Abstract
DNA-damaging agents can induce premature senescence in cancer cells, which contributes to the static effects of cancer. However, senescent cancer cells may re-enter the cell cycle and lead to tumor relapse. Understanding the mechanisms that control the viability of senescent cells may be helpful in eliminating these cells before they can regrow. Treating human squamous cell carcinoma (SCC) cells with the anti-cancer compounds, resveratrol and doxorubicin, triggered p53-independent premature senescence by invoking oxidative stress-mediated DNA damage. This process involved the mTOR-dependent phosphorylation of SIRT1 at serine 47, resulting in the inhibition of the deacetylase activity of SIRT1. SIRT1 phosphorylation caused concomitant increases in p65/RelA NF-κB acetylation and the expression of an anti-apoptotic Bfl-1/A1. SIRT1 physically interacts with the mTOR-Raptor complex, and a single amino acid substitution in the TOS (TOR signaling) motif in the SIRT1 prevented Ser-47 phosphorylation and Bfl-1/A1 induction. The pharmacologic and genetic inhibition of mTOR, unphosphorylatable S47A, or F474A TOS mutants restored SIRT1 deacetylase activity, blocked Bfl-1/A1 induction, and sensitized prematurely senescent SCC cells for apoptosis. We further show that the treatment of UVB-induced SCCs with doxorubicin transiently stabilized tumor growth but was followed by tumor regrowth upon drug removal in p53+/−/SKH-1 mice. The subsequent treatment of stabilized SCCs with rapamycin decreased tumor size and induced caspase-3 activation. These results demonstrate that the inhibition of SIRT1 by mTOR fosters survival of DNA damage-induced prematurely senescent SCC cells via Bfl-1/A1 in the absence of functional p53.
Keywords: DNA Damage, mTOR, Reactive Oxygen Species (ROS), Resveratrol, Sirtuins, Premature Senescence
Introduction
Cellular senescence is a state of permanent replicative arrest that is linked to telomere erosion and dysfunction that engages at least two mechanisms, the p53 and the p16INK4a-pRB pathways (1, 2). In cancer cells, the presence of oncogenic mutations, chemotherapeutic drugs, and oxidative stress can cause an acutely inducible, telomere-independent, stress-responsive form of cellular senescence, termed premature senescence (PS)2 (3, 4). PS is considered a physiologic mechanism of DNA damage response occurring in chemotherapy (5–7), and the senescence induction could be an effective in vivo mechanism to limit tumor progression by preventing cancer cell proliferation or by blocking the cells at risk of neoplastic transformation (8). However, the physiological consequences of prematurely induced senescent (PIS) cancer cells remain elusive. PIS cancer cells have been shown to promote the growth of neighboring cells, and they are intrinsically resistant to chemotherapeutic agents (5, 9, 10). Importantly, cells in prematurely senescent tumors are capable of escaping growth arrest and re-entering the cell cycle, leading to tumor relapse (5, 9, 11). As for the mechanism underlying escape from DNA damage-induced senescence, overexpression of the cyclin-dependent kinase Cdc2 has been found in clones that bypassed replicative arrest in human non-small cell H1299 carcinoma (5) and in MCF-7 breast cancer cells (9). It was recently shown that survivin is the immediate downstream effector of Cdc2/Cdk1 and that phosphorylated survivin is necessary for the escape of senescent cells (12). Moreover, Twist1, which is involved in the metastatic dissemination of cancer cells, was shown to override oncogene-induced senescence by abrogating cell cycle inhibition by p21 and p16 (11, 13), leading to complete epithelial-mesenchymal transition and implicating a direct link between escape from senescence and the acquisition of invasive features by cancer (11, 13). These data collectively suggest that there exist mechanisms that foster survival of PIS cancer cells and promote escape of these cells from the senescent state, which are likely detrimental to the overall therapeutic efficacy of cancer treatment. Because the p53 gene is frequently inactivated in 50% of human cancers, including SCCs, this study investigated the regulatory mechanisms that control the survival of PIS SCC cells lacking functional p53.
EXPERIMENTAL PROCEDURES
Cell Culture
A431 human epidermoid squamous carcinoma cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, and kept in an atmosphere of 95% air, 5% CO2 in a 37 °C humidified incubator. Resveratrol (RES, trans-3, 4′, 5-trihydroxystilbene, C14H12O3) and sirtinol were purchased from LKT Laboratories Inc. (St Paul, MN) and Calbiochem, respectively. The copper (I) specific chelator (neocuproine), 2′,7′-dichlorofluorescein diacetate, and 4-methylumbelliferyl-β-d-galactopyranoside were purchased from Sigma. The antibodies against caspase-3, acetyl-NF-κB p65/RelA(K310), Chk1, Chk2, SIRT1, ATM, ATR, mTOR, Raptor, p70S6K, p-ATR(S428), p-ATM(S1981), γH2AX, p-SIRT1(S47), p-Chk1(S428), p-Chk2(T68), p-mTOR(S2448), and p-p70S6K(T389) came from Cell Signaling Technology (Danvers, MA), whereas p21WAF1, NF-κB p65/RelA, PML, mtHSP70, HP1γ, lamin B, and β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and SIRT1 was from Upstate Biotech Millipore (Lake Placid, NY).
siRNA Transfection
The human siRNAs (Bfl-1/A1, mTOR, Raptor, and NF-κB subunits) were purchased from Santa Cruz Biotechnology. On the day before transfection, 5 × 105 A431 cells were plated in 100-mm Petri plates and grown in 10 ml of DMEM supplemented with 10% fetal bovine serum. After 24 h in culture, 50 μl of 10 μm stock solution of siRNA duplexes was transfected into cells with a LipofectamineTM RNAiMAX transfection reagent according to the manufacturer's protocol (Invitrogen). Following 24–36 h of incubation, the cells were treated with RES and maintained in culture for the indicated times before analysis.
Senescence-associated β-Galactosidase (SA-β-gal) Staining and Quantification
In situ staining of SA-β-gal was performed using a senescence β-galactosidase staining kit (Cell Signaling Technology, Danvers, MA). SA-β-gal activities present in cell extracts were measured by the rate of conversion of 4-methylumbelliferyl-β-d-galactopyranoside to the fluorescent hydrolysis product 4-methylumbelliferone at pH 6.0, as described previously (14). All experiments were performed in triplicate, and the results are expressed as the mean ± S.D. of three independent experiments.
Intracellular Reactive Oxygen Species (ROS)
Intracellular ROS were measured as described previously using a cell-permeable fluorogenic dye, 2′,7′-dichlorofluorescein diacetate, which detects hydrogen peroxide (15). The results were obtained as arbitrary absorbance units/mg of protein. All experiments were performed in triplicate, and the results are expressed as the mean ± S.D. of the three independent experiments.
Cell Proliferation Assay
A431 cells were treated with RES and/or other reagents for 48 h, and the cell proliferation was measured using 5-bromo-2′-deoxyuridine (BrdU) cell proliferation assay kit (Roche Applied Science).
Total RNA Preparation and RT-PCR
Total RNA was prepared, and RT-PCR was performed as described previously (16). Bfl-1/A1 primers were as follows: forward, 5′-GCTCAAGACTTTGCTCTCCACC-3′; reverse, 5′-GGAGTGTCCTTTCTGGTCAACAG-3′. GAPDH primers were as follows: forward, 5′-TCATTGACCTCAACTACATGGTTTAC-3′; reverse, 5′-GGCATGGATGTGGTCATGAGTC-3′.
Western Blotting, Immunoprecipitation, Immunohistochemical, and Immunofluorescence Staining
Western blotting, immunoprecipitation, immunohistochemical, and immunofluorescence staining were performed as described previously (15, 17).
SIRT1 Deacetylase Assay
The extracts (500 μg) were first incubated with the monoclonal anti-SIRT1 antibody at 4 °C for 2 h, immunoprecipitated using protein G-agarose for 4 h at 4 °C, and washed, and the SIRT1 deacetylase activity was assayed by a SIRT1 assay kit using 2′-O-acetyl-ADP-ribose as a substrate (Sigma). The net fluorescence signal was then determined by fluorometer at excitation 360 nm and emission 450 nm, respectively.
TUNEL Assay
The TUNEL assay was performed by in situ cell death detection kit, fluorescein (Roche Applied Science) in transduced A431 cells treated with RES and/or rapamycin (0.5 μm) for 24 h. DNA fragmentation was detected using the Tecan GENios Plus system (Männedorf, Switzerland) according to the manufacturer's instructions.
Construction of SIRT1 Mutants and Stable Cell Lines
Wild-type hSIRT1 was cloned into pBABE-puro vector at the BamHI and SalI sites. The pBABEpuro-hSIRT1 plasmid was then used as the backbone for the construction of mutants. hSIRT1F474A was created by site-directed mutagenesis. For hSIRT1S47A and hSIRT1S47D, the plasmid was cut with XhoI and Sbf1. The following synthetic oligonucleotides containing the S47A or S47D mutations replaced the cut fragment: S47A, 5′-TCGAGCGGGCCCCGGGCGAGCCCGGTGGGGCGGCCCCAGAGCGTGAGGTGCCGGCGGCGGCCAGGGGCTGCCCGGGTGCGGCGGCGGCGGCGCTGTGGCGGGAGGCGGAGGCAGAGGCGGCGGCGGCAGGCGGGGAGCAAGAGGCCCAGGCGACTGCGGCGGCTGGGGAAGGAGACAATGGGCCGGGCCTGCA-3′; S47D, 5′-TCGAGCGGGATCCGGGCGAGCCCGGTGGGGCGGCCCCAGAGCGTGAGGTGCCGGCGGCGGCCAGGGGCTGCCCGGGTGCGGCGGCGGCGGCGCTGTGGCGGGAGGCGGAGGCAGAGGCGGCGGCGGCAGGCGGGGAGCAAGAGGCCCAGGCGACTGCGGCGGCTGGGGAAGGAGACAATGGGCCGGGCCTGCA-3′. The integrity of the corresponding sequences was checked by sequencing. Construction of stable cell lines was performed according to the guidelines of the Phoenix retroviral expression system (Orbigen Inc., San Diego, CA).
In Vitro mTOR Kinase Assay
A431 cells were incubated in the presence or absence of 30 μm RES for 48 h and lysed in Tween 20-radioimmune precipitation buffer; 500 μg of total cell lysate was immunoprecipitated with anti-Raptor antibody. Immunoprecipitates were then subjected to in vitro kinase assays using recombinant maltose-binding protein (MBP)-SIRT1 or MBP-SIRT1F474A, purified by the pMAL purification system (New England Biolabs, Ipswich, MA), as substrates. To generate MBP-hSIRT1F474A, the pBABEpuro-hSIRT1F474A construct was digested with PacI and SalI and ligated into the same sites of the pMAL-hSIRT1.
Generation of p53+/−/SKH-1 Hairless Mice and UV Light Source
Generation of p53+/−/SKH-1 hairless mice and UV light source are as described previously (16).
Statistical Analyses
Statistical analyses were done using Student's t test (two-tailed); p < 0.05 is considered statistically significant.
RESULTS
RES Induces PS in p53-deficient Human SCC Cells
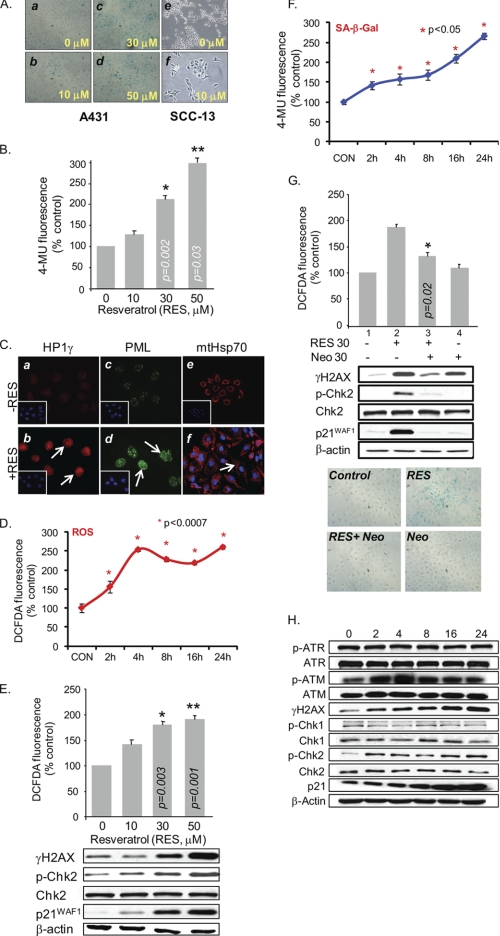
We have shown previously that A431 cells treated with RES accumulate in the G1 phase of the cell cycle (19). In this study, we show that A431 cells treated with RES at concentrations between 10 and 50 μm for 48 h became enlarged and flattened and showed increased activity of SA-β-gal, a marker of senescence induction (Fig. 1A, panels b–d). On the other hand, most of the untreated control cells were SA-β-gal-negative (Fig. 1A, panel a). The SA-β-gal activity increased in a dose-dependent manner (Fig. 1B). The punctate nuclear accumulations of HP1γ and PML and the pancytoplasmic distribution of mtHsp70, which are known to be associated with senescence, were evident in RES-treated A431 cells (Fig. 1C). Senescence-associated features were also observed in other human SCC cells, such as SCC-13 (Fig. 1A, panel f). However, confluence-induced quiescent cells were not SA-β-gal-positive (data not shown).
FIGURE 1.
RES induces copper-dependent PS via ROS-mediated DNA damage in p53-deficient A431 cells. A, RES treatment increases SA-β-gal activity. A431 (panels a–d) and SCC-13 (panels e and f) cells were treated with 0–50 μm or 0 and 10 μm RES, respectively, for 48 h. Senescence induction was detected by staining the cells for SA-β-gal activity and was visualized by light microscopy. B, quantitative SA-β-gal measurement in RES-treated A431 cells. Extracts of A431 cells, treated as in A, were incubated with 4-methylumbelliferyl-β-d-galactopyranoside (4-MU) at 37 °C. Data were normalized to total protein per assay to correct for differences in extract concentrations and expressed as percentages of fluorescence levels in untreated control cells. C, senescence-associated proteins mtHsp70, PML, and HP1γ are detected within A431 cells by indirect immunofluorescence after treatment with 0 (panels a, c, and e) or 30 μm (panels b, d, and f) RES for 24 h. Inset: DAPI staining of cell nuclei. D, ROS generation was measured in A431 cells treated with 30 μm RES for the indicated time periods. Data are expressed as percentages of fluorescence levels in untreated control (CON) cells. DCFDA, 2′,7′-dichlorofluorescein diacetate. E, dose-dependent generation of ROS and increases in the levels of γH2AX, p-Chk2(T68), and p21WAF1 in A431 cells following RES treatment. F, SA-β-gal activity in A431 cells treated with 30 μm RES for the indicated time periods. G, the copper (I) chelator neocuproine (Neo) blocks the RES-mediated DNA damage response. ROS generation and SA-β-gal activities were measured in A431 cells pretreated with 30 μm neocuproine for 2 h and then incubated for another 48 h in the presence or absence of 30 μm RES. H, activation of DNA damage response in A431 cells treated with 30 μm RES for the indicated time periods, determined by Western blotting. 50 μg of total protein was used per lane. β-Actin was used as an internal loading control. All assays were performed in triplicate, and the results are expressed as the means ± S.D. of the three independent experiments. Asterisks indicate significance at levels of p ≤ 0.05. P, phosphorylated.
RES Mediates PS Induction via ROS Generation
RES is considered an anti-oxidant. However, RES is also capable of inducing intracellular ROS production (20–22) and has been shown to promote oxidative DNA degradation during the Cu(II) to Cu(I) transition in lymphocytes (23). We tested whether RES produces ROS in A431 cells. RES treatment of A431 cells caused increases in intracellular ROS concentrations, which reached maximum levels at 4 h at 30 μm (Fig. 1D) and in a dose-dependent manner in response to RES treatments between 10 μm and 100 μm (responses to treatments between 10 μm and 50 μm are shown in Fig. 1E). The SA-β-gal activity also increased gradually over time in response to RES (30 μm) treatment (Fig. 1F). RES-mediated ROS generation and SA-β-gal staining could be blocked by pretreating cells with neocuproine, a membrane-permeable Cu(I)-specific chelator, prior to RES treatment (Fig. 1G). In the absence of RES, neocuproine treatment alone did not produce significant effects on ROS generation or on cell morphology and SA-β-gal staining (Fig. 1G).
The dose- and time-dependent induction of p21WAF1, a cell cycle inhibitor associated with senescence, was detected in RES-treated A431 cells (Fig. 1, E and H). This p21WAF1 induction was p53-independent because A431 cells contain mutant p53. Furthermore, phosphorylated histone H2AX, ATM, and Chk2 (but not ATR or Chk1) accumulated in PIS A431 cells (Fig. 1, E and H). Neocuproine pretreatment abolished the RES-induced expression of p21WAF1, p-Chk2, and γH2AX (Fig. 1G). These results indicate that RES-induced PS is mediated through ROS that are likely generated via the RES-coupled redox cycling of Cu(II) to Cu(I) and that this redox reaction involves activation of a DNA damage-response pathway.
Anti-apoptotic Bfl-1/A1 Is Important for the Survival of PIS Cells, and Its Induction Is Mediated by p65/RelA and p52 NF-κB
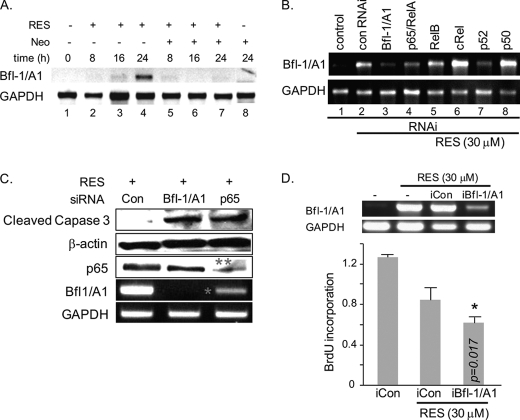
To gain insight into potential target genes regulated in RES-induced prematurely senescent A431 cells, a cDNA microarray was performed on A431 cells treated with 100 μm RES for 48 or 72 h. Analysis of the raw data, with filtering criteria of at least 2-fold changes and analysis of variance with p < 0.0001, indicates up-regulation of the cell cycle inhibitors p21WAF1 and p27KIP1 and down-regulation of cyclins D1, D2, and CDK4/5. These results were validated using real-time PCR (data not shown) and confirmed our previous findings (19). We also found that Bfl-1/A1, an anti-apoptotic Bcl-2 family gene, was dramatically up-regulated in PIS A431 cells when compared with control groups (supplemental Fig. S1). Expression of other Bcl2 family genes was not significantly altered in PIS A431 cells under these conditions, with the exception of BAD (Bcl2-antagonist of cell death) and BAK1 (Bcl2-antagonist/killer1), whose levels decreased slightly (supplemental Fig. S1). Bfl-1/A1 induction was detectable, slightly at 16 h and markedly at 24 h, following RES treatment, whereas pretreatment of cells with neocuproine blocked this induction (Fig. 2A).
FIGURE 2.
Bfl-1/A1 controls the survival of PIS A431 cells. A, neocuproine (Neo) blocks RES-mediated Bfl-1/A1 induction in A431 cells treated with RES (30 μm) and/or neocuproine (30 μm) for the indicated time periods, determined by RT-PCR. B, siRNA-mediated down-regulation of p65/RelA or p52 NF-κB suppresses Bfl-1/A1 induction in the presence of 30 μm RES. Each subunit of the NF-κB complex was down-regulated in the A431 cells by siRNA, after which the cells were treated with 30 μm RES for 48 h. con RNAi, control RNAi. C, down-regulation of either Bfl-1/A1 or p65/RelA NF-κB increases the level of cleaved active caspase-3 in PIS A431 cells, as determined by Western blot analysis. 50 μg of total protein was used per lane. β-Actin was used as an internal loading control. The knockdown efficiency of Bfl-1/A1 and p65 is indicated with * and **, respectively. D, the siRNA-mediated down-regulation of Bfl-1/A1 suppresses BrdU incorporation. A431 cells were transformed with control (iCon) or Bfl-1/A1-targeted (siBfl-1/A1) siRNA constructs. Bfl-1/A1 expression levels were assessed by RT-PCR (top panel). The cells were treated with 30 μm RES for 48 h followed by BrdU incorporation assay.
Bfl-1/A1 is a direct transcription target of NF-κB (24, 25). The NF-κB family includes p50, p52, p65/RelA, RelB, and c-Rel, which form homomeric or heteromeric dimers with one another. The involvement of p65/RelA and p52 in RES-induced Bfl-1/A1 expression was demonstrated, in which siRNA constructs targeted to either p65/RelA or p52 (but not other subunits) blocked Bfl-1/A1 expression (Fig. 2B; the knockdown efficiency is shown in supplemental Fig. S2). The levels of active caspase-3 increased following knockdown of either Bfl-1/A1 or p65/RelA (Fig. 2C). The knockdown of Bfl-1/A1 also attenuated BrdU incorporation (Fig. 2D). These data support an anti-apoptotic role for Bfl-1/A1 and indicate that RES-mediated induction of Bfl-1/A1 plays an important role in the survival of PIS A431 cells.
The Post-translational Modification of SIRT1 Regulates Bfl-1/A1 Induction
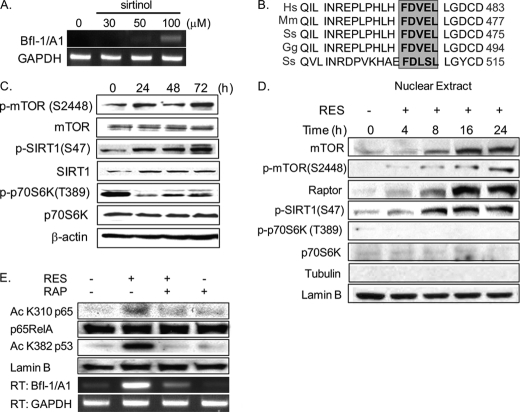
A431 cells treated with various concentrations of sirtinol, a SIRT1 inhibitor, showed enhanced Bfl-1/A1 induction (Fig. 3A), suggesting that SIRT1 may directly regulate Bfl-1/A1 expression. Interestingly, a highly conserved TOS (TOR signaling) motif (FDVEL) occurs in similar regions of the SIRT1 protein in various species (Fig. 3B, gray box). The TOS motif allows the mTOR substrate to bind to mTOR through Raptor and is present in S6K1 and 4EBP1 (26, 27). Additionally, we identified a putative mTOR phosphorylation site at serine 47 of SIRT1, suggesting the possible post-translational modification of SIRT1 by mTOR. As shown in Fig. 3C, Ser-2448 phosphorylation of mTOR, which reflects mTOR activation, increased during induction of PS by RES. However, we were surprised by the finding that phosphorylation of p70S6K, a cytoplasmic substrate of mTOR, was significantly reduced, whereas levels of phosphorylated SIRT1 (at Ser-47) increased (Fig. 3C). This discrepancy in phosphorylation pattern prompted us to examine the subcellular distribution of mTOR in PIS A431 cells. We found that mTOR, p-mTOR, Raptor, and p-SIRT1(S47) accumulated over time in cell nuclei following the onset of RES-induced PS, whereas both the total and the phosphorylated forms of p70S6K were undetectable (Fig. 3D). Because Ser-2448 is known to be a p70S6K site, our data suggest a yet-to-be-identified regulatory mechanism of mTOR in the nucleus. The SIRT1 protein is known to inhibit NF-κB-dependent transcription by deacetylating p65/RelA at Lys-310 (28). We observed an accumulation of acetyl p65/RelA(K310) in PIS A431 cells, and treating these cells with rapamycin, an mTOR inhibitor, abolished the acetylation of p65/RelA(K310) and substantially reduced Bfl-1/A1 mRNA levels (Fig. 3E). We also assessed the acetylation of p53 as a control. Although p53 is non-functional in the A431 cell line, acetylation of p53 Lys-382 was detectable in PIS A431 cells and was blocked in the presence of rapamycin (Fig. 3E). These data suggest the importance of nuclear mTOR activity in the induction of Bfl-1/A1 expression in PIS A431 cells.
FIGURE 3.
Bfl-1/A1 induction involves the nuclear accumulation of mTOR and the post-translational modification of SIRT1. A, up-regulation of Bfl-1/A1 expression in A431 cells treated with sirtinol for 24 h, as determined by RT-PCR. B, the SIRT1 protein sequence contains a TOS motif (gray box). Hs, Homo sapiens; Mm, Mus musculus; Ss, Sus scrofa; Gg, Gallus gallus. C, increases in p-mTOR(S2448) and p-SIRT1(S47) during PS induction in A431 cells treated with 30 μm RES for the indicated time periods, assessed by Western blotting. 50 μg of total protein was used per lane. β-Actin was used as an internal loading control. D, nuclear accumulation of mTOR, p-mTOR, Raptor, p-SIRT1(S47), p-p70S6K, and p70S6K in A431 cells treated with 30 μm RES for the indicated time periods, assessed by Western blotting. Lamin B and tubulin were used as internal loading controls. E, rapamycin (RAP) suppresses both the acetylation of p65/RelA NF-κB(K310) and Bfl-1/A1 induction. Nuclear fractions prepared from A431 cells treated with 30 μm RES for 24 h were assessed by Western blotting.
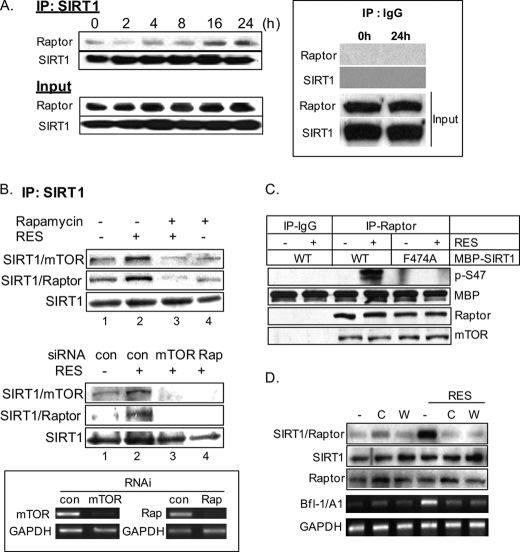
The mTOR-Raptor Complex Physically Interacts with SIRT1, and TOS Motif Is Important for Ser-47 SIRT1 Phosphorylation
Because of the presence of a putative TOS motif in SIRT1, we investigated whether SIRT1 physically interacts with Raptor during the induction of PS. Fig. 4A shows the time-dependent accumulation of the SIRT1-Raptor complex in total extracts prepared at different time points following RES treatment. This interaction between SIRT1 and mTOR-Raptor was markedly reduced in cells that were pretreated with rapamycin (Fig. 4B, lane 3, upper panel) or by siRNA-mediated down-regulation of either mTOR or Raptor (Fig. 4B, lanes 3 and 4, middle panel). A substitution of the phenylalanine residue at amino acid 474 in the TOS motif to alanine prevented Ser-47 phosphorylation, as determined by in vitro mTOR-dependent kinase assay using recombinant MBP-tagged wild-type SIRT1 and SIRT1F474A proteins as the kinase substrate (Fig. 4C). These data indicate that the presence of TOS in SIRT1 is required for Ser-47 phosphorylation by the rapamycin-sensitive mTOR complex 1 (mTORC1) in response to RES. Furthermore, the SIRT1-Raptor complex formation and Bfl-1/A1 induction were blocked in the presence of ATM inhibitors (caffeine and wortmannin), confirming involvement of a DNA damage response (Fig. 4D).
FIGURE 4.
SIRT1 physically interacts with an mTOR complex, and TOS motif is important for Ser-47 SIRT1 phosphorylation. A, SIRT1 and Raptor form a complex during PS induction. Whole cell lysates from A431 cells treated with 30 μm RES for the indicated time periods were immunoprecipitated with anti-SIRT1 antibody followed by Western blotting with anti-Raptor antibody and with anti-SIRT1 antibody (upper panel, IP:SIRT1). The original cell lysates were also analyzed with the same antibodies (lower panel, Input). The representative immunoprecipitation control with normal IgG (IP: IgG) is shown in the inset. B, Western blotting analysis of nuclear immunoprecipitates prepared from cells treated with RES in the presence or absence of rapamycin (upper panel) or from cells that had been transfected with control constructs (con) or siRNA constructs targeted to either mTOR or Raptor (Rap) (middle panel). The knockdown efficiency of mTOR and Raptor is shown in the inset. *, p < 0.05 when compared with control + RES. C, the TOS motif is required for in vitro Ser-47 phosphorylation. Total extracts prepared from A431 cells treated or not treated with RES were immunoprecipitated with anti-Raptor antibody. Immunoprecipitates were then subjected to an in vitro mTOR-dependent kinase assay using recombinant wild-type MBP-SIRT1 or mutant MBP-SIRT1F474A as kinase substrates followed by Western blotting for p-SIRT1 (S47), MBP, Raptor, and mTOR. The immunoprecipitation control with normal IgG is shown (IP-IgG). The MBP lane indicates the recombinant substrates present in kinase reactions. D, caffeine (C, 1 mm) or wortmannin (W, 5 μm) blocks the formation of the SIRT1-Raptor complex in A431 cells in the presence of RES at 24 h. Total lysates (500 μg) were immunoprecipitated with anti-SIRT1 antibody followed by Western blotting with anti-Raptor antibody. Total levels of SIRT1 and Raptor in the lysates were assessed by Western blotting. Bfl-1/A1 levels were assessed by RT-PCR.
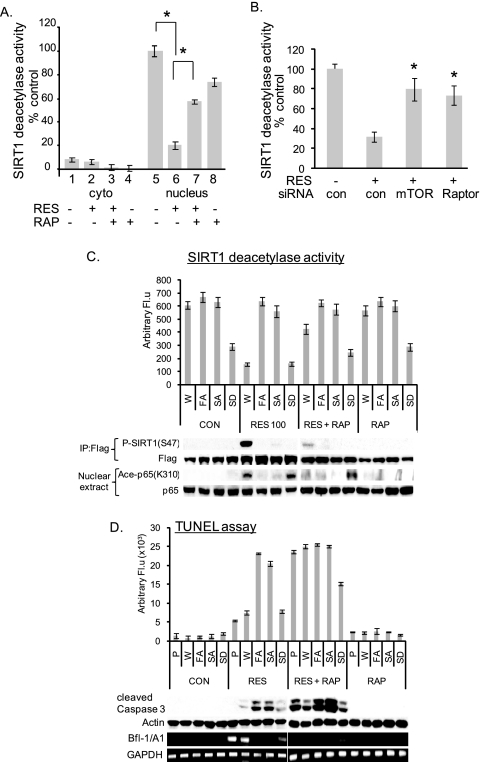
Ser-47 Phosphorylation Directly Affects SIRT1 Deacetylase Activity and the Sensitivity of Cells for Apoptosis
We further tested the effect of mTOR on SIRT1 deacetylase activity. We used 2′-O-acetyl-ADP-ribose as a substrate in an assay to measure SIRT1 deacetylase activity. SIRT1 deacetylase activity was mostly detected in the nuclear fractions of proliferating A431 cells (Fig. 5A, lane 1 versus lane 5), whereas its activity decreased in RES-treated A431 cells (Fig. 5A, lane 6). Treating cells with rapamycin or siRNA-mediated knockdown of either mTOR or Raptor partially restored SIRT1 deacetylase activity in the presence of RES (Fig. 5, A and B). We also observed that SIRT1 activity was inhibited when cells were treated with rapamycin alone (Fig. 5A, lane 5 versus lane 8). Because the rapamycin-sensitive mTORC1 is known to interact with the translation initiation complex and regulate translation initiation and ribosome biogenesis (29), the observed inhibitory effect of rapamycin on SIRT1 activity may be due to the inhibitory effects of rapamycin on SIRT1 translation. Although this requires further investigation, slight decreases in the levels of SIRT1 protein were observed in cells treated with rapamycin (Fig. 5C, CON versus RAP, FLAG Western blot). In support, sirolimus (rapamycin) was shown to induce cellular senescence in endothelial cells by down-regulating SIRT1 (30). To determine directly the effects of Ser-47 phosphorylation on SIRT1 deacetylase activity, A431 cells stably expressing unphosphorylatable SIRT1S47A, phosphomimetic SIRT1S47D, or a TOS mutant (SIRT1F474A) protein were treated with RES for 48 h. The presence of mutant SIRT1S47A or SIRT1F474A restored SIRT1 deacetylase activity, whereas phosphomimetic SIRT1S47D did not (Fig. 5C). The Ser-47 phosphorylation observed in response to RES and in the presence of wild-type SIRT1 was undetectable in the presence of SIRT1S47A or SIRT1F474A, and this decrease in Ser-47 phosphorylation correlated with decreased p65/RelA acetylation (Fig. 5C). Furthermore, the presence of SIRT1S47A or SIRT1F474A or the treatment of WT SIRT1-expressing PIS A431 cells with rapamycin decreased the levels of Bfl-1/A1; meanwhile, we observed increases in the number of TUNEL-positive cells and the levels of cleaved caspase-3 (Fig. 5D). These data indicate that Ser-47 phosphorylation has a direct inhibitory effect on SIRT1 deacetylase activity, leading to cell survival in PIS A431 cells.
FIGURE 5.
Ser-47 phosphorylation directly affects SIRT1 deacetylase activity and the sensitivity of cells for apoptosis. A, SIRT1 deacetylase activity is inhibited in PIS A431 cells. SIRT1 was immunoprecipitated from the nuclear and cytoplasmic (cyto) extracts of A431 cells treated or not treated with RES and/or rapamycin (RAP) using the anti-SIRT1 antibody. SIRT1 enzymatic activity was then measured. B, the SIRT1 deacetylase activity in PIS A431 cells is partially rescued following the siRNA-mediated knockdown of mTOR or Raptor. Experiments were performed as in Fig. 4B. con, control. C, the deacetylase activity of SIRT1 mutants was determined in FLAG immunoprecipitates. Extracts prepared from A431 cells stably expressing FLAG-tagged wild-type SIRT1 (W), SIRT1F474A (FA), SIRT1S47A (SA), or SIRT1S47D (SD) constructs and treated or not treated with RES for 48 h were immunoprecipitated with anti-FLAG antibody. Each bar represents the mean of three independent experiments, p < 0.05 when compared with wild-type SIRT1. The levels of P-SIRT1 (S47), FLAG, acetyl-p65, and p65 were detected by Western blotting. D, mutations in Phe-474 and Ser-47 sensitized RES-treated A431 cells for apoptosis. Quantification of apoptotic cells was by TUNEL assay. Each bar represents the mean of three independent experiments, p < 0.02 (W versus FA and W versus SA). Bfl-1/A1 and GAPDH expression was determined by RT-PCR, and the levels of cleaved caspase-3 were determined by Western blotting. The experiments in C and D were performed as in A, in the presence or absence of rapamycin. W, pBABEpuro-FlagSIRT1-wild type; FA, pBABEpuro-FlagSIRT1F474A mutants; SA, pBABEpuro-FlagSIRT1S47A mutants; SD, pBABEpuro-FlagSIRT1S47D mutants. Arbitrary Fl.u, arbitrary fluorescence units.
Rapamycin Sensitizes DNA Damage-induced PIS SCC Cells for Apoptosis
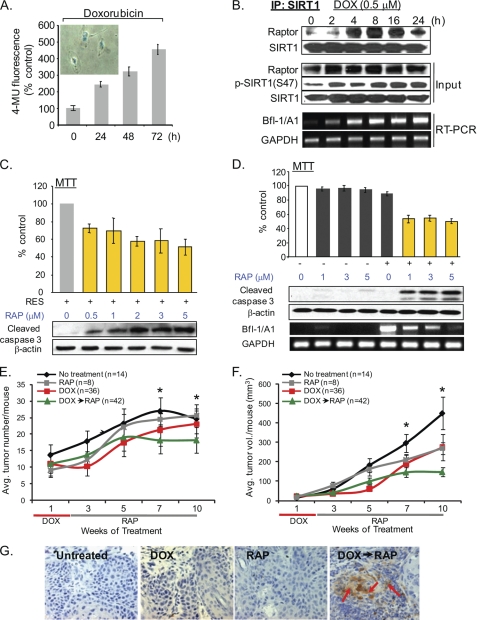
To investigate whether other anti-cancer drugs exhibit a similar cell survival mechanism, we employed sublethal doses of the anti-cancer drug doxorubicin (DOX) to assess its effects on the induction of PS (Fig. 6A). Gradual increases in the formation of the SIRT1-Raptor complex and SIRT1(S47) phosphorylation, along with increased Bfl-1/A1 expression, were detected in DOX-induced PIS A431 cells (Fig. 6B). Rapamycin treatment of the RES- or DOX-induced PIS A431 cells resulted in dose-dependent increases in active caspase-3 levels and decreases in Bfl-1/A1 expression (Fig. 6, C and D). Rapamycin alone did not affect the viability of the proliferating A431 cells (Fig. 6D). These data indicate that mTOR-mediated SIRT1 modification leading to the survival of PIS A431 cells is not exclusive to RES.
FIGURE 6.
The rapamycin treatment sensitizes RES- and DOX-induced PIS cancer cells for apoptosis. A, DOX induces PS in A431 cells. A431 cells treated with 0.5 μm DOX show time-dependent increases in SA-β-gal activity. Inset, SA-β-gal staining of A431 cells following DOX treatment. 4-MU, 4-methylumbelliferone. B, SIRT1 phosphorylation and its interaction with Raptor increases during the DOX-mediated induction of PS. A431 cells were treated with 0.5 μm DOX for the indicated time periods, and then cell lysates (500 μg) were prepared and immunoprecipitated with anti-SIRT1 antibody. The immunoprecipitates were analyzed by Western blotting with the anti-Raptor and SIRT1 antibodies. The levels of Raptor, p-SIRT1(S47), and SIRT1 were also analyzed in the original lysates (Input). C and D, the levels of Bfl-1/A1 expression in the same cell samples were analyzed by RT-PCR. RES (C)- or DOX (D)-induced PIS A431 cells were treated with various doses of rapamycin (RAP, 0–5 μm) for 24 h. Cell viability was assessed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays (histogram), and the levels of cleaved caspase-3 were analyzed by Western blotting. 50 μg of total protein was used per lane. β-Actin was used as an internal loading control. The levels of Bfl-1/A1 expression in the same cell samples were analyzed by RT-PCR. E–G, the rapamycin treatment suppresses the growth and induces apoptosis of DOX-treated SCCs in vivo. Tumors generated in p53+/−/SKH-1 mice following chronic UVB irradiation were treated with DOX (1 mg/kg, i.p., 5 days/week for 2 weeks) followed by rapamycin treatment (10 mg/kg, i.p., biweekly for an additional 8 weeks. The number (E) and size (F) of SCCs were measured. Asterisks indicate p < 0.05 between DOX→rapamycin (RAP) versus other groups. G, immunohistochemical detection of active caspase-3 in SCCs harvested from non-treated, rapamycin-, DOX-, or DOX/rapamycin-treated mice. 200× magnification. Avg. tumor, average tumor; vol., volume.
We then determined the in vivo effects of mTOR inhibition in p53+/−/SKH-1 mice. These mice develop cutaneous SCCs in response to chronic UV irradiation at a much faster rate than the wild-type littermates, and many of their tumors exhibit a highly invasive tumor phenotype (31). In addition, tumors induced in this murine model carry one mutant and one deleted p53 allele. Thus, these animals provide a unique opportunity to investigate the p53-independent biological responses. For this study, 100 p53+/−/SKH-1 mice were irradiated with 180 mJ/cm2 UVB twice per week for 25 weeks until the number of SCCs reached an average of 10 per mouse. These tumor-bearing mice were then divided into four groups. Group 4 mice were treated with DOX (1 mg/kg, i.p., 5 days/week) for 2 weeks to induce senescence followed by rapamycin treatment (10 mg/kg, i.p., biweekly) for an additional 8 weeks. Mice in Groups 2 and 3 only received rapamycin or doxorubicin, respectively. Group 1 mice served as vehicle controls. The DOX treatment initially stabilized tumor growth; however, the tumor growth eventually resumed upon DOX withdrawal (Fig. 6, E and F). Rapamycin treatment alone did not have significant effects on the number of tumors (Fig. 6E), but it decreased tumor size by 40% when compared with untreated control mice (Fig. 6F). Notably, rapamycin treatment of DOX-treated tumors further reduced both tumor numbers and sizes (Fig. 6, E and F). Consistent with our in vitro data (Fig. 6D), at week 10, caspase-3 activation was detected only in tumors treated with DOX followed by rapamycin, but not in tumors treated with only DOX or rapamycin (Fig. 6G).
DISCUSSION
Our observation that the survival of DNA damage-induced prematurely senescent A431 cells involves phosphorylation-dependent inhibition of SIRT1 deacetylase activity provides a novel regulatory loop of SIRT1 activity. Our results indicate that mTOR is a cellular regulator of SIRT1 activity in response to DNA damage. mTOR is predominantly a cytoplasmic protein; it primarily regulates proteins involved in translation. However, a small fraction of mTOR is detected in the nucleus under normal conditions, and mTOR shuttles between the cytoplasm and nucleus (32). An intrinsic nuclear import signal has been identified in mTOR, and its cytoplasmic-nuclear shuttling appears to be important for its cytoplasmic regulation of S6K1 (33). mTOR has been shown to accumulate in the nucleus following hypoxia. Moreover, PML was shown to physically interact with mTOR, favoring mTOR nuclear accumulation and resulting in reduced tumor angiogenesis (34, 35). However, the signals and mechanisms controlling the nuclear translocation of mTOR, and its functional status in the nucleus, remain largely unknown. In particular, the role of nuclear mTOR in the context of PS has not previously been addressed. In PIS A431 cells, the nuclear accumulation of mTOR corresponded to a marked decrease in the phosphorylation levels of its cytoplasmic targets and was accompanied by the up-regulation of a cell survival gene, Bfl-1/A1. These data suggest that mTOR has a yet-to-be-identified regulatory mechanism in the nucleus, in addition to playing a role in enhancing cytoplasmic signaling. The mechanisms underlying the nuclear translocation of mTOR, such as whether the process requires catalytic activity, are not known. In the presence of p53, genotoxic stress was suggested to inhibit mTOR via the p53-regulated genes Sestrin1 and Sestrin2 (36), thus providing a mechanistic link between genotoxic stress and mTOR. A direct role for mTOR signaling in DNA damage response in p53 mutant cancer cells is not clear. Our results indicate that DNA damage-dependent activation of mTOR is critically important for the survival of PIS A431 cells, in which the mTOR complex physically interacts with SIRT1, likely via the TOS motif. The importance of the TOS motif was further demonstrated as SIRT1-Raptor interaction was substantially reduced by the F474A mutation (supplemental Fig. S3). However, because the knockdown of mTOR also abolished the interaction of SIRT1 and Raptor (Fig. 4B), our data raise the possibility that the physical interaction between SIRT1 and mTOR-Raptor may not be entirely mediated via the TOS motif. The exact mechanism(s) of SIRT1 interaction with mTORC1, and whether additional mechanism(s) exist, is not clear at the present time. Of interest, PRAS40, an mTORC1 subunit and substrate, also binds to Raptor via a TOS motif in PRAS40 (37, 38). Binding of PRAS40 to Raptor inhibits mTORC1 signaling as it blocks access of TOS-mediated substrates (e.g. 4EBP1) to Raptor (39, 40). A release of PRAS40 from the substrate binding site of Raptor is believed to allow substrate presentation to mTORC1 and subsequent phosphorylation (39). Although mTOR knockdown has been shown to disrupt the mTOR-PRAS40 interaction, the total endogenous levels of PRAS40 were not affected (39). It is tempting to speculate that in the absence of mTOR, and therefore in the absence of mTOR-dependent phosphorylation of PRAS40, PRAS40 could remain bound to Raptor, thereby blocking SIRT1 access to Raptor, in which case the knockdown of mTOR would likely prevent SIRT1-Raptor interaction. Moreover, the incubation of mTORC1 with rapamycin leads to a complete disintegration of mTORC1, which has been shown to exist as an obligate dimer with a central cavity and with Raptor, PRAS40, and mLSt8 bound to mTOR (38, 41). This obligate dimeric structure may be necessary for performing multiple phosphorylations, as may be the case for PRAS40 (37). In this scenario, destabilization of mTORC1 by rapamycin could block PRAS40 release from Raptor, which may explain the lack of SIRT1-Raptor interaction in cells treated with RES and rapamycin (Fig. 4B).
The interaction between mTOR complex and SIRT1 appears to be a prerequisite for Ser-47 phosphorylation. SIRT1 is a phosphoprotein (42), and several studies show the implication of the post-translational regulation of SIRT1 for its biological functions. SIRT1 phosphorylation by JNK2 at serine 27 was shown to increase SIRT1 protein stability, resulting in elevated SIRT1 protein levels in human cancer cells (43). SIRT1 was also shown to be phosphorylated by cyclin B/Cdk1 at threonine 530 and serine 540 (32) and by the dual specificity, tyrosine phosphorylation-regulated kinase 1A (DYRK1A) and another pro-survival member of the DYRK family, DYRK3 at Thr-522, leading to cell survival in HEK293T cells (44). Unlike our data, those of Nasrin et al. (45) report that phosphorylations of three residues (Ser-27, Ser-47, and Thr5–30) by JNK1 increase its enzymatic activity; however, their study did not determine the effect of Ser-47 phosphorylation alone on its activity (45). It is interesting to note that multiple phosphorylations can lead to quite distinct phenotypes. These data implicate the complexity of SIRT1 regulation as its activity is not only distinctively regulated by various kinases but may also be differentially regulated by various stimuli in a context-dependent manner.
The factors regulating the reversibility of PS remain to be defined. However, several in vitro studies have shown that senescent cells (p53-null lung cancer) can escape the senescent state and re-enter the cell cycle (5, 9). Similarly, cancer cells (p53-null) following oxidative stress are arrested in a senescence-like state, but they eventually re-enter the cell cycle (46). Our DOX-treated, growth-stabilized SCCs in p53+/−/SKH-1 mice also resumed growth upon DOX withdrawal. Our study raises questions about the biological consequences of drug therapies that induce prematurely senescent, but viable, cancer cells. These cells could escape from senescence and ultimately contribute to tumor regrowth and recurrence. The therapeutic efficacy of these treatments is likely to depend on whether senescence is followed by cell death (18). Our data provide the first functional link between mTOR and SIRT1 in response to genotoxic stress and reveal a p53-independent mechanism that controls the survival of PIS SCC cells. These results further suggest that inhibition of mTOR signaling could eliminate tumors containing senescent cells and block tumor recurrences.
Supplementary Material
Acknowledgment
We are grateful to Dr. David R. Bickers for insightful and valuable comments.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES015323 (to M. A.) and K01-AR048582 and R03 CA125855 (to A. L. K.). This work was also supported by an Irving Scholar Award and Skin Cancer Foundation grant (to A. L. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PS
- premature senescence
- PIS
- prematurely induced senescent
- ATM
- ataxia telangiectasia, mutated
- ATR
- ATM and Rad3-related
- Bfl-1/A1
- Bcl2-related protein A1
- Chk1/2
- checkpoint kinase 1/2
- DOX
- doxorubicin
- DYRK
- the dual specificity, tyrosine phosphorylation-regulated kinase
- MBP
- maltose-binding protein
- mTOR
- mammalian target of rapamycin
- mTORC1
- the rapamycin-sensitive mTOR complex 1
- PML
- promyelocytic leukemia protein
- RES
- resveratrol
- ROS
- reactive oxygen species
- SA-β-gal
- senescence-associated β-galactosidase
- SCC
- squamous cell carcinoma
- SIRT1
- sirtuin 1
- hSIRT1
- human SIRT1
- mtHSP70
- mitochondrial HSP70
- TOS
- TOR signaling.
REFERENCES
- 1. Campisi J. (2005) Cell 120, 513–522 [DOI] [PubMed] [Google Scholar]
- 2. Jacobs J. J., de Lange T. (2005) Cell Cycle 4, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 3. Campisi J. (2005) Science 309, 886–887 [DOI] [PubMed] [Google Scholar]
- 4. Roninson I. B. (2003) Cancer Res. 63, 2705–2715 [PubMed] [Google Scholar]
- 5. Roberson R. S., Kussick S. J., Vallieres E., Chen S. Y., Wu D. Y. (2005) Cancer Res. 65, 2795–2803 [DOI] [PubMed] [Google Scholar]
- 6. te Poele R. H., Okorokov A. L., Jardine L., Cummings J., Joel S. P. (2002) Cancer Res. 62, 1876–1883 [PubMed] [Google Scholar]
- 7. Saretzki G. (2010) Curr. Pharm. Des. 16, 79–100 [DOI] [PubMed] [Google Scholar]
- 8. Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., Beach D., Serrano M. (2005) Nature 436, 642–642 [DOI] [PubMed] [Google Scholar]
- 9. Elmore L. W., Di X., Dumur C., Holt S. E., Gewirtz D. A. (2005) Clin. Cancer Res. 11, 2637–2643 [DOI] [PubMed] [Google Scholar]
- 10. Ling Y. H., Zou Y., Perez-Soler R. (2000) Anticancer Res. 20, 693–702 [PubMed] [Google Scholar]
- 11. Evan G. I., d'Adda di Fagagna F. (2009) Curr. Opin. Genet. Dev. 19, 25–31 [DOI] [PubMed] [Google Scholar]
- 12. Wang Q., Wu P. C., Roberson R. S., Luk B. V., Ivanova I., Chu E., Wu D. Y. (2011) Int. J. Canc. 128, 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ansieau S., Bastid J., Doreau A., Morel A. P., Bouchet B. P., Thomas C., Fauvet F., Puisieux I., Doglioni C., Piccinin S., Maestro R., Voeltzel T., Selmi A., Valsesia-Wittmann S., Caron de Fromentel C., Puisieux A. (2008) Cancer Cell 14, 79–89 [DOI] [PubMed] [Google Scholar]
- 14. Gary R. K., Kindell S. M. (2005) Anal. Biochem. 343, 329–334 [DOI] [PubMed] [Google Scholar]
- 15. Rezvani H. R., Kim A. L., Rossignol R., Ali N., Daly M., Mahfouf W., Bellance N., Taïeb A., de Verneuil H., Mazurier F., Bickers D. R. (2011) J. Clin. Invest. 121, 195–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim K. H., Back J. H., Zhu Y., Arbesman J., Athar M., Kopelovich L., Kim A. L., Bickers D. R. (2011) J. Invest. Derm. 131, 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim A. L., Athar M., Bickers D. R., Gautier J. (2002) J. Invest. Derm. 118, 818–824 [DOI] [PubMed] [Google Scholar]
- 18. Gewirtz D. A., Holt S. E., Elmore L. W. (2008) Biochem. Pharmacol. 76, 947–957 [DOI] [PubMed] [Google Scholar]
- 19. Kim A. L., Zhu Y., Zhu H., Han L., Kopelovich L., Bickers D. R., Athar M. (2006) Exp. Dermatol. 15, 538–546 [DOI] [PubMed] [Google Scholar]
- 20. Galati G., Sabzevari O., Wilson J. X., O'Brien P. J. (2002) Toxicology 177, 91–104 [DOI] [PubMed] [Google Scholar]
- 21. Murias M., Jäger W., Handler N., Erker T., Horvath Z., Szekeres T., Nohl H., Gille L. (2005) Biochem. Pharmacol. 69, 903–912 [DOI] [PubMed] [Google Scholar]
- 22. Tyagi A., Singh R. P., Agarwal C., Siriwardana S., Sclafani R. A., Agarwal R. (2005) Carcinogenesis 26, 1978–1987 [DOI] [PubMed] [Google Scholar]
- 23. Azmi A. S., Bhat S. H., Hanif S., Hadi S. M. (2006) FEBS. Lett. 580, 533–538 [DOI] [PubMed] [Google Scholar]
- 24. Wang C. Y., Guttridge D. C., Mayo M. W., Baldwin A. S., Jr. (1999) Mol. Cell. Biol. 19, 5923–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H., Kim Y. N., Kim H., Kim C. W. (2005) Oncogene 24, 1252–1261 [DOI] [PubMed] [Google Scholar]
- 26. Schalm S. S., Blenis J. (2002) Curr. Biol. 12, 632–639 [DOI] [PubMed] [Google Scholar]
- 27. Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 28. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 30. Ota H., Eto M., Ako J., Ogawa S., Iijima K., Akishita M., Ouchi Y. (2009) J. Am. Coll. Cardiol. 53, 2298–2305 [DOI] [PubMed] [Google Scholar]
- 31. van Kranen H. J., Westerman A., Berg R. J., Kram N., van Kreijl C. F., Wester P. W., de Gruijl F. R. (2005) Mutat. Res. 571, 81–90 [DOI] [PubMed] [Google Scholar]
- 32. Kim J. E., Chen J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14340–14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bachmann R. A., Kim J. H., Wu A. L., Park I. H., Chen J. (2006) J. Biol. Chem. 281, 7357–7363 [DOI] [PubMed] [Google Scholar]
- 34. Paglin S., Lee N. Y., Nakar C., Fitzgerald M., Plotkin J., Deuel B., Hackett N., McMahill M., Sphicas E., Lampen N., Yahalom J. (2005) Cancer Res. 65, 11061–11070 [DOI] [PubMed] [Google Scholar]
- 35. Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J., Cordon-Cardo C., Simon M. C., Rafii S., Pandolfi P. P. (2006) Nature 442, 779–785 [DOI] [PubMed] [Google Scholar]
- 36. Budanov A. V., Karin M. (2008) Cell 134, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fonseca B. D., Smith E. M., Lee V. H., MacKintosh C., Proud C. G. (2007) J. Biol. Chem. 282, 24514–24524 [DOI] [PubMed] [Google Scholar]
- 38. Oshiro N., Takahashi R., Yoshino K., Tanimura K., Nakashima A., Eguchi S., Miyamoto T., Hara K., Takehana K., Avruch J., Kikkawa U., Yonezawa K. (2007) J. Biol. Chem. 282, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L., Harris T. E., Roth R. A., Lawrence J. C., Jr. (2007) J. Biol. Chem. 282, 20036–20044 [DOI] [PubMed] [Google Scholar]
- 40. Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 41. Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A. (2010) Mol. Cell 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ford J., Ahmed S., Allison S., Jiang M., Milner J. (2008) Cell Cycle 7, 3091–3097 [DOI] [PubMed] [Google Scholar]
- 44. Guo X., Williams J. G., Schug T. T., Li X. (2010) J. Biol. Chem. 285, 13223–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) PLoS. One 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Macip S., Kosoy A., Lee S. W., O'Connell M. J., Aaronson S. A. (2006) Oncogene 25, 6037–6047 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.