Abstract
This study describes mitochondrial behaviour during the C2C12 myoblast differentiation program and proposes a proteomic approach to mitochondria integrated with classical morphofunctional and biochemical analyses. Mitochondrial ultrastructure variations were determined by transmission electron microscopy; mitochondrial mass and membrane potential were analysed by Mitotracker Green and JC-1 stains and by epifluorescence microscope. Expression of PGC1α, NRF1α, and Tfam genes controlling mitochondrial biogenesis was studied by real-time PCR. The mitochondrial functionality was tested by cytochrome c oxidase activity and COXII expression. Mitochondrial proteomic profile was also performed. These assays showed that mitochondrial biogenesis and activity significantly increase in differentiating myotubes. The proteomic profile identifies 32 differentially expressed proteins, mostly involved in oxidative metabolism, typical of myotubes formation. Other notable proteins, such as superoxide dismutase (MnSOD), a cell protection molecule, and voltage-dependent anion-selective channel protein (VDAC1) involved in the mitochondria-mediated apoptosis, were found to be regulated by the myogenic process. The integration of these approaches represents a helpful tool for studying mitochondrial dynamics, biogenesis, and functionality in comparative surveys on mitochondrial pathogenic or senescent satellite cells.
1. Introduction
Skeletal muscle represents an important model for studying mitochondrial behaviour during cell growth and differentiation. Myoblasts cultured in vitro, if induced by cell confluence and serum deprivation, follow a myogenic program, which includes an active proliferation, withdrawal from the cell cycle, synthesis of muscle-specific proteins, and fusion into multinucleated myotubes [1, 2]. This event is accomplished by the activation of specific myogenic regulatory factors (MRFs) [3–5].
Recent studies suggest that mitochondria are involved in the regulation of the skeletal muscle physiology and play a critical role in cell growth, cell proliferation, cell death, and cell differentiation [6–13]. In particular, mitochondrial activity is involved in the regulation of myoblast differentiation through myogenin expression, the activity of myogenic factors, and by control of c-Myc expression [8, 14, 15]. Furthermore, differentiation appears to be a program which is dependent on both mitochondrial function and mitochondrial biogenesis, as indicated by the rapid increase in mitochondrial mass/volume, mtDNA copy number, mitochondrial enzyme activities, and mRNA levels within the first 48 hrs of myoblast differentiation [6, 7]. Mitochondrial DNA transcription and replication are key events in cellular differentiation, which requires interaction between the nucleus and the mitochondrion [16].
Several aging theories are associated with mitochondrial damage or with a decline in mitochondrial energy production in which links between mitochondria genome expression and senescence symptoms are not always recognized [17–19]. Our interest is particularly focused on the role that mitochondria may play in the proliferative and differentiation capacity of satellite stem cells. It is well documented that with aging, satellite stem cells lose both mitogenic and myogenesis abilities and may decrease in numbers in both mice and humans [20–23]. The C2C12 cell line satellite myoblasts could offer a suitable model for studying mitochondrial behaviour during the differentiation program.
In this study, we combined a morphological and bio-molecular approach to analyze changes in mitochondrial phenotype, ultrastructure, biogenesis, and functional activity during C2C12 myoblast differentiation. Although the contribution of the proteomic profile of mitochondria during the myogenesis program is significant, it has not been described in the literature. In this paper we aim to better define the involvement of mitochondria in the regulation of muscle cell differentiation and discover new proteins potentially involved in the crosstalk between nuclei and mitochondria.
2. Material and Methods
2.1. Cell Line
Mouse C2C12 myoblasts were grown in flasks in the presence of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine at 37°C, and 5% CO2. To induce myogenic differentiation, when 80%–90% confluence was obtained, the medium was changed to DMEM supplemented with 1% FBS. Cells were analyzed at the undifferentiated stage and at the early-, middle-, and late-differentiation stage. In order to eliminate divergences in the differentiation time points analyzed, we assessed several differentiation markers. The cells, grown in the presence of 10% fetal calf serum until 80% cell confluence, were considered undifferentiated cells, corresponding to day 0 of the differentiation process (T0). To induce differentiation, cells at T0 were switched to differentiation medium. They were analyzed in the early-differentiation stage, 24 h after serum removal (T1), in mid-differentiation, 3–5 days after serum removal, when myotubes containing one of two nuclei appeared (T3–5), and in the late-differentiation stage, that is, 7–10 days after serum removal, in the presence of long multinucleated myotubes (T7-10).
2.2. Estimation of Myoblast Fusion
Myoblasts and myotubes were methanol fixed and air dried under different experimental conditions. They were then stained with water 1 : 10 May Grunwald-Giemsa solution, washed, and mounted to evaluate cell fusion. Cells were considered fused if they contained two nuclei within one cytoplasmic continuity as reported by Ferri et al. [5]. The fusion percentage was evaluated as the number of nuclei in myotubes divided by the total number of nuclei in myoblasts and myotubes magnified by 100 (×40 objective) using a TE 2000-S Nikon reverted microscope (RM) with a digital Nikon DN100 acquisition system. Twenty optical fields were randomly chosen. Data were expressed as means ± S.E.M.
2.3. Mitochondrial Ultrastructure
Undifferentiated and differentiated cell monolayers were washed and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer for 15 min, gently scraped, and centrifuged at 1200 rpm.
Cell pellets, as well as purified mitochondria, were further fixed by glutaraldehyde for 1 h. All specimens were OsO4 postfixed, alcohol dehydrated, and embedded in araldite, as previously described [24]. Thin sections were stained with uranyl acetate and lead citrate and analysed with a Philips CM10 electron microscope. Mitochondrial density was calculated in 20 different areas of 10 × 15 cm at 28000 magnification. Mitochondrial sizes were evaluated at 28000 magnification using the Philips CM10 microscope and Megaview software system.
2.4. Mitochondrial Mass and Membrane Potential
The fluorescent dye Mito Tracker Green FM (Molecular Probes), which covalently binds to mitochondrial proteins by reacting with free thiol groups of cysteine residues regardless of membrane potential (DWm) and JC-1 (Molecular Probes), a mitochondrial membrane potential sensor, were used to monitor mitochondrial mass and membrane potential respectively [25, 26]. The medium was removed from the culture dish and replaced with prewarmed growth medium containing 100 nM Mito Tracker Green or 2 μg/mL JC-1. After incubation for 20 min at 37°C, cells were immediately washed twice in cold PBS and analyzed using a Zeiss LSM 510 metaconfocal microscope. The variation of JC-1 signals was also analyzed by Zeiss LSM Image Examiner software.
2.5. Nucleic Acid Extraction and cDNA Synthesis
At each differentiation step, plates (n = 3) were washed with PBS, and nucleic acids were isolated. Total DNA and total RNA were extracted using QIAmp DNA kit (Qiagen, Chatsworth, Calif, USA) and RNeasy Mini Kit (Qiagen, Chatsworth, Calif, USA), respectively, following the manufacturer's instructions. Nucleic acid concentrations were estimated spectrophotometrically (DU-640; Beckman Instruments, Milan, Italy) at 260 nm. One microgram of DNase-treated total RNA was reverse transcribed using Omniscript RT (Qiagen, Chatsworth, Calif, USA) and random hexamers in a final volume of 20 μL as suggested in the manufacturer's protocol.
2.6. Construction of the Reference Plasmid pDGC
To construct the reference plasmid pDGC, a 98 bp amplicon of the mouse GAPDH, Acc. no. NM_008084, and a 100 bp sequence of mouse mtDNA located within the COXII, Acc. no. NP_904331.1, were inserted into the TA cloning and HindIII restriction sites, respectively, of the polylinker region of pDrive (Qiagen, Chatsworth, Calif, USA). The resultant dual-insert plasmid of 4048 bp, renamed pDGC, was purified by using DNA plasmid purification Kit (Qiagen, Chatsworth, Calif, USA) and was verified as having only one copy of each insert by restriction enzyme digestion as well as DNA sequencing. Plasmid concentration was estimated spectrophotometrically (DU-640; Beckman Instruments, Milan, Italy) at 260 nm and was adjusted to give a stock solution of 1 × 109 molecule/μL. Further 10-fold serial dilutions down to a concentration of 1 × 101 molecule/μL were prepared.
2.7. Determination of mtDNA Content and mRNA Expression Levels by Quantitative Real-Time PCR
All quantitative real-time PCR reaction were carried out in a Bio-Rad iCycler iQ Multi-Color Real-Time PCR Detection System using 2x Quantitect SYBR Green PCR kit (Qiagen). The PCR conditions were set up as follows: hot start at 95°C for 10 min then 40 cycles of the two steps at 95°C for 30 sec and at 60°C for 30 sec. Reaction mix (25 μL final volume) consisted of 12.5 μL Mix Hot-Start (Qiagen), total DNA (50 ng) or cDNA (1 μL) template, 2 μL SYBR Green, and 0.3 μM of each primer (Table 1). Threshold cycle (Ct) was determined on the linear phase of PCRs using the iCycler iQ Optical System software version 3 (BioRad, Milan, Italy). The specificity of the amplification products obtained was confirmed by examining thermal denaturation plots, by sample separation in a 3% DNA agarose gel and by sequencing. A precise determination of mitochondrial DNA (mtDNA) copy number was determined amplifying both COXII and GAPDH as mtDNA and nDNA targets, respectively. Quantification of mtDNA was performed by reference to a single recombinant plasmid (pDGC) containing a copy of each target DNA sequence (mitochondrial and nuclear). COXII and GAPDH gene copy number were determined by interpolating the threshold cycle (Ct) from standard curves that were obtained using serial dilution of the recombinant plasmid pDGC. The mtDNA/nDNA ratio was obtained, relating the mitochondrial and nuclear DNA quantities. The relative expression of Tfam, PGC1- α transcription factors, and COXII were quantified using 1 μL of cDNA template and the PCR condition already described above. The amount of each target transcript was related to that of the reference gene (the ribosomal protein S16) using the method described by Pfall [28]. In fact, previous experiments have shown that S16 mRNA is stable during the differentiation process [5]. All oligonucleotide primers were designed using Primer Express version 1.0 (Perkin-Elmer Applied Biosystem) from the GenBank database and are listed in Table 1.
Table 1.
List of primer pairs.
| Genes | Primers (forward) | Primers (reverse) | References |
|---|---|---|---|
| Mouse COXII | 5′-CATCTGAAGACGTCCTCCACTCAT-3′ | 5′-TCGGTTTGATGTTACTGTTGCTTGAT-3′ | this study |
| Mouse TfamA | 5′-GGGAGCTACCAGAAGCAGAA-3′ | 5′-CTTTGTATGCTTTCCACTCAGC-3′ | this study |
| Mouse PGC1-α | 5′-CGGAAATCATATCCAACCAG-3′ | 5′-TGAGGACCGCTAGCAAGTTTG-3′ | [27] |
| Mouse S16 | 5′-TGAAGGGTGGTGGACATGTG-3′ | 5′-AATAAGCTACCAGGGCCTTTGA-3′ | [5] |
| Mouse GAPDH | 5′-TGACGTGCCGCCTGGAGAAA-3′ | 5′-AGTGTAGCCCAAGATGCCCTTCAG-3′ | [27] |
2.8. Preparation of Mitochondria for Enzymatic and Proteomic Analyses
About 3 × 107 cells were harvested and washed with 1 × PBS buffer. The pellet was resuspended in 5 mL of an ice-cold solution containing 5 mM K+-Hepes, pH 7.4, 210 mM mannitol, 1 mM EGTA, 70 mM sucrose, and 55 μg/mL digitonin and homogenized by 10 strokes in an ice-cold glass homogenizer. Nonlysed cells and nuclei were pelleted by centrifugation at 750 g for 20 min at 4°C, and the supernatant was centrifuged again at 8000 g for 15 min at 4°C. The resulting mitochondrial pellet was resuspended in 1 mL of 5 mM K+-Hepes, pH 7.4, 210 mM mannitol, and 70 mM sucrose at 37°C and treated for cytochrome oxidase activity and proteomic analysis as described below.
2.9. Enzymatic Activity of Cytochrome c Oxidase
Cytochrome c oxidase activity was determined spectrophotometrically using the Cytochrome c Oxidase Assay Kit (Sigma, MO, USA). Reactions were started by the addition of ferrocytochrome c. The difference in extinction coefficients (ΔεmM) between ferrocytochrome c and ferricytochrome c is 21.84 at 550 nm. One unit of enzyme will oxidize 1.0 μmole of ferrocytochrome c per minute at pH 7.0 at 25°C. The proteins were determined according to the method of Lowry et al. [29] using bovine serum albumin as the standard.
2.10. Proteomic Analysis
Mitochondria were resuspended in urea lysis buffer (8 M urea, 4% CHAPS, 65 mM DTE, and 40 mM Tris base) and sonicated for 5 s on ice. Following centrifugation at 21000 g, protein concentration was determined by Bradford assay [30]. Aliquots were then stored at −80°C until use. Two dimensional electrophoresis (2-DE) was carried out as previously described [31]. Briefly, isoelectric focusing was made on Immobiline strips providing a nonlinear pH 3–10 gradient (GE Healthcare Italy, Milan, Italy) using an IPGphore system (GE Healthcare) and applying an increasing voltage from 200 V to 3500 V during the first 3 h, then stabilized at 5000 V for 20 h. After IPG strip equilibration, the second dimension was carried out in a Laemmli system on 9%–16% polyacrylamide linear gradient gels (18 cm × 20 cm × 1.5 mm) at 40 mA/gel constant current, until the dye front reached gel bottom. Forty-five μg (analytical runs) or 500 μg (semipreparative runs) of proteins were used for each electrophoretic run.
Analytical gels were stained with silver nitrate [32], while semipreparative gels for mass spectrometry analysis were stained with Brilliant Blue G-Colloidal (Sigma- Aldrich, Saint Louis, USA) according to the manufacturer's procedure. Gel images were acquired by Fluor-S MAX multi-imaging system (BioRad Laboratories Italy, Segrate, Italy), and the data were analysed with ImageMaster 2D Platinum software. To test the significant differences in the relative protein levels for each spot, a paired Student's t-test statistic was applied at a significant level of P < .05.
The gel digestion procedure was adapted from Shevchenko et al. [33] as previously described [34].
LC-ESI-MS/MS analysis was performed using a Q-TOF microTM mass spectrometer (Micromass, Manchester, UK) equipped with a Z-spray nanoflow electrospray ion source and a CapLC system. The sample was analyzed using a Symmetry C18 nano column (Waters, Milford, Mass, USA) as an analytical column. For protein identification, MS/MS spectra were searched by MASCOT (Matrix science, www.matrixscience.com, UK) using the database of NCBI nr. For unmatched peptides, however, good quality MS/MS spectra were manually sequenced using de novo sequencing process (carried out by PepSeq of the Masslynx 4.0 software, Micromass), and the obtained sequence was subsequently used in Expasy TagIdent.
2.11. Statistical Analysis
Unless noted otherwise, the results were expressed as mean values ± S.E.M. for the indicated number of measurements. Results from PCR real-time analysis were compared with the ANOVA test, followed by a post hoc test using Tukey's multiple comparison test. The threshold of significance for the ANOVA and the Tukey's test was fixed at P ≤ .05.
3. Results
3.1. Cell Differentiation
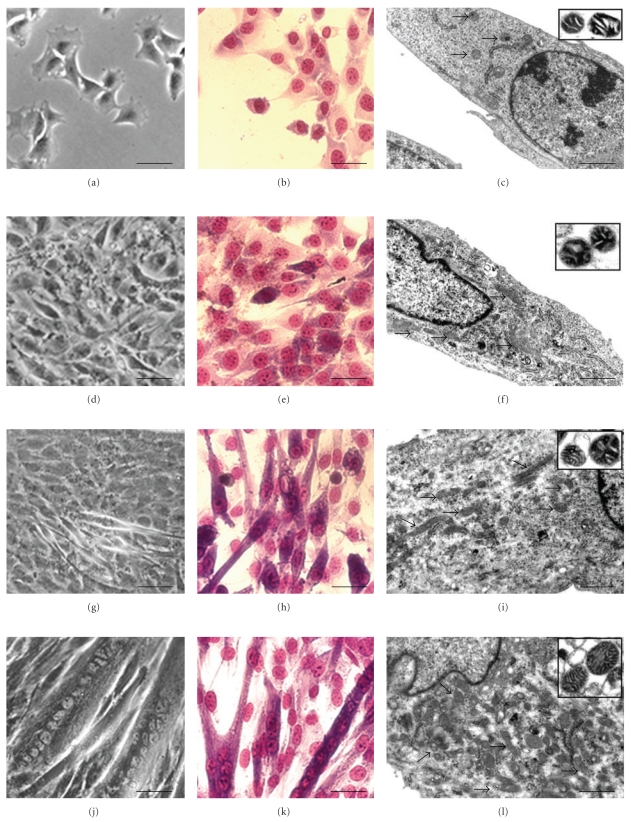
The monolayer organization, as directly analysed at RM and by means of Giemsa staining, deeply changes from undifferentiated myoblasts to myotubes. In the undifferentiated condition (Figures 1(a), 1(b), and 1(c)), myoblasts appear as fusiform or star-shaped cells, mostly flattened and closely adherent to the substrate. At the initial differentiation stage (Figures 1(d), 1(e), and 1(f)), intercellular spaces disappear, cells progressively align, and, occasionally, elongate. Four days after differentiation induction (Figures 1(g), 1(h), and 1(i)), early myotubes, with 2 or more centrally located nuclei, appear (T = 4, fusion index = 38 ± 3.4%). The late differentiation condition (7 days) is characterized by the presence of highly structured myotubes (Figures 1(j), 1(k), and 1(l)). These are 100–600 μm syncytia and contain even more than 20 nuclei, mainly centrally located or, occasionally, aligned in parallel rows (T = 7, fusion index 84.6 ± 6%).
Figure 1.
Undifferentiated (a, b, c), early differentiation (d, e, f), intermediate differentiation (g, h, i) and late differentiation stages (j, k, l), are indicated by RM (a, d, g, j), Giemsa staining (b, e, h, k), and TEM (c, f, i, l). Mitochondrial morphology is further detailed by the correspondent insets, showing TEM analysis of isolated mitochondria. C2C12 cell differentiation morphological progression is evident, as well as mitochondrial behaviour in the various stages. (a, b, d, e, g, h, j, k): Bar = 20 μm; (c, f, i, l): Bar = 0.5 μm; insets, Bar = 0.1 μm.
3.2. Morphofunctional Changes in Mitochondrial Content
Changes in mitochondrial ultrastructure were determined by transmission electron microscopy (TEM). Figure 1 shows the progression of C2C12 cell differentiation and the related mitochondrial behaviour. Their number per area significantly increases from the undifferentiated condition (c), through the initial (f) and the intermediate (i) differentiation stages, to the final phase, characterized by myotubes, which show the maximal mitochondrial content (l). Conversely, the size of single mitochondria, appears to change throughout differentiation. It increases in the undifferentiated stage (c) reaching maximal values at initial differentiation condition (f). It then steadily decreases (f, i), showing minimal values in the late differentiation stage (l). TEM of isolated mitochondria further highlights mitochondrial changes. Table 2 represents mitochondrial number and area variability during differentiation. They undergo a progressive rounding from 0 (Figure 1, inset c) to 7 day (Figure 1, inset l) after differentiation induction.
Table 2.
Mitochondrial area and number variability during differentiation by means of ultrastructural observations of resin-embedded sections.
| Differentiation day | Δ cell mitochondria area/10 × 15 cm total surface | Δ isolated mitochondria area/10 × 15 cm total surface | Number of mitochondria/10 × 15 cm total area |
|---|---|---|---|
| T = 0 | 3.30E−02 ± 0.005 | 6.90E−02 ± 0.009 | 6 ± 0.89 |
| T = 1 | 9.30E−02 ± 0.008 | 9.80E−02 ± 0.008 | 10 ± 1.14 |
| T = 4 | 8.20E−02 ± 0.004 | 8.10E−02 ± 0.004 | 13 ± 0.91 |
| T = 7 | 3.40E−02 ± 0.008 | 5.40E−02 ± 0.005 | 15 ± 0.86 |
Analysis of mitochondria suggests a numerical increase of mitochondrial cristae from the undifferentiated to differentiated condition (Figure 1, insets: c, f, i, and l) probably correlated with the reported increase in enzymatic activities [6].
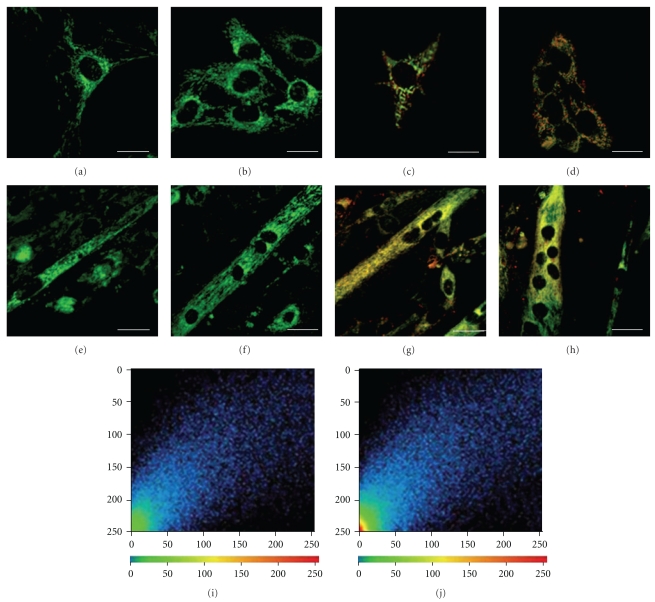
Figure 2 describes mitochondrial characteristics during differentiation, analysed by confocal microscopy, after Mito Tracker green (a–d) and JC-1 (e–h) staining, both specific mitochondrial dyes. The first covalently binds to mitochondrial proteins and is generally considered an available indicator of mitochondrial mass. The second undergoes characteristic fluorescence changes according to the mitochondrial membrane ΔΨ, thus revealing functional mitochondrial alterations. In myoblasts (a, b, c, and d), both fluorescent probes show a perinuclear mitochondrial distribution. Indeed, at initial differentiation stages, numerous mitochondria can be identified as clearly distinguishable single organelles. Moreover, after differentiation induction, mitochondrial mass increased appearing uniform in myotubes (e and f). Mitochondrial membrane potential also increased, highlighted by JC-1 main red staining (g), still more evident in late differentiation condition shown in (h). Graphs of lower panel show the increasing level of red fluorescence JC-1 intensity from myoblasts (i) to late myotubes (j).
Figure 2.
Confocal microscopy of C2C12 myoblasts (a–d) and late myotubes (e–h), after Mito Tracker (a, b, e, f) and JC-1 (c, d, g, h) staining. Graphs of lower panel show the different fluorescence JC-1 intensity in myoblasts (i) and late myotubes (j). (a–h): Bar = 20 μm.
3.3. mtDNA Content
To ensure accurate quantification of mtDNA, we applied a PCR-based assay using a dual-insert reference plasmid, containing both mtDNA and nuclear DNA targets [35]. In this work, pDrive plasmid was used to construct the reference plasmid pDGC, containing a single copy of COXII and GAPDH segments, the mitochondrial and nuclear target genes, respectively.
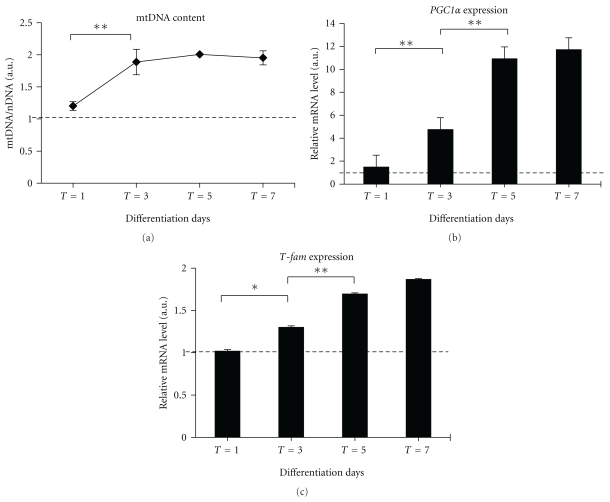
As shown in Figure 3(a), twenty-four hours after differentiation induction, the relative amount of mtDNA undergoes a 2-fold increment at the intermediate period of differentiation (T = 3) reaching a plateau level at the final stage of maturation (T = 7).
Figure 3.
Evaluation of mitochondrial biogenesis during myoblast differentiation. In (a), determination by real-time PCR of mtDNA content expressed as mtDNA/nDNA ratio (COXII/GAPDH), as described in Section 2. In (b), quantitative analyses of PGC-1 α and T-fam by real-time PCR. The amount of each target transcript was related to that of the reference gene (the ribosomal protein S16). Data are expressed as the mean ± SEM of three experiments; all samples were analyzed in triplicate. Results from PCR real-time analysis were compared with the ANOVA test, followed by a post hoc test using Tukey's multiple comparison test. The threshold of significance for the ANOVA and the Tukey's test was fixed at *P ≤ .05; **P ≤ .01.
3.4. mRNA Expression Level of Mitochondrial Biogenesis “Master” Genes
An increase in mitochondrial biogenesis reflects an enhanced expression of nuclear and mitochondrial genes [36–38]. Two master genes involved in the mitochondrial biogenesis, the nuclear transcriptional coactivator peroxisome proliferative activated receptor, gamma, coactivator 1 alpha (PGC1 α), and the mitochondrial transcription factor A (Tfam) were quantified using RT real-time PCR during differentiation. PGC1 α induces mitochondrial biogenesis by interacting with several nuclear transcription factors [36–39], and Tfam is involved in the mitochondrial genome transcription [40, 41], replication [42], and it is also crucial for maintaining mitochondrial DNA [43].
As shown in Figure 3(b), PGC-1 α expression does not change during the first 24 h from the induction of differentiation while progressively increasing up to 9.2-fold in differentiated myotubes on the 7th day compared to the myoblasts at time T0.
The Tfam expression level during the myoblasts differentiation is slightly shifted compared to the PGC-1 α expression; in fact, it increased significantly between days 3–7 (Figure 3(c)).
3.5. Cytochrome c Oxidase Activity and COXII Expression Level
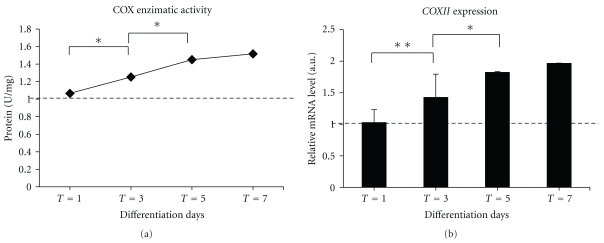
The mitochondrial enzymatic activities of cytochrome oxidase reflecting the respiratory chain activities were significantly higher in myoblasts able to differentiate (Figure 4(a)).
Figure 4.
Time course change of cytochrome oxidase (COX) enzymatic activity and transcription level of cytochrome oxidase subunit II (COXII) gene at progressive differentiation stages. (a) Quantitative analysis enzymatic activity. (b) The expression level of COXII is related to S16 mRNA gene level. Results from PCR real-time analysis were compared with the ANOVA test, followed by a post hoc test using Tukey's multiple comparison test. The threshold of significance for the ANOVA and the Tukey's test was fixed at *P ≤ .05; **P ≤ .01.
In addition, we evaluated the expression level of the corresponding gene coding for the subunit II of mitochondrial cytochrome c oxidase (COXII), which represents a target gene for mitochondrial transcriptional activity [27, 44, 45]. On days 3–7, the mitochondrial COXII transcript levels were significantly higher than in proliferating myoblasts (Figure 4(b)).
3.6. Changes in Mitochondrial Proteomic Profile
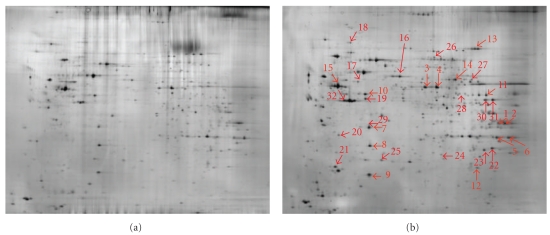
To highlight significant changes in mitochondrial proteome during differentiation, we performed a 2D page on mitochondria isolated from C2C12 myoblasts over a 7-day time span differentiation. A total of 994 spots (mean) could be resolved on a silver-stained large 2DE gel, where we loaded 45 μg of total protein. A larger amount of protein per spot was necessary for protein identification, thus we used preparative gels stained with Brilliant Blue G-Colloidal. To evaluate the possible presence of cellular contaminants, we compared the mitochondrial map with that of the whole cellular lysate in which we had previously identified several cytosolic and membrane proteins [46]. The comparison of 2D maps of mitochondria and whole cell lysate allowed us to state that the preparation of mitochondria contained little or no cellular contaminants.
The study of quantitative changes of individual proteins in a purified mitochondrial fraction showed that 32 mitochondrial proteins increased significantly in abundance (Figure 5). The proteins showing the greatest expression changes were also characterized by electrospray ionisation (ESI) tandem mass spectrometry. In particular, the major changes occurred between T1 and T4 time of differentiation, while fewer differences were shown between T0-T1 and T4–T7 (Table 3).
Figure 5.
Image of a silver-stained 2-DE gel of 45 μg purified mitochondrial proteins from C2C12 myoblasts at 0 (a) and 7 (b) days of differentiation time. Differentially expressed spots are indicated by arrows and numbered according to Table 3.
Table 3.
Identification of mitochondrial protein differentially expressed during myogenesis.
| No. | Protein | Score | NCBI nr | Peptides | MW | PI | Localization |
|---|---|---|---|---|---|---|---|
| 1 | Malate dehydrogenase, mitochondrial (MDH2) | 162 | DEMSMM | IFGVTTLDIVR, | 35589 | 8.93 | Mitochondrial matrix |
| VDFPQDQLATLTGR, | |||||||
| IQEAGTEVVK | |||||||
| 2 | Malate dehydrogenase precursor (MDH2) | 345 | DEMSMM | VAVLGASGGIGQPLSLLLK, | 35589 | 8.93 | Mitochondrial matrix |
| IFGVTTLDIVRANTFVAELK, | |||||||
| VDFPQDQLATLTGRIQEAGTEVVK, | |||||||
| MIAEAIPELK | |||||||
| 3 | Aldehyde dehydrogenase 2, mitochondrial (Aldh2); putative uncharacterized protein | 143 | Q3TVM2_MOUSE | TIPIDGDFFSYTR, | 56560 | 7.03 | Mitochondrial matrix |
| VAEQTPLTALYVANLIK, | |||||||
| EAGFPPGVVNIVPGFGPTAGAAIASHEGVDK | |||||||
| 4 | Aldehyde dehydrogenase precursor, mitochondrial | 411 | I48966 | TIPIDGDFFSYTR, | 56502 | 7.53 | Mitochondrial matrix |
| LGPALATGNVVVMK, | |||||||
| TFVQENVYDEFVER, | |||||||
| TEQGPQVDETQFK, | |||||||
| GYFIQPTVFGDVK, | |||||||
| TIEEVVGR, | |||||||
| YGLAAAVFTK | |||||||
| 5 | Voltage-dependent anion channel 1 (VDAC1) | 161 | VDAC1_MOUSE | LTFDSSFSPNTGK, | 32331 | 8.55 | Mitochondrial outer membrane |
| VTQSNFAVGYK, | |||||||
| LTLSALLDGK | |||||||
| 6 | Voltage-dependent anion channel 1 (VDAC1) | 344 | VDAC1_MOUSE | GYGFGLIK, | 32331 | 8.55 | Mitochondrial outer membrane |
| WTEYGLTFTEK, | |||||||
| LTFDSSFSPNTGK, | |||||||
| VTQSNFAVGYK, | |||||||
| VNNSSLIGLGYTQTLKPGIK, | |||||||
| LTLSALLDGK | |||||||
| 7 | Pyruvate dehydrogenase (lipoamide) beta. (Pdhb protein) | 288 | Q99LW9_MOUSE | TYYMSAGLQPVPIVFR, | 34814 | 5.63 | Mitochondrial matrix |
| DFLIPIGK, | |||||||
| IMEGPAFNFLDAPAVR, | |||||||
| VTGADVPMPYAK, | |||||||
| VLEDNSVPQVK | |||||||
| 8 | Prohibitin | 396 | A39682 | DLQNVNITLR, | 29802 | 5.57 | Mitochondrial intermembrane space |
| ILFRPVASQLPR, | |||||||
| IYTSIGEDYDER, | |||||||
| VLPSITTEILK, | |||||||
| FDAGELITQR, | |||||||
| AAIISAEGDSK, | |||||||
| AAELIANSLATAGDGLIELR, | |||||||
| NITYLPAGQSVLLQLPQ | |||||||
| 9 | ATP synthase D chain, mitochondrial (ATP5H) | 131 | ATP5H_MOUSE | ANVAKPGLVDDFEK, | 18607 | 5.52 | Mitochondrial inner membrane |
| YTALVDQEEKEDVK | |||||||
| 10 | Ubiquinol-cytochrome c reductase core protein 1 | 135 | Q3THM1_MOUSE | TDLTDYLNR, | 52806 | 5.89 | Mitochondrial inner membrane |
| IQEVDAQMLR | |||||||
| 11 | Fumarate hydratase precursor, mitochondrial (FH) | 170 | UFRT | AAAEVNQEYGLDPK, | 54429 | 9.06 | Mitochondrial |
| AIEMLGGELGSK, | |||||||
| VAALTGLPFVTAPNK | |||||||
| 12 | Superoxide dismutase precursor | 72 | DSRTN | GDVTTQVALQPALK | 24659 | 8.96 | Mitochondrial matrix |
| 13 | Aconitase 2, mitochondrial (ACO2) | 340 | Q3UDK9_MOUSE | DINQEVYNFLATAGAK, | 85376 | 8.08 | Mitochondrial matrix |
| SQFTITPGSEQIR, | |||||||
| NTIVTSYNR, | |||||||
| FNPETDFLTGK, | |||||||
| NAVTQEFGPVPDTAR, | |||||||
| WVVIGDENYGEGSSR | |||||||
| 14 | Dihydrolipoamide dehydrogenase (DLDH) | 139 | Q99LD3_MOUSE | ADGSTQVIDTK, | 54238 | 7.99 | Mitochondrial matrix |
| EANLAAAFGHPINF | |||||||
| 15 | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit (ATP5B) | 998 | Q3TFD7_MOUSE | LVLEVAQHLGESTVR, | 56207 | 5.25 | Mitochondrial inner membrane |
| TIAMDGTEGLVR, | |||||||
| VLDSGAPIK, | |||||||
| IPVGPETLGR, | |||||||
| IMNVIGEPIDER, | |||||||
| VVDLLAPYAK, | |||||||
| IGLFGGAGVGK, | |||||||
| TVLIMELINNVAK, | |||||||
| EGNDLYHEMIESGVINLK, | |||||||
| VALVYGQMNEPPGAR, | |||||||
| VALTGLTVAEYFR, | |||||||
| FTQAGSEVSALLGR, | |||||||
| AIAELGIYPAVDPLDSTSR, | |||||||
| IMDPNIVGNEHYDVAR, | |||||||
| ILQDYK, | |||||||
| FLSQPFQVAEVFTGHMGK | |||||||
| 16 | Protein disulfide isomerase A3 (Pdia3) | 94 | PDIA3_MOUSE | DASVVGFFR, | 56472 | 5.88 | Endoplasmic reticulum, also present in mitochondria (see discussion). |
| GFPTIYFSPANK, | |||||||
| ELNDFISYLQR | |||||||
The main mitochondrial proteins which could be detected in fully differentiated syncytia were involved in the citric acid cycle (malate dehydrogenase: MDH2, fumarate hydratase: FH, and aconitase 2: ACO2) or belong to the pyruvate dehydrogenase complex (pyruvate dehydrogenase, lipoamide beta: PDHB, dihydrolipoamide dehydrogenase), complex III (ubiquinol-cytochrome c reductase core protein 1: UQCRC1) and complex V (ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit: ATP5B, and ATP synthase d chain: ATP5H) of the respiratory chain.
This was also interesting for the superoxide dismutase (MnSOD), a voltage-dependent anion-selective channel protein 1 (VDAC1), and the protein disulfide-isomerase A3 (Pdia3) that were differentially expressed during differentiation.
4. Discussion
In this study, we described temporal mitochondrial changes during the myogenic program of C2C12 myoblasts by analyzing complementary key parameters for mitochondrial dynamics, biogenesis, and functionality. Of particular interest is the contribution of the proteomic approach to better define the pattern of mitochondrial protein expression accompanying differentiation in myotubes and potentially involved in the crosstalk between nuclei and mitochondria.
Morphological analysis performed by fluorescence microscopy with markers of mitochondrial mass/volume and ΔΨ, as well as ultrastructural analysis, allowed us to acquire more information regarding the mitochondrial organization and dynamics in C2C12 myoblast differentiation.
Mitochondrial organization in myoblasts was perinuclear, and it was possible to discriminate individual mitochondrion by both MitoTracker Green and JC1 staining. This type of mitochondrial distribution is described in the literature for other cell types including fibroblasts [47], pancreatic acinar cells [48, 49], astrocytes, and neurons [50].
On the contrary, in myotubes, morphological observation by epifluorescence did not allow us to discriminate individual mitochondrion, showing homogeneous staining, representative of the mitochondrial network, well described in skeletal muscle tissue [51, 52]. TEM analysis showed a mitochondrial remodeling during differentiation and alignment of organelles along the myotubes.
At this level, we cannot show the formation of a network equal to that which is found in skeletal muscle fibers, where mitochondria are arranged in crystal structures closely related to the sarcoplasm [51, 52]. Indeed, the sarcomeres of myotubes are only sketched [53], but they may support the development of a mitochondrial network during myotube maturation.
The mitochondrial counting per area of cell surface, obtained by TEM, showed that the number of mitochondria increased from undifferentiated to differentiated conditions. Mature myotubes contained approximately 2-fold more mitochondria than myoblasts. However, in the first 24 hours after induction of differentiation, the mitochondria increased in size up to 3-fold gradually decreasing in size only after the intermediate phases to reach the same size observed in myoblasts at T = 0, in mature myotubes (T = 7). This observation suggests that mitochondria first undergo fusion and then fission, which allows their distribution during syncytia formation as previously reported [54]. Nevertheless, the mitochondria in myotubes showed a greater extension of mitochondrial cristae than mitochondria in myoblasts. Marked stimulation of the biosynthesis of the phospholipid cardiolipin during the differentiation phases has been observed in previous studies on L6E9 myoblasts and other cells [54, 55]. It is probably necessary to supply the proper amount of functional mitochondrial inner membrane for the respiratory chain proteins involved in oxidative metabolism [7].
All the parameters observed through morphological analysis confirm a linear increase in mitochondrial biogenesis during differentiation. The morphological analysis corroborates the progression of the myogenic process and the increase in biochemical markers such as the transcription factors of mitochondrial biogenesis PGC-1 α and Tfam.
Of particular interest was the timing of mtDNA replication compared to mitochondrial biogenesis. Although mitochondrial biogenesis increased linearly during differentiation, mtDNA content increased significantly from the early days of differentiation already reaching a plateau at the intermediate stage. Hence, our investigation shows a slight difference in timing between DNA replication and mitochondrial biogenesis. This shift could be explained by the biological cycle of mitochondria [56]. Mitochondrial fission is preceded by an extension of the organelles and the mtDNA replication phase. Although there is a slight shifting, the correlation between the number of copies of mtDNA and mitochondrial biogenesis is positive (r2 = 0.85, data not shown).
In several studies, the measure of mtDNA copy number has been considered proportional to the number of mitochondria, a golden star for mitochondrial density [57–60]. However, changes in mitochondrial abundance regardless of the mtDNA copy number may occur, especially in peculiar conditions such as during alterations in the rates of intracellular ROS generation [61]. Franko et al., investigating C2F3 mouse myoblasts, showed that an increment in mtDNA does not always correlate with the proliferation of mitochondria or with their activity [62]. In this investigation, the mtDNA copy number of C2C12 myoblasts significantly increased during the early-intermediate differentiation phases (T = 1 and T3) up to 2-fold remaining constant during the myotube maturation. Likewise, over the course of myoblast differentiation in rat cell line L6, a small but significant increase in mitochondrial DNA copy number was observed by [27]. Furthermore, in a recent study on the regulation of mitochondrial biogenesis during myogenesis, mtDNA copy number was determined as a marker for mitochondrial density using QPCR, and the mtDNA copy number was 4-fold higher in fully differentiated myotubes than it was in myoblasts [60].
Interestingly, during differentiation, an increased mtDNA transcriptional activity and oxidative metabolism correspond to an enhanced mitochondrial biogenesis, as highlighted by the upregulation of COXII mRNA levels and cytochrome c oxidase activity (r2 = 0.83 and r2 = 0.97, resp., data not shown).
In our investigation, we integrated the mitochondrial changes observed by multiple key determinants with proteomic analysis.
In the literature, mitochondrial proteomic maps of differentiating myoblasts are not available; hence, this work presents the first proteomic profile of mitochondria during the myogenesis program. Previously, proteome-based investigations have been carried out to provide a description of the myogenic differentiation program [46, 63, 64]. We employed a proteomic approach using two dimensional electrophoresis, particularly helpful for investigating the subset of cellular proteins, such as organellar proteins, due to the reduced complexity of the protein sample [65].
In particular, analyzing the differentially expressed proteins in the mitochondrial proteome map during the myogenic process, we observed that also the enzymes involved in cellular respiration, such as pyruvate dehydrogenase, MDH2, FH, ACO2, and more markedly HB and 5B ATP synthase subunits, representative of oxidative phosphorylation, increase linearly with the mitochondrial biogenesis showing a positive correlation (r2 = 0.915, data not shown). These findings are consistent with the differentiating cells' greater reliance on aerobic metabolism compared to the glycolytic metabolism that characterizes the undifferentiated myoblasts [7].
Moreover, our observations are in agreement with Moyes and coworkers who demonstrated an mRNA increment for pyruvate dehydrogenase, citrate synthase, isocitrate dehydrogenase, cytochrome c oxidase, and NADH dehydrogenase [6]. The increment of Krebs cycle and respiratory chain proteins supports the augmented mitochondrial functionality also confirmed by the COX enzymatic activity during myoblast differentiation [6, 7].
The data obtained using the proteomic approach are consistent with the increase in mitochondrial function and membrane depolarization highlighted by JC-1 and in agreement with the increase of mitochondrial cristae observed by TEM. These data support the evidence described by Sauvanet (2010) assuming that mitochondrial bioenergetics and dynamics are linked and that mitochondrial morphology reflects their functional status [66].
Proteomic analysis revealed other notable proteins involved in the crosstalk between nuclei and mitochondria, such as MnSOD, a recently described cell protection molecule whose role is the maintenance of myoblast mitochondrial function and the preservation of the potential of myoblast stem cell differentiation [67]. In our model, the increment of MnSOD expression is highlighted during myotube formation (T = 4–T = 7), as recently documented by Kislinger and collaborators [64].
Another interesting protein associated with the progressive myoblast differentiation is the protein transport voltage-dependent anion-selective channel protein 1. It has been reported that its gene expression changes during myoblast differentiation [68], and it has been recognized as a key protein in mitochondria-mediated apoptosis, since it is a target for the pro- and antiapoptotic Bcl2-family of proteins, and for its function in releasing apoptotic proteins located in the intermembrane space [69]. Apoptosis is considered to be essential for normal skeletal muscle development by eliminating cells with defects or undergoing damage during differentiation [70–73]. The detection of mitochondrial myogenesis-correlated proteins, known to play a role in apoptosis, supports the link between differentiation and this type of cell death [46, 74].
Another protein induced during differentiation belongs to the protein disulfide isomerase (PDI) family: disulfide-isomerase A3 (Pdia3). These molecules are best known for their role as chaperones in protein-folding reactions in the endoplasmic reticulum [75]. However, mitochondrial localization has been documented in the outer membrane of rat liver mitochondria [76–78], bovine liver mitochondria [79], in a mitochondrial fraction of Arabidopsis [80] and a chloroplast isoform in Chlamydomonas [81]. PDI may act in mitochondria in several ways: enhancing protein folding of newly synthesized proteins, reducing disulphides required to activate proteins, controlling mitochondrial membrane permeability, and playing a role in the assembly and function of some enzymatic systems [76, 77].
In conclusion, this investigation demonstrates that the proteomic approach, integrated with the classical morpho-functional and biochemical analyses, provides a complete scenario of mitochondrial dynamics, biogenesis and functionality useful in comparative surveys of mitochondrial pathogenic or senescent satellite cells.
Acknowledgments
The authors thank Dr. Rosa Curci (Orthopedic Rizzoli Institute, Bologna, Italy) for providing confocal microscope images. They wish to thank Professor Timothy Bloom, Centro Linguistico di Ateneo of the University of Urbino, for a critical reading of the paper.
References
- 1.Pownall ME, Gustafsson MK, Emerson CP., Jr. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annual Review of Cell and Developmental Biology. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 2.Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clinical Genetics. 2000;57(1):16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 3.Dedieu S, Mazères G, Cottin P, Brustis JJ. Involvement of myogenic regulator factors during fusion in the cell line C2C12. International Journal of Developmental Biology. 2002;46(2):235–241. [PubMed] [Google Scholar]
- 4.Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Developmental Biology. 2000;219(2):287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- 5.Ferri P, Barbieri E, Burattini S, et al. Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. Journal of Cellular Biochemistry. 2009;108(6):1302–1317. doi: 10.1002/jcb.22360. [DOI] [PubMed] [Google Scholar]
- 6.Moyes CD, Mathieu-Costello OA, Tsuchiya N, Filburn C, Hansford RG. Mitochondrial biogenesis during cellular differentiation. American Journal of Physiology. 1997;272(4):C1345–C1351. doi: 10.1152/ajpcell.1997.272.4.C1345. [DOI] [PubMed] [Google Scholar]
- 7.Kraft CS, LeMoine CMR, Lyons CN, Michaud D, Mueller CR, Moyes CD. Control of mitochondrial biogenesis during myogenesis. American Journal of Physiology. 2006;290(4):C1119–C1127. doi: 10.1152/ajpcell.00463.2005. [DOI] [PubMed] [Google Scholar]
- 8.Rochard P, Rodier A, Casas F, et al. Mitochondrial activity is involved in the regulation of myoblast differentiation through myogenin expression and activity of myogenic factors. The Journal of Biological Chemistry. 2000;275(4):2733–2744. doi: 10.1074/jbc.275.4.2733. [DOI] [PubMed] [Google Scholar]
- 9.Hoppeler H, Flock M. Plasticity of skeletal muscle mitochondria: structure and function. Medicine and Science in Sports and Exercise. 2003;35(1):95–104. doi: 10.1249/01.MSS.0000043292.99104.12. [DOI] [PubMed] [Google Scholar]
- 10.Duguez S, Sabido O, Freyssenet D. Mitochondrial-dependent regulation of myoblast proliferation. Experimental Cell Research. 2004;299(1):27–35. doi: 10.1016/j.yexcr.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Auré K, Fayet G, Leroy JP, Lacène E, Romero NB, Lombès A. Apoptosis in mitochondrial myopathies is linked to mitochondrial proliferation. Brain. 2006;129(5):1249–1259. doi: 10.1093/brain/awl061. [DOI] [PubMed] [Google Scholar]
- 12.Hood DA, Irrcher I, Ljubicic V, Joseph AM. Coordination of metabolic plasticity in skeletal muscle. Journal of Experimental Biology. 2006;209(12):2265–2275. doi: 10.1242/jeb.02182. [DOI] [PubMed] [Google Scholar]
- 13.Toledo FGS, Watkins S, Kelley DE. Changes induced by physical activity and weight loss in the morphology of intermyofibrillar mitochondria in obese men and women. Journal of Clinical Endocrinology and Metabolism. 2006;91(8):3224–3227. doi: 10.1210/jc.2006-0002. [DOI] [PubMed] [Google Scholar]
- 14.Miner JH, Wold BJ. c-myc Inhibition of MyoD and myogenin-initiated myogenic differentiation. Molecular and Cellular Biology. 1991;11(5):2842–2851. doi: 10.1128/mcb.11.5.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seyer P, Grandemange S, Busson M, et al. Mitochondrial activity regulates myoblast differentiation by control of c-Myc expression. Journal of Cellular Physiology. 2006;207(1):75–86. doi: 10.1002/jcp.20539. [DOI] [PubMed] [Google Scholar]
- 16.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes and Development. 2004;18(4):357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 17.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual Review of Genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 19.Mammucari C, Rizzuto R. Signaling pathways in mitochondrial dysfunction and aging. Mechanisms of Ageing and Development. 2010;131(7-8):536–543. doi: 10.1016/j.mad.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopinath SD, Rando TA. Stem cell review series: aging of the skeletal muscle stem cell niche. Aging Cell. 2008;7(4):590–598. doi: 10.1111/j.1474-9726.2008.00399.x. [DOI] [PubMed] [Google Scholar]
- 21.Di Iorio A, Abate M, Di Renzo D, et al. Sarcopenia: age-related skeletal muscle changes from determinants to physical disability. International Journal of Immunopathology and Pharmacology. 2006;19(4):703–719. doi: 10.1177/039463200601900401. [DOI] [PubMed] [Google Scholar]
- 22.Cerletti M, Shadrach JL, Jurga S, Sherwood R, Wagers AJ. Regulation and function of skeletal muscle stem cells. Cold Spring Harbor Symposia on Quantitative Biology. 2008;73:317–322. doi: 10.1101/sqb.2008.73.054. [DOI] [PubMed] [Google Scholar]
- 23.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464(7288):520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Emilio A, Biagiotti L, Burattini S, et al. Morphological and biochemical patterns in skeletal muscle apoptosis. Histology and Histopathology. 2010;25(1):21–32. doi: 10.14670/HH-25.21. [DOI] [PubMed] [Google Scholar]
- 25.Pendergrass W, Wolf N, Pool M. Efficacy of MitoTracker Green™ and CMXRosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry Part A. 2004;61(2):162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 26.Szilágyi G, Simon L, Koska P, Telek G, Nagy Z. Visualization of mitochondrial membrane potential and reactive oxygen species via double staining. Neuroscience Letters. 2006;399(3):206–209. doi: 10.1016/j.neulet.2006.01.071. [DOI] [PubMed] [Google Scholar]
- 27.Mikula M, Dzwonek A, Hennig EE, Ostrowski J. Increased mitochondrial gene expression during L6 cell myogenesis is accelerated by insulin. International Journal of Biochemistry and Cell Biology. 2005;37(9):1815–1828. doi: 10.1016/j.biocel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9):p. e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 30.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 31.Sestili P, Barbieri E, Martinelli C, et al. Creatine supplementation prevents the inhibition of myogenic differentiation in oxidatively injured C2C12 murine myoblasts. Molecular Nutrition and Food Research. 2009;53(9):1187–1204. doi: 10.1002/mnfr.200800504. [DOI] [PubMed] [Google Scholar]
- 32.Sinha P, Poland J, Schnölzer M, Rabilloud T. A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics. 2001;1(7):835–840. doi: 10.1002/1615-9861(200107)1:7<835::AID-PROT835>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 34.Guescini M, Guidolin D, Vallorani L, et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Experimental Cell Research. 2010;316(12):1977–1984. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Research. 2003;31(11):p. e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 37.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Molecular and Cellular Biology. 2000;20(5):1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Molecular and Cellular Biology. 2005;25(4):1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Molecular and Cellular Biology. 2001;21(11):3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252(5008):965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 41.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature Genetics. 2002;31(3):289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 42.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annual Review of Biochemistry. 1997;66:409–436. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 43.Kanki T, Ohgaki K, Gaspari M, et al. Architectural role of mitochondrial transcription factor a in maintenance of human mitochondrial DNA. Molecular and Cellular Biology. 2004;24(22):9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hood DA. Invited review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. Journal of Applied Physiology. 2001;90(3):1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- 45.Kim K, Lecordier A, Bowman LH. Both nuclear and mitochondrial cytochrome c oxidase mRNA levels increase dramatically during mouse postnatal development. Biochemical Journal. 1995;306(2):353–358. doi: 10.1042/bj3060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casadei L, Vallorani L, Gioacchini AM, et al. Proteomics-based investigation in C2C12 myoblast differentiation. European Journal of Histochemistry. 2009;53(4):261–268. doi: 10.4081/ejh.2009.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaffe MP. The machinery of mitochondrial inheritance and behavior. Science. 1999;283(5407):1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- 48.Johnson PR, Dolman NJ, Pope M, et al. Non-uniform distribution of mitochondria in pancreatic acinar cells. Cell and Tissue Research. 2003;313(1):37–45. doi: 10.1007/s00441-003-0741-1. [DOI] [PubMed] [Google Scholar]
- 49.Bruce JIE, Giovannucci DR, Blinder G, Shuttleworth TJ, Yule DI. Modulation of [Ca2+] signaling dynamics and metabolism by perinuclear mitochondria in mouse parotid acinar cells. The Journal of Biological Chemistry. 2004;279(13):12909–12917. doi: 10.1074/jbc.M309070200. [DOI] [PubMed] [Google Scholar]
- 50.Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. The EMBO Journal. 2002;21(7):1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vendelin M, Béraud N, Guerrero K, et al. Mitochondrial regular arrangement in muscle cells: a “crystal- like” pattern. American Journal of Physiology. 2005;288(3):C757–C767. doi: 10.1152/ajpcell.00281.2004. [DOI] [PubMed] [Google Scholar]
- 52.Bo H, Zhang Y, Ji LL. Redefining the role of mitochondria in exercise: a dynamic remodeling. Annals of the New York Academy of Sciences. 2010;1201:121–128. doi: 10.1111/j.1749-6632.2010.05618.x. [DOI] [PubMed] [Google Scholar]
- 53.Burattini S, Ferri R, Battistelli M, Curci R, Luchetti F, Falcieri E. CC murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. European Journal of Histochemistry. 2004;48(3):223–233. [PubMed] [Google Scholar]
- 54.Jahnke VE, Sabido O, Freyssenet D. Control of mitochondrial biogenesis, ROS level, and cytosolic Ca2+ concentration during the cell cycle and the onset of differentiation in L6E9 myoblasts. American Journal of Physiology. 2009;296(5):C1185–C1194. doi: 10.1152/ajpcell.00377.2008. [DOI] [PubMed] [Google Scholar]
- 55.Menshikova EV, Ritov VB, Ferrell RE, Azuma K, Goodpaster BH, Kelley DE. Characteristics of skeletal muscle mitochondrial biogenesis induced by moderate-intensity exercise and weight loss in obesity. Journal of Applied Physiology. 2007;103(1):21–27. doi: 10.1152/japplphysiol.01228.2006. [DOI] [PubMed] [Google Scholar]
- 56.Mouli PK, Twig G, Shirihai OS. Frequency and selectivity of mitochondrial fusion are key to its quality maintenance function. Biophysical Journal. 2009;96(9):3509–3518. doi: 10.1016/j.bpj.2008.12.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rochard P, Cassar-Malek I, Marchal S, Wrutniak C, Cabello G. Changes in mitochondrial activity during avian myoblast differentiation: influence of triiodothyronine or v-erb A expression. Journal of Cellular Physiology. 1996;168(2):239–247. doi: 10.1002/(SICI)1097-4652(199608)168:2<239::AID-JCP2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 58.Hamai N, Nakamura M, Asano A. Inhibition of mitochondrial protein synthesis impaired C2C12 myoblast differentiation. Cell Structure and Function. 1997;22(4):421–431. doi: 10.1247/csf.22.421. [DOI] [PubMed] [Google Scholar]
- 59.Medeiros DM. Assessing mitochondria biogenesis. Methods. 2008;46(4):288–294. doi: 10.1016/j.ymeth.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 60.Remels AHV, Langen RCJ, Schrauwen P, Schaart G, Schols AMWJ, Gosker HR. Regulation of mitochondrial biogenesis during myogenesis. Molecular and Cellular Endocrinology. 2010;315(1-2):113–120. doi: 10.1016/j.mce.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 61.Igosheva N, Abramov AY, Poston L, et al. Maternal diet-induced obesity alters mitochondrial activity and redox status in mouse oocytes and zygotes. PLoS ONE. 2010;5(4) doi: 10.1371/journal.pone.0010074. Article ID e10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franko A, Mayer S, Thiel G, et al. CREB-1α is recruited to and mediates upregulation of the cytochrome c promoter during enhanced mitochondrial biogenesis accompanying skeletal muscle differentiation. Molecular and Cellular Biology. 2008;28(7):2446–2459. doi: 10.1128/MCB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tannu NS, Rao VK, Chaudhary RM, et al. Comparative proteomes of the proliferating C2C12 myoblasts and fully differentiated myotubes reveal the complexity of the skeletal muscle differentiation program. Molecular and Cellular Proteomics. 2004;3(11):1065–1082. doi: 10.1074/mcp.M400020-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Kislinger T, Gramolini AO, Pan Y, Rahman K, MacLennan DH, Emili A. Proteome dynamics during C2C12 myoblast differentiation. Molecular and Cellular Proteomics. 2005;4(7):887–901. doi: 10.1074/mcp.M400182-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Xie J, Techritz S, Haebel S, et al. A two-dimensional electrophoretic map of human mitochondrial proteins from immortalized lymphoblastoid cell lines: a prerequisite to study mitochondrial disorders in patients. Proteomics. 2005;5(11):2981–2999. doi: 10.1002/pmic.200401191. [DOI] [PubMed] [Google Scholar]
- 66.Sauvanet C, Duvezin-Caubet S, di Rago JP, Rojo M. Energetic requirements and bioenergetic modulation of mitochondrial morphology and dynamics. Seminars in Cell and Developmental Biology. 2010;21(6):558–565. doi: 10.1016/j.semcdb.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 67.Lee S, Van Remmen H, Csete M. Sod2 overexpression preserves myoblast mitochondrial mass and function, but not muscle mass with aging. Aging Cell. 2009;8(3):296–310. doi: 10.1111/j.1474-9726.2009.00477.x. [DOI] [PubMed] [Google Scholar]
- 68.Sterrenburg E, Turk R, ’T Hoen PAC, et al. Large-scale gene expression analysis of human skeletal myoblast differentiation. Neuromuscular Disorders. 2004;14(8-9):507–518. doi: 10.1016/j.nmd.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Current Pharmaceutical Design. 2006;12(18):2249–2270. doi: 10.2174/138161206777585111. [DOI] [PubMed] [Google Scholar]
- 70.Burattini S, Battistelli M, Falcieri E. Morpho-functional features of in-vitro cell death induced by physical agents. Current Pharmaceutical Design. 2010;16(12):1376–1386. doi: 10.2174/138161210791033941. [DOI] [PubMed] [Google Scholar]
- 71.Huppertz B, Tews DS, Kaufmann P. Apoptosis and syncytial fusion in human placental trophoblast and skeletal muscle. International Review of Cytology. 2001;205:215–253. doi: 10.1016/s0074-7696(01)05005-7. [DOI] [PubMed] [Google Scholar]
- 72.Sandri M, Carraro U. Apoptosis of skeletal muscles during development and disease. International Journal of Biochemistry and Cell Biology. 1999;31(12):1373–1390. doi: 10.1016/s1357-2725(99)00063-1. [DOI] [PubMed] [Google Scholar]
- 73.Walsh K. Coordinate regulation of cell cycle and apoptosis during myogenesis. Progress in Cell Cycle Research. 1997;3:53–58. doi: 10.1007/978-1-4615-5371-7_5. [DOI] [PubMed] [Google Scholar]
- 74.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(17):11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freedman RB, Hirst TR, Tuite MF. Protein disulphide isomerase: building bridges in protein folding. Trends in Biochemical Sciences. 1994;19(8):331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 76.Rigobello MP, Donella-Deana A, Cesaro L, Bindoli A. Isolation, purification, and characterization of a rat liver mitochondrial protein disulfide isomerase. Free Radical Biology and Medicine. 2000;28(2):266–272. doi: 10.1016/s0891-5849(99)00237-3. [DOI] [PubMed] [Google Scholar]
- 77.Rigobello MP, Donella-Deana A, Cesaro L, Bindoli A. Distribution of protein disulphide isomerase in rat liver mitochondria. Biochemical Journal. 2001;356(2):567–570. doi: 10.1042/0264-6021:3560567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozaki T, Yamashita T, Ishiguro SI. ERp57-associated mitochondrial μ-calpain truncates apoptosis-inducing factor. Biochimica et Biophysica Acta. 2008;1783(10):1955–1963. doi: 10.1016/j.bbamcr.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 79.Kimura T, Horibe T, Sakamoto C, et al. Evidence for mitochondrial localization of P5, a member of the protein disulphide isomerase family. Journal of Biochemistry. 2008;144(2):187–196. doi: 10.1093/jb/mvn057. [DOI] [PubMed] [Google Scholar]
- 80.Sweetlove LJ, Heazlewood JL, Herald V, et al. The impact of oxidative stress on Arabidopsis mitochondria. Plant Journal. 2002;32(6):891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- 81.Trebitsh T, Meiri E, Ostersetzer O, Adam Z, Danon A. The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. The Journal of Biological Chemistry. 2001;276(7):4564–4569. doi: 10.1074/jbc.M005950200. [DOI] [PubMed] [Google Scholar]