their exponential growth phase were diluted 1:10 in fresh complex medium and incubated for 1 h at 37 °C with agitation. For motility defined as in the dark, cell trajectories in nonactinic light at wavelengths of >720 nm were captured as real time AVI files using a Flashbus Spectrim Lite® video capture PCI card on a Dell Dimension 8300 running VirtualDub 1.6.19.0 AVI encoder software for video capture. VirtualDub was set to record 10 frames/s during 30 s of cell swimming. Means of reversals of the cells were measured by tracking each cell captured in the AVI files for 30 s to count the numbers of swimming reversals. Means ± S.D. were calculated from 20 to 30 cells. Illumination was delivered from a Nikon 100-watt He/Xe short arc lamp beam passed through either 500 ± 20 nm (green light), 590 ± 20 nm (orange-red light) interference filters, or a broadband filter (CS600 cyan-subtractive dichroic filter, Corion, Franklin, MA) transmitting wavelengths between 380 and 600 nm (“white light” stimulus). For measurements of spontaneous reversal frequencies of cells in the light, recording began after 1 min of light exposure.
Purification and Spin Labeling of SRII and SRII-HtrI1157 in Detergent Solution
E. coli membranes containing free SRII or SRII-HtrII157 and harvested as described previously (31) were suspended with 2.0% n-dodecyl-β-d-maltopyranoside (DDM) in the binding buffer (0.3 m NaCl, 10 mm imidazole, 25 mm Tris-HCl (pH 7.0)) and allowed to solubilize overnight. The solubilized fraction was combined with Qiagen nickel-nitrilotriacetic acid-agarose His-binding resin with an anticipated binding efficiency of ∼3.5 mg/ml (protein/resin) and gently shaken at 4 °C for 2 h. The resin was washed with a 4× bed volume of 1.0% DDM in the binding buffer and subsequently suspended in the spin labeling buffer containing 0.01% DDM, 200 mm (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (Toronto Research Chemicals, North York, Ontario, Canada), 0.3 m NaCl, and 25 mm Tris-HCl (pH 7.0) by gently shaking it at 4 °C overnight. Excess labeling reagent was removed by washing with a 10× bed volume of the binding buffer containing 0.01% DDM. The purified labeled hexahistidine-tagged proteins were eluted with buffer containing 1.0% n-octyl-β-d-glucopyranoside (OG) (0.3 m NaCl, 250 mm imidazole, 25 mm Tris (pH 7.0)). To 1 ml of the 20 μm eluted protein, 1 mg of halobacterial polar lipid dissolved in 10% OG was combined, followed by dialysis to remove the imidazole and OG against buffer (0.3 m NaCl, 25 mm Tris-HCl (pH 7.0)). The proteoliposome containing ∼40 nmol of the protein was pelleted by centrifugation, suspended in 30 μl of the buffer, and loaded into a glass capillary (KIMAX-51, Kimble Glass Inc., Vineland, NJ).
Spin Labeling of SRI-HtrI Cysteine Mutants in the Native Membrane
Membranes were prepared by sonication of H. salinarum cells grown to stationary phase in complex medium containing 1 mg/ml mevinolin and harvested as described previously (28). Before the spin labeling of membranes, DTT was added to the membrane samples (10 mm concentration) for 1 h. The samples were then washed via pelleting by ultracentrifugation (147,000 × g for 15 min, Beckman OptimaTM Ultracentrifuge) four times with resuspension in 25 mm MES (pH 6.0), 4 m NaCl. Then excess (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (final concentration 200 μm) was added to the membrane suspension (spin label to SRI-HtrI ratio of 10:1), and labeling was allowed to proceed overnight to ensure quantitative labeling. Double labeling produced approximately twice the total label compared with monocysteine samples in each case. The excess reagent was removed by pelleting by ultracentrifugation (147,000 × g for 15 min) and resuspending in the same MES buffer six times.
Laser Flash Photolysis
Flash-induced absorption changes of membrane samples containing spin-labeled double-cysteine mutant SRI-HtrI samples used for the EPR measurements were acquired with a laboratory-constructed cross-beam laser flash photolysis system as described previously (14).
EPR Spectroscopy
EPR spectra were recorded on a Bruker EMX X-band spectrometer using a modulation amplitude of 2.0 G, a modulation frequency of 100 kHz, and a microwave power of 1 milliwatt. The temperature was controlled with a heater and cold nitrogen gas provided through a silver-coated double-jacketed glass transfer line, and a BVT3000 temperature controller was used. Data acquisition and analyses were conducted using WinEPR. Illumination of the sample was from an illuminator (FOI-250, Titan Tool Supply Inc., Buffalo, NY) equipped with a 250-watt tungsten-halogen lamp transmitted through a light guide and a 500 ± 20-nm interference filter or a long pass filter >520 nm through the front window of 4119HS resonator. In free SRII and SRII-HtrII, M states were accumulated at 5 °C upon continuous illumination with 500 nm light, whereas in SRI-HtrI (for both wild-type and HtrIE56Q) the M states were accumulated at 23 °C by illumination with >520 nm light. The protein sample was transferred into 1.0-mm inner diameter capillaries, placed at the center of a 5-mm outer diameter quartz EPR tube (KIMAX-51, Gerreshimer, Quretaro, Mexico).
RESULTS
Kinase Modulation by Dark- and Light-adapted Wild-type and Mutant SRI-HtrI Complexes
The frequency of spontaneous swimming reversals in H. salinarum cells reflects the activity of the histidinyl kinase CheA (32, 33), which is modulated by changes in chemotaxis effector concentrations and light intensity (8). As in E. coli chemotaxis (34), changes in CheA kinase activity in H. salinarum following changes in conditions are transient, causing brief responses to a change in light intensity after which the cells adapt to the new condition. Therefore, in wild-type H. salinarum cells under natural conditions, the steady-state swimming reversal frequency is the same in the dark and in the light (35). The transient nature of phototaxis responses to a step-up or step-down in light intensity was accomplished by adaptation machinery involving a methylation system in the cells that counteracts the effects of the light intensity change. At the low natural wild-type SR-Htr complex concentrations, the adaptation reactions reset the kinase activity, and consequently the swimming reversal frequency, to its prestimulus value (35). However, when the receptor-transducer complexes are overexpressed in the cells to >10-fold levels, the adaptation system is not sufficient to reset the kinase activation level (29, 36). This effect allows us to use the spontaneous reversal frequency to compare the kinase modulation of different complexes under constant conditions, specifically darkness or continuous light.
The model with two photointerconvertible conformers proposes that SRI-inverted signaling mutants (37) are constitutively activated forms of the wild-type SRI receptor. Because wild-type SRI mediates an attractant response (i.e. a step-up in light intensity reduces CheA activity and suppresses reversals), the inverting mutations are therefore predicted to reduce the spontaneous reversal frequency in the dark and increase it in continuous light. The predicted reduction was observed for the inverted mutant SRI-HtrIE56Q in the dark (16).
Here, we present a more comprehensive analysis of a large number of inverting mutants and suppressor mutations in the dark (Fig. 2) and in continuous light (Fig. 3). These mutants were isolated previously and have been extensively characterized in terms of phototaxis response phenotypes (28, 37). Inverting mutations in the SRI-HtrI complex invert orange light attractant signaling into repellent signaling, and suppressor mutations, when added to an inverting mutation, were shown to revert the inversion to wild-type attractant signaling (37). The residues that when mutated cause the phenotypic conversions presented in Figs. 2 and 3 are located near the interaction surface of SRI with the transmembrane domain of HtrI and in the membrane-proximal HAMP domain, which connects the transmembrane domain and the distal cytoplasmic domain that binds the CheA kinase. For the purposes of this study, the important property of these mutants is that they can be grouped into three response behavior phenotypic classes as follows: inverted (i.e. repellent signaling) mutants, mutants with wild-type behavior (inverting mutation + suppressor mutation), and mutants with stronger attractant responses than wild type (28, 37). The spontaneous reversal frequencies in the dark for each mutant are displayed in Fig. 2 as white, gray, or black bars.
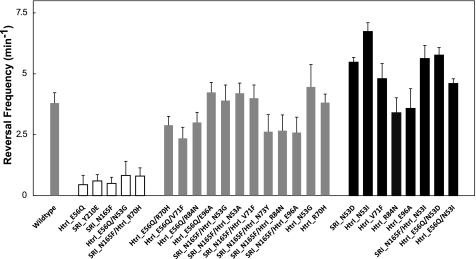
FIGURE 2.
Spontaneous reversal frequencies of Pho81 cells in the dark expressing wild-type and mutant SRI-HtrI. White bars, omniphobic phenotypic mutants; gray bars, wild-type phenotypic mutants; black bars, omniphilic phenotypic mutants. Error bars indicate +1 S.D.
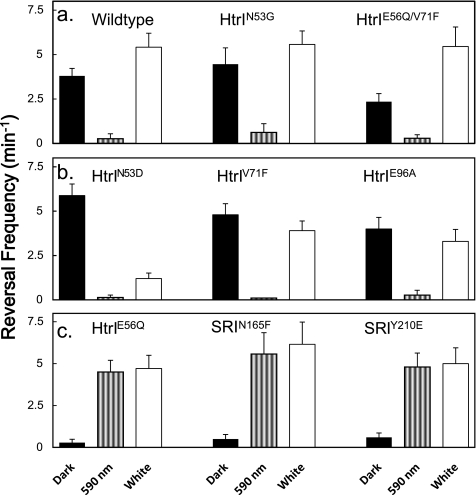
FIGURE 3.
Spontaneous reversal frequencies of Pho81 cells expressing wild-type and mutant SRI-HtrI in the light and in the dark. The mutants were classified by the following phenotypic classes. a, wild-type phenotypes, wild-type, HtrIN53G, and HtrIE56Q/V71F; b, omniphilic phenotypes, HtrIN53D, HtrIV71F, and HtrIE96A; and c, omniphobic phenotypes, HtrIE56Q, SRIN165F, and SRIY210E. Black bars, dark condition; striped bars, 590-nm illumination; white bars, white light illumination. Error bars indicate +1 S.D.
According to the model, the three classes of response phenotypes should exhibit predictable differences in their dark spontaneous reversal frequencies, because these frequencies are determined by the activity of the CheA kinase, postulated to be controlled by the two conformers. The predicted correlation is evident in Fig. 2, namely cells expressing the wild-type SRI-HtrI and mutants with the wild-type phenotype (gray bars) exhibit an intermediate spontaneous reversal frequency in the dark, mutants with an inverted phenotype (white bars) uniformly exhibit a greatly reduced dark reversal frequency, and mutants with strengthened attractant behavior (black bars) have on the average higher dark reversal frequencies than wild type.
This behavior confirms the prediction from the model, which proposes that opposite responses are due to the presence in the dark of conformers with opposite effects on the CheA kinase. Moreover, the spontaneous dark reversal frequency, an in vivo measure of the CheA kinase activity, closely correlates with the response phenotypes in the manner expected.
The correlation is evident as well with closer examination of individual sets of related mutants. Each of the three different inverting mutations in the SRI-HtrI complex, E56Q in HtrI and Y210E or N165F in SRI, reduces the dark spontaneous reversal frequency by 6.3–8.4-fold compared with wild type. The mutation N53G in HtrI suppresses the SRIN165F signal inversion, restoring SRIN165F to wild-type (attractant) signaling function,3 and it also restores the dark spontaneous reversal frequency to that of cells with wild-type SRI (Fig. 2). N53G in HtrI does not suppress the inverting mutation E56Q in HtrI, i.e. HtrIE56Q/N53G exhibits repellent signaling like HtrIE56Q,3 and it also does not restore the wild-type reversal frequency (Fig. 2). The same correlation between suppression of the particular inverting mutation signaling defect and spontaneous reversal frequency alteration is observed with HtrIR70H.
Suppresser mutations as single mutations did not alter the orange light responses, but some of them inverted the repellent responses to a white light pulse that activates the two-photon cycle of SRI in wild type into attractant responses (37). The latter behavior therefore has been denoted as the omniphilic phenotype, and the inverted mutants have been denoted as exhibiting the omniphobic phenotype (28). We interpret the latter behavior as resulting from a strengthened attractant response to the first photon, which overcomes the repellent signal from the second photon absorption event in the SRI two-photon cycle (37). Using these definitions, in Fig. 2 the white, gray, and black bars in the histogram represent the omniphobic, wild type, and omniphilic behavioral phenotypes, respectively.
The changes in spontaneous reversal frequency in the light further confirm the model. We measured the spontaneous reversal frequencies of three mutants with wild type (Fig. 3a, wild type, HtrIN53G, and HtrIE56Q/V71F), omniphobic (Fig. 3b, HtrIN53D, HtrIV71F, and HtrIE96A), and omniphilic (Fig. 3c, HtrIE56Q, SRIN165F, and SRIY210E) phenotypes in darkness, under illumination with 590-nm light, and in white light (see “Experimental Procedures”), shown in black, striped, and white bars, respectively. In the omniphilic and wild-type phenotype mutants, in a background of 590 nm light, an attractant for these mutants, the frequency is reduced to near zero (Fig. 3, a and b). Our interpretation is that the SRI-HtrI complex in the photostationary state shifts into the CheA-inhibiting conformer.
In contrast, in the omniphobic mutants, the 590-nm light stimuli elevated the frequency from the low dark value to a high frequency comparable with those of omniphilic mutants in the dark. In terms of the model, light shifts the kinase-inhibiting conformer into the kinase-activating conformer. This result provides evidence of photointerconvertibility of SRI-HtrI, which we earlier deduced from the effects of mutations and light on the chromophore properties (16).
White light produces a two-photon elicited state(s) from photointermediate photoexcitation that functions as a repellent signaling state(s) (37, 38). The higher spontaneous reversal frequencies of the wild-type phenotype mutant cells in white light relative to those in the dark are as expected from the model, which proposes accumulation of the CheA-activating conformer. The decrease in the frequency of omniphilic mutants under white light illumination is expected from the model because the net effect of the white light used is an attractant response in these mutants. As noted above, a likely explanation of the omniphilic phenotype is that the suppressor mutations strengthen the one-photon attractant signal to such an extent that it overwhelms the repellent effect of the second photon stimulation. This would occur if the mutation biased the equilibrium in the dark so far toward the kinase-activating conformer that even after two-photon stimulation a net increase in the one-photon product is produced (i.e. the two-photon activation by near-UV light contained in the “white” photostimulus does not overcome the attractant effect of the one-photon product).
Structural Changes in Attractant and Repellent Signaling by SRI-HtrI Complexes
The availability of attractant and repellent complexes strongly biased into either the kinase-activating or kinase-inhibiting conformer enabled us to apply site-directed spin labeling and EPR spectroscopy to monitor structural changes in the complexes as they switch from one conformer to the other. We emphasized the displacement of helix F with respect to helix G near the cytoplasmic surface of the photoreceptor because in BR the largest changes occur at that location (20, 22–24), the HtrII transducer subunit is tightly bound to helices F and G (39), and helix F displacement has been implicated in signaling by SRII (40, 41).
We applied the strategy by Xiao et al. (23) who demonstrated light-induced helix F outward displacement in BR by monitoring distance changes between two spin probes covalently attached to pairs of cysteine residues introduced on each of the helices near the cytoplasmic surface. A distance change between the probes results in broadening (decreasing distance) or sharpening (distance increases) of the line shape of the EPR spectrum due to spin-spin dipolar interaction (23).
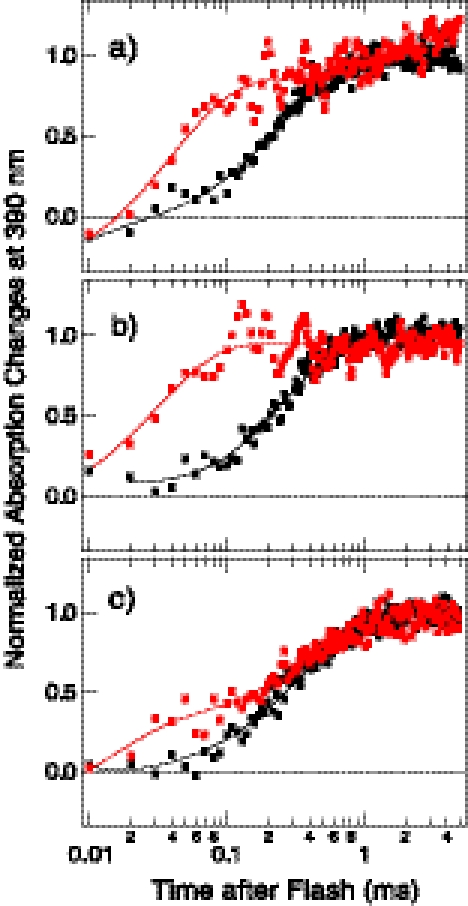
Pairs of positions (Fig. 4) were mutated to cysteine and spin-labeled. Prior to EPR measurements, we examined the double spin-labeled complexes in the membranes to assess whether the mutations or labeling procedure altered the equilibrium between the two conformers established from in vivo measurements. This was accomplished by taking advantage of the previous finding that the outwardly connected repellent receptor conformer exhibits an ∼10-fold higher rate of light-induced deprotonation of the Schiff base, which results in formation of the M intermediate, than that of the inwardly connected attractant receptor conformer. Repellent signaling receptors show M-rise time constants (τ) of <50 μs, whereas those in the attractant signaling receptors are of several hundred microseconds (16). The three samples made with wild-type HtrI each exhibit ∼90% amplitude of slow component M-rise, matching the value observed with the wild-type receptor established from photocurrent directions (16), showing that the introduction of the two cysteine mutations and the spin labeling procedure did not significantly alter the equilibrium between the two conformers (Fig. 5 and Table 1). The signal-inverting E56Q mutation in HtrI significantly increases the fraction of the molecules with fast component M-rise attributable to the repellent signaling photoreceptor in all three samples. The increase in proportion of the fast component M-rise in E56Q samples compared with wild type is consistent with the in vivo measurements in which the E56Q mutation increases the proportion of the fast component from ∼15 to ∼65% (16). The altered M-rise kinetics in SRI caused by the E56Q mutation in HtrI also confirms that HtrIE56Q exerts the same effect on SRI despite the cysteine mutations, purification of the complex, production of proteoliposomes, and the spin labeling procedure.
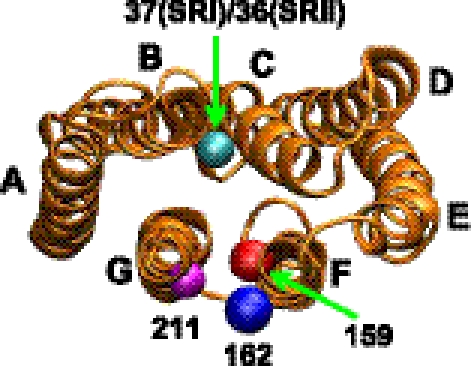
FIGURE 4.
Molecular structure of the photoreceptor based on the SRII crystal structure (Protein Data Bank code 1JGJ (49)) viewed from the cytoplasmic side. Residues used for the spin labeling are Ala37 in SRI (Tyr36 in SRII) (positions shown with a cyan ball) on helix B, Leu159 in SRI/SRII (positions shown with a red ball), Leu162 in SRI (Arg162 in SRII) (positions shown with a blue ball) on helix F, and Phe211 in SRI/SRII (the positions shown with a pink ball) on helix G. Pairs of residues were mutated to cysteine and the spin probes covalently attached through disulfide linkages.
FIGURE 5.
Normalized absorbance changes at 390 nm monitoring M formation in engineered SRI double cysteine mutants double-labeled with the spin probe. The labeling positions are 37/159 (a), 162/211 (b), and 159/211 (c). HtrI in complex with each of the photoreceptors is either wild-type HtrI (black points) or E56Q HtrI (red points). Two-exponential fits of each curve are shown.
TABLE 1.
Percentage of the total absorption change amplitude for the fast (τ1 = 3–44 μs) and slow (τ2 = 211–434 μs) component rise of 390-nm absorbance (τ1 and τ2, respectively) due to deprotonation of the Schiff base induced by a laser flash (532 nm, 6-ns pulse) of SRI double labeled with the spin probe
The positions of the residues replaced with cysteine and labeled with the probe are shown in the left column. The fractions of the two kinetic components are compared between each of the engineered SRI with HtrI either with (HtrIE56Q) or without (HtrIWT) the E56Q mutation.
| Labeling positions | HtrIWT | HtrIE56Q | |
|---|---|---|---|
| % | % | ||
| 37/159 | τ1 | 6 | 73 |
| τ2 | 94 | 27 | |
| 159/211 | τ1 | 6 | 30 |
| τ2 | 94 | 70 | |
| 162/211 | τ1 | 12 | 100 |
| τ2 | 88 | 0 |
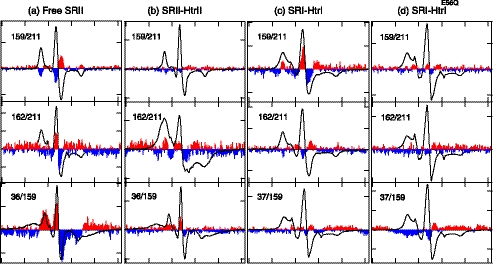
As a control, EPR spectra were recorded in the dark and in a photostationary state consisting primarily of the M photointermediate in transducer-free SRII, which was examined previously with this method by another group (40). Large increases in intensity evident in the light-dark difference spectra in the same direction as the absolute spectrum show sharpening of the line shape in the 36/159 pair of probes, which indicates the separation between helices B and F upon M formation (Fig. 6a). The probe at position 211 on helix G exhibited line shape broadening with a probe at position 159 on helix F and line shape sharpening with a probe at position 162 (Fig. 6a), which indicates that position 159 on helix F approaches helix G position 211, and position 162 on helix F moves away from the helix G position 211. These changes indicate that helix F movement contains a counterclockwise rotation component and/or undergoes outward displacement. All of these changes in transducer-free SRII are in the same directions as those observed for by the same technique for BR (23) and for SRII (40), which have been attributed to helix counterclockwise rotation and outward helix F displacement.
FIGURE 6.
EPR spectra of spin probes doubly labeled at the indicated two positions in the photoreceptors. a, free SRII; b, SRII in complex with HtrII157; c, SRI in complex with HtrI147; and d, SRI in complex with HtrI147 with the E56Q mutation. EPR spectra measured in the dark are shown with black solid lines. The light-minus-dark difference spectra subtracting the dark spectra from those measured during the illumination with 500 nm of light for SRII and >520 nm of light for SRI are appended with 10-fold magnification to each panel with the positive and the negative peaks filled with red and blue, respectively. The width of the abscissa axis in each of the panels represents 150 G.
A key result is that light-dark difference EPR spectra of SRII in complex with HtrII show that transducer binding greatly diminishes the motion of helix F (Fig. 6b). The reduced change of distance between the pair of the spin labels at position 36/159 and the nearly undetectable change at position 159/211 indicate that the helix F movement that occurs in free SRII at these positions is inhibited by the bound transducer. We conclude that the helix F displacement that occurs in the free SRII is restricted by the bound HtrII. This observation confirms the suggestion of such an inhibitory effect made from Fourier transform infrared (FTIR) light-dark difference spectra, which showed suppression of the large amide-I changes seen in free SRII when it is complexed with HtrII (42). The 162/211 pair shows stronger dipolar coupling in both the dark and light conditions evident from the broadened outermost bands in the complexed SRII compared with the free SRII. A possible explanation is that HtrII interaction at the outer surface of helix F alters the orientation of the spin probe to bring the two probes closer to one another than in free SRII.
A second key observation is that SRI in complex with HtrI exhibits changes in the difference EPR spectra for the pairs 159/211 and 162/211 (Fig. 6c) opposite to that of SRII, namely line shape sharpening and broadening, respectively, attributable to separation and approaching of the two spin labels. The changes are ascribable to inward helix F movement with respect to helix G and/or clockwise rotation of helix F with respect to helix G, which is the opposite of the helix F displacement observed above in free SRII upon M formation and in BR upon M or N formation (23) in which outward helix F movement and a counterclockwise rotation were observed.
To confirm that the opposite light-induced movement of helix F between SRII and wild-type SRI is correlated with their opposite signaling, we examined the SRI-HtrI complex with the inverting HtrI mutation E56Q, which shows repellent responses to photostimuli like the SRII-HtrII complex. In SRI-HtrIE56Q, the difference EPR spectra for the pairs 159/211 and 162/211 are greatly diminished (Fig. 6d) indicating that, as in the repellent signaling SRII-HtrII, the movement of helix F with respect to helix G is restrained by the bound Htr transducer. A particular difference in SRI with the mutated HtrI complex is the line shape broadening upon illumination in the 37/159 pair (Fig. 6d), which showed little if any change in wild-type SRI-HtrI. This broadening indicates light-induced decrease of the distance between helix B and helix F, possibly a separate effect of the E56Q mutation on helix B.
Note that the inverting mutations are located in SRI on helix F (N165F and H166S likely near the outer surface) and helix G (D201N facing the photoactive site and Y210E on the outer surface) and in HtrI (E56Q) in the junction region between the second transmembrane helix (TM2) and the following HAMP domain. Most of these mutations are located in the interface constituting the binding site of SRI and HtrI and are likely to alter the structure of TM2 of HtrI, which then switches the conformation of the membrane-proximal cytoplasmic HAMP domain which in turn alters the more distal cytoplasmic domain to inhibit CheA. The suppressor mutations are found in the HAMP domain of HtrI, with the exception of R215W, which is on the cytoplasmic surface of SRI on helix G. These mutations indicate reversibility of the signaling path from the photoreceptor to the transducer, because mutations in the HAMP domain alter the structure of the SRI photoactive site and the SRI conformation.
Absence of Conformer Photointerconvertibility in the SRII-HtrII Complex
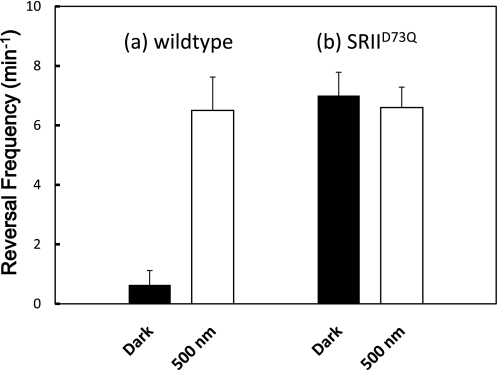
The question arises whether the SRII-HtrII complex also exhibits photointerconvertibility of a kinase-activating and kinase-inhibiting state. The wild-type SRII-HtrII complex behaves like the repellent mutant SRI-HtrIE56Q in that photon absorption produces a CheA kinase-activating conformer. One way to describe the photointerconvertibility of the two conformers of SRI-HtrI is that each conformer acts as a photoreceptor mediating in one case an attractant and the other case a repellent response, and each conformer is a constitutively activated form of the other. The constitutively activated mutant SRIID73N was identified and shown to elevate cell reversal frequencies by 3.1-fold over cells expressing wild-type SRII (29, 36). We tested here another mutation at the same position in SRIID73Q-HtrII, and we observes a 10.2-fold increase in spontaneous reversal frequency in the dark over the wild-type value (Fig. 7). Wild-type SRII under continuous saturating 500-nm illumination produces the same reversal frequency as SRIID73Q in the dark, indicating SRIID73Q appears to be fully constitutively activated. Yet illumination of the SRIID73Q-HtrII complex does not produce an attractant response (36), and in fact, the mutant complex exhibits the same elevated reversal frequency in the light as in the dark (Fig. 7). Therefore, the photointerconvertibility of the two signaling conformers appears to be a property only of SRI, likely deriving from its unique one- and two-photon dual signaling mechanism.
FIGURE 7.
Spontaneous reversal frequencies of Pho81 cells expressing either wild-type SRII-HtrII or SRIID73Q-HtrII. The mean frequency values of the cells in the dark or constant 500 nm of light are presented with black or white bars, respectively. Error bars indicate +1 S.D.
DISCUSSION
The model with two-photointerconvertible conformers was proposed on the basis of the observation of two conformers of SRI with opposite signaling that have the Schiff base connectivity directed toward opposite sides of the protein (15), and the light-induced conversion of each conformer into the other (16). Three key aspects of that model are that attractant signaling by SRI-HtrI and repellent signaling by its inverted mutants as well as by SRII-HtrII are (i) attributed to two conformers with opposite effects on the CheA kinase (activating versus inhibiting), (ii) that one or the other of these conformers predominate in the dark state, and (iii) that each SRI conformer is photoconverted into the other. The measurements reported here of spontaneous reversal frequencies in the dark and in the light, an in vivo measure of kinase activity, confirm these three aspects of the model for a large number of mutants and mutant combinations, adding compelling evidence for the model by direct measurement of phototaxis behavior.
The BR proton-pumping mechanism and SR signaling mechanism, although distinctly different in terms of molecular outcome (electrogenic proton transport versus activation of Htr by protein-protein interaction), have been shown nevertheless to result from very similar chemical processes (3, 43). Most dramatically, only three mutations in BR and its attachment to HtrII are necessary to convert BR into a robust repellent-signaling phototaxis receptor (14). Therefore, an intriguing question raised by the two-conformer mechanism is whether the two well established conformers responsible for alternating proton access during the BR proton pumping are the two conformers used for SR signaling. If this were the case, a strong prediction is that the conformer of SRI in the wild-type SRI-HtrI complex in the dark would be equivalent to the BR conformer in the light. Such equivalence was suggested by our finding Schiff base connectivity in dark attractant signaling SRI opposite to that of dark BR (15, 16). The EPR data presented here show that light-induced displacement/rotation of helix F during generation of the SRI-HtrI attractant signaling state M is indeed in the opposite direction as it is in the light-induced M state during the BR pumping cycle. A clear picture emerges that evolution has modified the BR pumping cycle to use the conformational change that occurs in BR for repellent signaling by SRII and to switch the conformers to opposite positions in the photocycle for attractant signaling by SRI.
An example of inward helix F movement upon photoactivation was observed previously in BR with F42C and D85N mutations, which stabilize an N-like state with a 13-cis-retinylidene chromophore, and cytoplasmic channel conformation implied by FTIR spectroscopy from the amide-I band in the dark. This N-like state is photoconverted transiently to an O-like species with an all-trans-chromophore (44). Our data show that the protein conformation of SRI in the dark in the SRI-HtrI-complex is similar to that of the BR double mutant F42C/D85N. However, in SRI the dark conformer, unlike the mutant BR N-like state, contains an all-trans-retinylidene chromophore (45–47). The nearly indistinguishable absorption spectra of the two opposite signaling SRI-HtrI conformers deduced from their independent titration in wild-type and inverted SRI-HtrIE56Q complexes suggested that both conformers of SRI in complex with HtrI contain primarily or exclusively all-trans-retinylidene chromophore (16). Our recent FTIR results in collaboration with Kandori and co-workers (50) show that the repellent signaling conformer in the inverted mutant SRI-HtrIE56Q undergoes all-trans-photoisomerization to 13-cis, compellingly confirming this suggestion. The M intermediate of SRI (also known as S373 (8)), the natural repellent receptor form in the wild-type SRI-HtrI complex, contains primarily the 13-cis-retinylidene chromophore. Therefore, in natural conditions, the AR conformer predominates in the dark state all-trans pigment, and the RR conformer predominates in the 13-cis photoproduct M. In contrast, the mechanism by which the same light-induced isomeric configuration change of the retinylidene chromophore in wild type and the E56Q mutant results in opposite conformational changes is intriguing, and it represents an unexpected flexibility in coupling of the chromophore isomerization to the protein conformational change. The versatility of the conformational changes of microbial rhodopsins is likely to be a significant factor enabling the evolution of diverse functionality of this protein family.
This work was supported, in whole or in part, by National Institutes of Health Grant R37GM27750 (to J. L. S.) and Grant RO1HL095820 (to A.-L. T.). This work was also supported by Department of Energy Grant DE-FG02-07ER15867 and Endowed Chair AU-0009 from the Robert A. Welch Foundation (to J. L. S.).
J. Sasaki and J. L. Spudich, unpublished data.
- BR
- bacteriorhodopsin
- HR
- halorhodopsin
- SRI and SRII
- sensory rhodopsin I and II, respectively
- DDM
- n-dodecyl-β-d-maltopyranoside
- OG
- n-octyl-β-d-glucopyranoside
- AR
- attractant receptor conformer
- RR
- repellent receptor conformer.
REFERENCES
- 1. Spudich J. L., Yang C. S., Jung K. H., Spudich E. N. (2000) Annu. Rev. Cell Dev. Biol. 16, 365–392 [DOI] [PubMed] [Google Scholar]
- 2. Nagel G., Szellas T., Kateriya S., Adeishvili N., Hegemann P., Bamberg E. (2005) Biochem. Soc. Trans. 33, 863–866 [DOI] [PubMed] [Google Scholar]
- 3. Spudich J. L. (2006) Trends Microbiol. 14, 480–487 [DOI] [PubMed] [Google Scholar]
- 4. Jung K. H. (2007) Photochem. Photobiol. 83, 63–69 [DOI] [PubMed] [Google Scholar]
- 5. Klare J. P., Chizhov I., Engelhard M. (2008) Results Probl. Cell Differ. 45, 73–122 [DOI] [PubMed] [Google Scholar]
- 6. Lanyi J. K. (1990) Physiol. Rev. 70, 319–330 [DOI] [PubMed] [Google Scholar]
- 7. Oesterhelt D., Tittor J., Bamberg E. (1992) J. Bioenerg. Biomembr. 24, 181–191 [DOI] [PubMed] [Google Scholar]
- 8. Hoff W. D., Jung K. H., Spudich J. L. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 223–258 [DOI] [PubMed] [Google Scholar]
- 9. Sasaki J., Spudich J. L. (2000) Biochim. Biophys. Acta 1460, 230–239 [DOI] [PubMed] [Google Scholar]
- 10. Bogomolni R. A., Stoeckenius W., Szundi I., Perozo E., Olson K. D., Spudich J. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10188–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmies G., Engelhard M., Wood P. G., Nagel G., Bamberg E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sudo Y., Iwamoto M., Shimono K., Sumi M., Kamo N. (2001) Biophys. J. 80, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudo Y., Furutani Y., Kandori H., Spudich J. L. (2006) J. Biol. Chem. 281, 34239–34245 [DOI] [PubMed] [Google Scholar]
- 14. Sudo Y., Spudich J. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16129–16134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sineshchekov O. A., Sasaki J., Phillips B. J., Spudich J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sineshchekov O. A., Sasaki J., Wang J., Spudich J. L. (2010) Biochemistry 49, 6696–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki J., Spudich J. L. (1998) Biophys. J. 75, 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chizhov I., Schmies G., Seidel R., Sydor J. R., Lüttenberg B., Engelhard M. (1998) Biophys. J. 75, 999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kamo N., Shimono K., Iwamoto M., Sudo Y. (2001) Biochemistry 66, 1277–1282 [DOI] [PubMed] [Google Scholar]
- 20. Vonck J. (1996) Biochemistry 35, 5870–5878 [DOI] [PubMed] [Google Scholar]
- 21. Kamikubo H., Kataoka M., Váró G., Oka T., Tokunaga F., Needleman R., Lanyi J. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1386–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luecke H., Schobert B., Richter H. T., Cartailler J. P., Lanyi J. K. (1999) Science 286, 255–261 [DOI] [PubMed] [Google Scholar]
- 23. Xiao W., Brown L. S., Needleman R., Lanyi J. K., Shin Y. K. (2000) J. Mol. Biol. 304, 715–721 [DOI] [PubMed] [Google Scholar]
- 24. Subramaniam S., Henderson R. (2000) Nature 406, 653–657 [DOI] [PubMed] [Google Scholar]
- 25. Shibata M., Yamashita H., Uchihashi T., Kandori H., Ando T. (2010) Nat. Nanotechnol. 5, 208–212 [DOI] [PubMed] [Google Scholar]
- 26. Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sineshchekov O. A., Spudich J. L. (2004) Photochem. Photobiol. Sci. 3, 548–554 [DOI] [PubMed] [Google Scholar]
- 28. Sasaki J., Phillips B. J., Chen X., Van Eps N., Tsai A. L., Hubbell W. L., Spudich J. L. (2007) Biophys. J. 92, 4045–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spudich E. N., Zhang W., Alam M., Spudich J. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4960–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X., Spudich J. L. (2002) Biochemistry 41, 3891–3896 [DOI] [PubMed] [Google Scholar]
- 31. Inoue K., Sasaki J., Spudich J. L., Terazima M. (2007) Biophys. J. 92, 2028–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rudolph J., Oesterhelt D. (1995) EMBO J. 14, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trivedi V. D., Spudich J. L. (2003) Biochemistry 42, 13887–13892 [DOI] [PubMed] [Google Scholar]
- 34. Hazelbauer G. L., Falke J. J., Parkinson J. S. (2008) Trends Biochem. Sci. 33, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spudich J. L., Bogomolni R. A. (1988) Annu. Rev. Biophys. Biophys. Chem. 17, 193–215 [DOI] [PubMed] [Google Scholar]
- 36. Sasaki J., Nara T., Spudich E. N., Spudich J. L. (2007) Mol. Microbiol. 66, 1321–3130 [DOI] [PubMed] [Google Scholar]
- 37. Jung K. H., Spudich J. L. (1998) J. Bacteriol. 180, 2033–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spudich J. L., Bogomolni R. A. (1984) Nature 312, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gordeliy V. I., Labahn J., Moukhametzianov R., Efremov R., Granzin J., Schlesinger R., Büldt G., Savopol T., Scheidig A. J., Klare J. P., Engelhard M. (2002) Nature 419, 484–487 [DOI] [PubMed] [Google Scholar]
- 40. Wegener A. A., Chizhov I., Engelhard M., Steinhoff H. J. (2000) J. Mol. Biol. 301, 881–891 [DOI] [PubMed] [Google Scholar]
- 41. Klare J. P., Gordeliy V. I., Labahn J., Büldt G., Steinhoff H. J., Engelhard M. (2004) FEBS Lett. 564, 219–224 [DOI] [PubMed] [Google Scholar]
- 42. Bergo V., Spudich E. N., Scott K. L., Spudich J. L., Rothschild K. J. (2000) Biochemistry 39, 2823–2830 [DOI] [PubMed] [Google Scholar]
- 43. Sasaki J., Spudich J. L. (2008) Photochem. Photobiol. 84, 863–868 [DOI] [PubMed] [Google Scholar]
- 44. Dioumaev A. K., Brown L. S., Needleman R., Lanyi J. K. (1998) Biochemistry 37, 9889–9893 [DOI] [PubMed] [Google Scholar]
- 45. Yan B., Nakanishi K., Spudich J. L. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9412–9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rath P., Spudich E., Neal D. D., Spudich J. L., Rothschild K. J. (1996) Biochemistry 35, 6690–6696 [DOI] [PubMed] [Google Scholar]
- 47. Furutani Y., Takahashi H., Sasaki J., Sudo Y., Spudich J. L., Kandori H. (2008) Biochemistry 47, 2875–2883 [DOI] [PubMed] [Google Scholar]
- 48. Wegener A. A., Klare J. P., Engelhard M., Steinhoff H. J. (2001) EMBO J. 20, 5312–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luecke H., Schobert B., Lanyi J. K., Spudich E. N., Spudich J. L. (2001) Science 293, 1499–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sasaki J., Takahashi H., Furutani Y., Kandori K., Spudich J. L. (2011) Biophys. J., doi: 10.1016/j.bpj.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]