Abstract
Methylation of histone H3 lysine 4 (H3K4me), a mark associated with gene activation, is mediated by SET1 and the related mixed lineage leukemia (MLL) histone methyltransferases (HMTs) across species. Mammals contain seven H3K4 HMTs, Set1A, Set1B, and MLL1–MLL5. The activity of SET1 and MLL proteins relies on protein–protein interactions within large multisubunit complexes that include three core components: RbBP5, Ash2L, and WDR5. It remains unclear how the composition and specificity of these complexes varies between cell types and during development. Caenorhabditis elegans contains one SET1 protein, SET-2, one MLL-like protein, SET-16, and single homologs of RbBP5, Ash2L, and WDR5. Here we show that SET-2 is responsible for the majority of bulk H3K4 methylation at all developmental stages. However, SET-2 and absent, small, or homeotic discs 2 (ASH-2) are differentially required for tri- and dimethylation of H3K4 (H3K4me3 and -me2) in embryos and adult germ cells. In embryos, whereas efficient H3K4me3 requires both SET-2 and ASH-2, H3K4me2 relies mostly on ASH-2. In adult germ cells by contrast, SET-2 serves a major role whereas ASH-2 is dispensable for H3K4me3 and most H3K4me2. Loss of SET-2 results in progressive sterility over several generations, suggesting an important function in the maintenance of a functional germ line. This study demonstrates that individual subunits of SET1-related complexes can show tissue specificity and developmental regulation and establishes C. elegans as a model to study SET1-related complexes in a multicellular organism.
Keywords: chromatin, epigenetics, mortal germ line
Histone H3 lysine 4 (H3K4) methylation and the proteins involved in its implementation are evolutionarily conserved marks of active and potentially active genes in all eukaryotes examined (1, 2). In yeast, Set1 is found in a multiprotein complex known as COMPASS, which is solely responsible for all histone H3K4 methylation (3–5). In mammalian cells, seven family members have been characterized: SET1a and SET1b (orthologs of yeast Set1) (6) and five mixed lineage leukemia (MLL) family members, MLL1–5, that share only limited similarity with yeast Set1 beyond the SET domain (7–11). Human SET1a/SET1b mediate the bulk of H3K4 trimethylation (H3K4me3) in mammalian cell extracts (12). Members of the MLL family of proteins do not appear to contribute significantly to bulk changes in H3K4 methylation, but serve both unique and overlapping tissue- and developmental stage-specific functions. Loss of either MLL1 or MLL2 results in embryonic lethality in mice (13, 14), whereas MLL3, -4, and -5 gene knockout mice are viable but have distinct developmental defects (15–18). Caenorhabditis elegans contains one SET1 protein, SET-2, and one MLL-like protein, SET-16 (19, 20). Whereas both SET-2 and SET-16 are required for global H3K4 methylation, SET-2 plays a predominant role (19–21).
The enzymatic activity of SET1/MLL family members is regulated by interactions with a number of other proteins, including Swd3/WDR5, Swd1/RbBP5, Bre2/Ash2, and Sdc1/hDPY30 (7–11). Drosophila absent, small, or homeotic discs 2 (ASH2) is a member of the trithorax group of positive regulators of homeotic genes (22) whose targets also include genes associated with cell proliferation (23). C. elegans ASH-2 has been implicated in vulval development and longevity (19, 21).
Recent evidence suggests that H3K4 methylation may also be important in regulating the unique transcription program in germ cells. During C. elegans embryonic development, the primordial germ cells (PCGs) undergo rapid loss of H3K4me2 soon after their birth, a process partially mediated by the SPR-5 histone demethylase (24, 25). Whether regulation of H3K4 histone methyltransferase (HMT) activity also contributes to proper levels of H3K4 methylation in the germ line was not investigated in these studies. Using newly isolated loss-of-function alleles of C. elegans set-2 and ash-2, we investigated how SET-2 and ASH-2 contribute to H3K4 methylation during C. elegans development. We show that SET-2 is responsible for most bulk H3K4 methylation at all developmental stages tested and that SET-2–dependent H3K4me3 peaks at the 5′ end of ubiquitously expressed genes. The contribution of ASH-2 to H3K4 methylation varies between tissues and stages. In embryos, ASH-2 serves a major role, whereas in adult germ cells, its contribution is minimal. This finding challenges the current view that Ash2 is an obligatory partner in Set1/MLL complexes across species. Consistent with SET-2 playing an important role in the germ line, we find that loss of SET-2 is associated with a mortal germ-line phenotype, in which animals become progressively sterile. Our data suggest that diverse SET-2 complexes are active at different stages and play distinct roles during development.
Results
Loss of SET-2 and ASH-2 Differentially Affects Global Levels of H3K4 Methylation.
A previously analyzed allele of set-2, ok952, encodes a SET-2L protein with an intact catalytic SET domain but missing a region between the RRM RNA-binding domain and the SET domain (20) and Fig. 1A). We isolated a second deletion allele, bn129, which removes 748 bp from exon 11 of SET-2L and from exon 3 of SET-2S. This deletion results in a frameshift after 885 aa of SET-2L and after 117 aa of SET-2S and a premature stop four codons later. To test the methylation activity of both alleles, we used quantitative Western blot analysis to measure global levels of H3K4me in extracts prepared from synchronized populations of mutant animals at different developmental stages (Fig. 2 A and B and Fig. S1). The ok952 allele did not cause a significant decrease in H3K4me3 levels in embryos and L1s and caused a 20–40% decrease in L3s and L4s (Fig. 2A). Little or no effect on H3K4me2 was observed for this allele at all stages. The bn129 allele caused a more pronounced effect on both H3K4me3 and -me2: a 70–80% decrease in H3K4me3 (Fig. 2A) and a 50–70% decrease in H3K4me2 (Fig. 2B) at all stages. The greater effect of the bn129 allele on global H3K4 levels, together with molecular analysis, suggests that bn129 is a null allele, whereas ok952 is likely to be a hypomorphic allele. In the remainder of this paper, we use the bn129 allele.
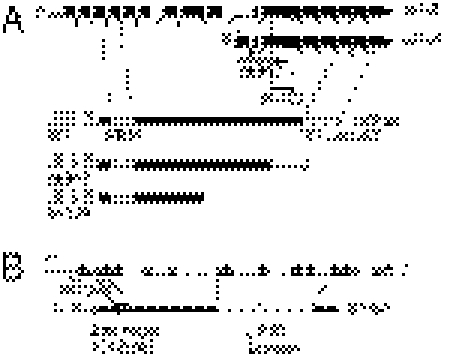
Fig. 1.
(A and B) Molecular characterization of set-2 and ash-2 deletion alleles. The extents of the deletion alleles used in this study are indicated by vertical dashed lines. Below the structure of each gene is shown a schematic representation of the corresponding protein product with conserved domains represented by shaded boxes. Thin dotted lines are drawn to the exons encoding the conserved domains.
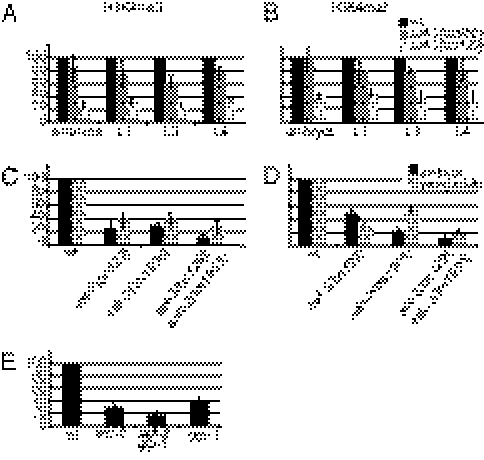
Fig. 2.
H3K4 global methylation is differentially affected by loss of SET-2 and ASH-2. The levels of histone H3K4me3 (A, C, and E) and H3K4me2 (B and D) were assayed using quantitative Western blots and normalized to levels of histone H3 and actin (Materials and Methods). (A–D) Total cell extracts were derived from synchronized populations of wild-type and mutant strains at the indicated developmental stages. (E) Levels of H3K4me3 were compared between wild type, set-2(bn129), set-2(bn129)glp-1, and glp-1 young adults. Bar graphs depict the combined results from three independent biological replicates; error bars indicate SD.
In previous studies, we showed that whereas reducing the activity of SET-2 and other predicted SET1/COMPASS subunits suppressed the growth defects caused by loss of the HP1 homolog of HPL-2 (20), reducing ASH-2 activity failed to do so. These results suggest that SET-2 and ASH-2 may have at least some independent functions and prompted us to test whether H3K4me levels are altered in ash-2 loss-of-function mutants. The tm1905 allele of ash-2 deletes 55 bp of upstream sequence and the first, the second, and part of the third exon (Fig. 1B). On the basis of molecular and genetic criteria, this is expected to be a strong loss-of-function or null allele. H3K4me3 was reduced to a similar degree, 60–80%, in ash-2 and set-2 mutant embryos and young adults (Fig. 2C). Surprisingly, however, H3K4me2 in ash-2 mutant embryos was reduced by 80%, compared with 50% in set-2 embryos. In young adults by contrast, H3K4me2 levels were decreased 50% in ash-2 mutants, as previously reported (21), and 70% in set-2 mutants (Fig. 2D). In ash-2;set-2 double mutants, both H3K4me3 and -me2 levels were further decreased with respect to either single mutant. Altogether, these results indicate that SET-2 strongly contributes to H3K4 methylation at all developmental stages and that SET-2 and ASH-2 contribute differently to H3K4me2 in embryos and young adults.
The above analysis does not allow us to distinguish between H3K4 methylation in the soma and germ line. Links between SET-2 and germ-line development include the finding that RNAi of set-2 enhances the maternal-effect sterile (Mes) phenotype of mes-3 and mes-4 mutants, and that immunostaining of SET-2 is enriched in the PGCs of L1s and the germ cells of adults, compared with somatic cells (26). To investigate the contribution of SET-2 to H3K4me3 in the germ line versus the soma, we compared methylation levels in wild-type, set-2, glp-1, and set-2 glp-1 double mutants synchronized as young adults. glp-1(e2141ts) mutants grown at the restrictive temperature lack a germ line and are sterile (27). Comparing wild type and glp-1 shows that ∼60% of global H3K4me3 is derived from the germ line. Comparing glp-1 and set-2 glp-1 shows that SET-2 is responsible for >50% of the H3K4me3 in the soma. With those values, comparing wild type and set-2 suggests that SET-2 is responsible for the majority (∼80–90%) of H3K4me3 in the germ line (see SI Materials and Methods for calculation). A different H3K4 HMT, perhaps the C. elegans MLL homolog SET-16 (19, 20), or a non-SET activity (28, 29) may catalyze the remaining H3K4me3 in the soma and germ line. As set-16 mutants are larval lethal (30), we were unable to test set-2set-16 double mutants for their effect on H3K4 methylation by Western blot analysis.
SET-2 Is Responsible for H3K4 Methylation at the 5′ End of Genes.
In most species, H3K4me3 is enriched around the transcription start sites (TSSs) of actively transcribed genes, whereas H3K4me2 displays a broader distribution (2, 31, 32). To examine the involvement of SET-2 in H3K4 methylation at TSSs of C. elegans genes, we carried out chromatin immunoprecipitation (ChIP) analysis on several ubiquitously expressed genes that in genome-wide studies emerged as having H3K4me3 peaks in their promoter regions in L3s (http://www.modencode.org/). For all three genes tested, pcaf-1, lin-13, and rpa-2, we reproducibly observed H3K4me3 and -me2 enrichment in a region overlapping with the TSS (Fig. 3). This signal was substantially reduced in ChIP experiments carried out on extracts from set-2(bn129) mutant animals at the same stage. No difference in total histone H3 occupancy was observed on any of the loci tested (Fig. S2). Therefore, SET-2 is responsible for most H3K4me3 and -me2 on these genes. Analysis of whole animals failed to show any significant difference in transcript levels from pcaf-1, lin-13, or rba-2 in set-2 mutant worms compared with wild type (Fig. S3).
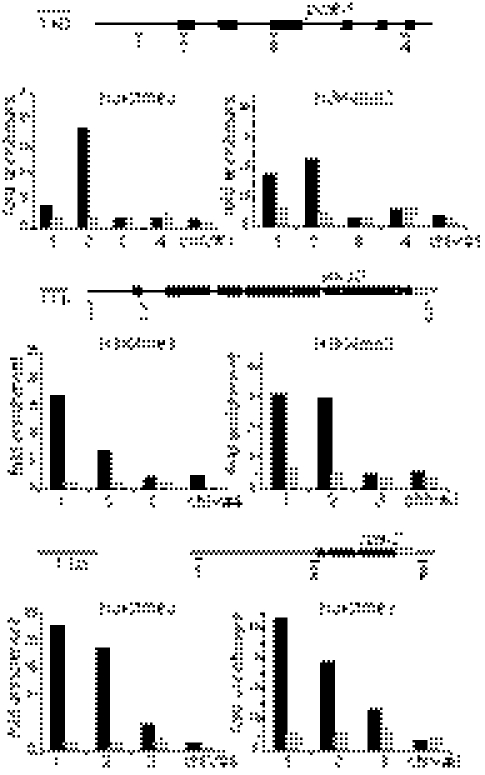
Fig. 3.
SET-2 is required for H3K4 methylation at transcript start sites. ChIP-qPCR analysis was carried out on extracts from synchronized L3 populations of wild-type (solid bars) or set-2(bn129) mutants (shaded bars), using H3K4me3- and H3K4me2-specific antibodies and a preimmune serum as a control. For each gene tested, a cartoon shows the position of the primers used for qPCR analysis and a histogram representing the fold enrichment from ChIP experiments, calculated as the ratio between signal from the antibodies and signal from preimmune serum, along the gene. chIV nos. 1, 4, and 6 are primers from an intergenic region on chromosome IV used as an internal control. The data are representative of at least three independent cultures.
SET-2 and ASH-2 Differentially Affect H3K4me in the Germ Line and Embryos.
To investigate the contribution of SET-2 and ASH-2 to H3K4 methylation in the germ line and embryos, we carried out immunostaining experiments (Fig. 4). In wild-type embryos, high H3K4me3 levels were observed in all cells except the two PGCs, Z2 and Z3 (Fig. 4A, arrows). A similar difference between somatic cells and the PGCs was previously reported for H3K4me2 (24). H3K4me3 was significantly reduced in the somatic cells of set-2, wdr-5, and ash-2 mutant embryos (Fig. 4 B–D) (20). Interestingly, residual signal was evident in the interphase nuclei of all mutant embryos in the form of distinct foci (Fig. 4 E–H and Fig. S4). These foci may represent relocalization of remaining H3K4me3 marks in these mutant contexts, or H3K4me3-enriched regions that are more easily detected when the overall nuclear signal is reduced.
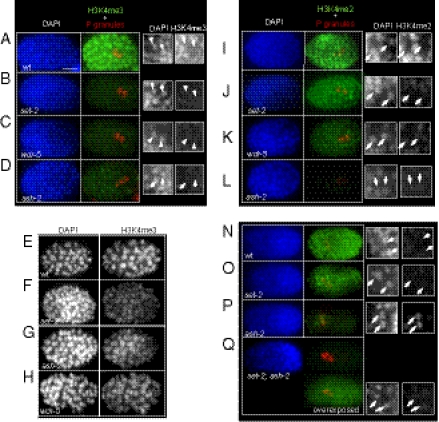
Fig. 4.
SET-2, WDR-5, and ASH-2 differentially influence H3K4 methylation in embryos. Fixed embryos of the given genotype were stained with anti-H3K4me3 (Abcam; ab8580, A–H) or H3K4me2 (Abcam; ab7766, I–P) (green), anti-PGL-1 [red; marking the primordial germ cells (PGCs) Z2 and Z3], and DAPI (blue). Small black and white panels show enlargements of H3K4me staining in PGCs (white arrows). Unless indicated, all images were acquired using the same exposure settings. In E–H, black and white images of H3K4me3 more clearly show residual H3K4me3 in foci in mutant embryos. (Scale bar, 10 μm.)
Staining with H3K4me2 antibodies gave different results (Fig. 4 I–P). In set-2 and wdr-5 mutant embryos, H3K4me2 levels were not as severely affected as H3K4me3 levels (compare Fig. 4 J and K to 4 B and C), consistent with Western blot analysis (Fig. 2 A and B). By contrast, H3K4me2 in ash-2 mutant embryos was greatly decreased under the same staining and exposure conditions used for set-2 and wdr-5 (Fig. 4L). In older ash-2 mutant embryos (at the end of gastrulation, ∼350-cell stage), H3K4me2 levels appeared to be elevated in the PGCs compared with surrounding somatic cells (Fig. 4O, arrows). In wild-type embryos at the same stage (Fig. 4M), H3K4me2 remained low in PGCs compared with the surrounding somatic cells; this mark reappears in wild type only after L1s hatch and feed (24). H3K4me2 signal was absent in set-2;ash-2 double mutants imaged with the same settings used on the single mutants (Fig. 4P). Overexposure showed that a very faint signal was equally present in PGCs and somatic cells. Therefore, SET-2 is responsible for both the accumulation of H3K4me2 in PGCs and the residual signal in the soma of ash-2 mutant embryos.
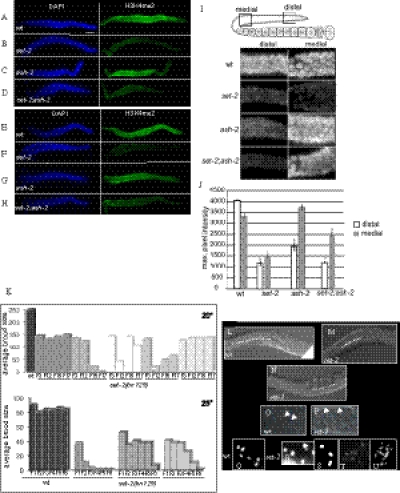
In the germ line of wild-type animals, robust levels of H3K4me3 and -me2 were detected by immunostaining in all germ nuclei in the mitotic and meiotic regions (Fig. 5 A and E). H3K4me3 and -me2 were strongly reduced in the germ line of set-2 mutant animals, particularly in the distal and medial germ line (Fig. 5 B and F). In contrast to set-2, germ lines from ash-2 mutants appeared to have wild-type levels of H3K4me3 and H3K4me2 in all regions (Fig. 5 C and G). Interestingly, in set-2ash-2 double mutants, the overall H3K4me2 signal in the germ line appeared higher than in set-2 single mutants (Fig. 5H). Higher magnification imaging and quantification of the fluorescence signal confirmed that in set-2 mutants, H3K4me2 is reduced in both distal and medial regions (Fig. 5 I and J). This analysis also revealed that in ash-2 mutants, H3K4me2 is specifically reduced in the distal region, although this decrease was not as substantial as in set-2 mutants. Altogether, these results suggest that SET-2 is broadly required for germ-line H3K4 methylation, whereas ASH-2 plays a more complex role in this tissue. SET-2 and ASH-2 may operate together to promote H3K4 methylation in the distal germ line, whereas in the more proximal germ line, SET-2 apparently operates independently of ASH-2. Restoration of some H3K4me2 signal in the medial region of double-mutant germ lines suggests that ASH-2 may normally repress the activity of the other HMT (perhaps SET-16) in this tissue.
Fig. 5.
SET-2 but not ASH-2 is required for H3K4 methylation in the distal germ line. (A–H) Gonads were dissected and stained with anti-H3K4me3 (Abcam; ab8580, A–D) or H3K4me2 (Abcam; ab7766, E–H) (green) and DAPI (blue). (Scale bar, 20 μm.) (I) Close-up images of the distal and medial region of gonads (boxed regions in cartoon) from wild-type and mutant animals. (J) Maximal pixel intensity values for images in I. Error bars show SEM, asterisks denote Mann–Whitney P values <0.05 when comparing the same region between different backgrounds. (K–U) Loss of SET-2 function impairs fertility. (K) set-2(bn129) mutants display progressive sterility. Representative data from one experiment are shown. For experiments at 20 °C, offspring from individual animals (n = 5) were counted at the third generation (F3) and then at F12, F15, and F17, giving the total brood size. For experiments at 25 °C, scoring was every generation starting at F1. Independent lines are depicted by different shades of gray. Similar results were obtained in at least five independent experiments. (L–U) Germ-line defects in set-2 mutant worms. (L) Wild-type proximal gonad; (M) reduced number of oocytes; (N) absence of fertilized embryos in the proximal gonad of set-2 animals (dotted lines); (P) abnormal nuclei in the distal germ line of set-2 animals (arrows) compared with wild type (O). (Q–U) Diakinetic nuclei showing six DAPI-stained bodies in wild type (Q) and nuclei with either fewer (R and S) or more (T and U) than six DAPI-stained bodies in set-2 mutant animals. In R, a DAPI-stained body with an abnormal shape is shown with an arrow.
SET-2–Dependent H3K4 Methylation Is Required for Normal Germ-Line Development and Fertility and Normal Growth Rate.
The above results suggest that SET-2–mediated H3K4 methylation may be important for germ-line function. Increased H3K4me2 retention in PGCs has been shown to correlate with progressive sterility over 15–30 generations (25). We previously showed that a null mutation in the SET1 complex subunit wdr-5/WDR5 also causes a mortal germ-line phenotype (Mrt) (20). However, we and others failed to observe any germ-line defects in set-2(ok952) mutant animals (20, 21). The set-2(bn129) null allele allowed us to reexamine the requirement for SET-2 in fertility.
set-2(bn129) mutant animals have smaller broods than wild type and display temperature-sensitive progressive sterility (Fig. 5K). At 20 °C, the initial average brood size of set-2 animals was 140 ± 20, compared with 250 ± 30 for wild type. In subsequent generations, we observed a variable decrease in brood size for some, but not all lines. Many mrt mutants become progressively sterile only when propagated at elevated temperature (25 °C) (33). At 25 °C, the average brood size of first-generation set-2 homozygous animals was only 40–50 progeny, and by the F6 generation, all animals became sterile. This progressive phenotype is quite distinct from the germ-line phenotype associated with ash-2 mutant animals, which show reduced fertility at permissive temperatures and are near sterile at 25 °C (Fig.S5). Furthermore, the fact that set-2;ash-2 double mutants show more severe fertility defects than either single mutant (Fig.S5) argues for independent functions of set-2 and ash-2 in germ-line maintenance.
Differential interference contrast (DIC) imaging showed that early-generation set-2 animals had fewer oocytes than wild type (compare Fig. 5M and 5L). At later generations, as animals neared sterility, more severe defects were observed, including the absence of fertilized eggs (Fig. 5N) and/or abnormal nuclei in the distal germ line (compare Fig. 5O and 5P). A number of defects can cause a Mrt phenotype, including telomere shortening, mutagenic load, and epigenetic loss of germ-line identity (25, 33–35). To investigate whether the Mrt phenotype of set-2 mutants is associated with gross chromosomal changes, the germ lines of set-2 animals were analyzed by DAPI staining. We observed defects in all lines examined starting at the F3–F4 generation. Unfertilized wild-type oocytes contain 6 DAPI-stained bivalents (Fig. 5Q). In set-2 animals approaching sterility, we observed 1–2% of oocytes (n = 450) with 5, 4 or 3 DAPI-stained bodies at diakinesis (Fig. 5 R and S), suggesting the occurrence of chromosome fusion events. An additional 1–2% of oocytes contained >6 DAPI-stained bodies (Fig. 5T), a defect commonly associated with errors in meiosis (36). In addition, we occasionally observed >12 DAPI-stained bodies (Fig. 5U). These bodies were sometimes smaller and less intensely stained and may represent chromosome fragments formed in the absence of set-2. Therefore, SET-2–mediated H3K4 methylation may be more generally required for genome stability. This result is supported by the observation that set-2 mutants are hypersensitive to UV-induced DNA damage (Fig. S6). The progressive sterility caused by the absence of SET-2 activity could reflect defects that arise from altered chromatin organization or altered gene expression.
To determine whether loss of SET-2 activity also affects somatic development, we examined growth rates. set-2 mutant worms show a slow growth phenotype that is most pronounced at cold temperatures (Fig. S7). This result suggests that changes in chromatin organization or gene expression caused by loss of SET-2 HMT activity impair overall developmental progression.
Discussion
SET-2 Is the Major HMT Responsible for Global H3K4me2 and -me3.
To begin to define the roles of SET1-related complexes in a developmental context, we studied C. elegans SET-2, the homolog of yeast and mammalian SET1, and ASH-2, the single homolog of ASH2. Through analysis of a null allele, bn129, we show that SET-2 is responsible for most H3K4me3 and -me2 in animals at all stages of development. The residual levels of H3K4 methylation observed in extracts prepared from set-2(bn129) mutants may be due to the activity of the MLL-related H3K4 HMT SET-16 (19), another of the 38 SET-domain proteins (30), or even a non-SET HMT complex (28, 29).
SET-2 and ASH-2 Differentially Contribute to H3K4me2 and -me3 in Embryos and the Germ Line.
Like set-2 and wdr-5, ash-2 is required for H3K4me3 in embryos. These results are consistent with SET-2, WDR-5, and ASH-2 acting as part of the same complex for H3K4me3 in embryos. Indeed, SET-2 and WDR-5 could be coimmunoprecipitated from embryonic extracts (Fig. S8). In embryos, loss of SET-2 has a larger effect on H3K4me3 than on H3K4me2, as observed in mammalian cells upon inactivation of SET1 (12). Surprisingly, however, we found that ASH-2 is required for most H3K4me2 in embryos. By contrast, in L4/young adults, global H3K4me2 was only partially reduced in the absence of ASH-2. This difference is likely due to the contribution made by the germ line to bulk H3K4 methylation in L4/young adults and the fact that H3K4me2 is only slightly decreased in the germ line of ash-2 mutant animals. Altogether, these results indicate that the role of ASH-2 in H3K4me2 and -me3 is developmentally regulated.
ASH-2 Influences H3K4me2 Independently of SET-2.
Mono-, di-, and trimethylation activities have been attributed to SET1 family complexes in vivo and in vitro, but conflicting reports exist on the enzymatic activity of purified complexes (37). Recently, a complex containing WDR5, RbBP5, Ash2L, and DPY-30, but lacking MLL1, was shown to possess HMT activity in the absence of a conserved SET domain (28, 29). Furthermore, ASH2, WDR5, and RBP5 have been identified in a complex in the absence of SET1 (38, 39). These data suggest that the substrate and product specificity of H3K4 HMT complexes may be regulated by specific protein–protein interactions in the cell. Our results suggest that in embryos, SET-2 and ASH-2 function together to mediate H3K4me3, but that ASH-2 functions independently of SET-2 to mediate H3K4me2.
Whereas in wild-type embryos H3K4me2 is undetectable in the PGCs by immunostaining, in late-stage ash-2 mutant embryos, H3K4me2 levels appear higher in PGCs compared with the surrounding somatic cells. Furthermore, ASH-2 activity is dispensable for H3K4me3 in the adult germ line, but is specifically required for H3K4me2 in the distal region. Because H3K4me2 is undetectable in PGCs of ash-2;set-2 double-mutant animals, ASH-2 may antagonize SET-2–dependent H3K4me2 in PGCs and perhaps in developing germ cells. One mechanism by which ASH-2 could antagonize SET-2 activity is through the recruitment of an H3K4 demethylase activity, as reported in yeast and Drosophila (40, 41). Alternatively, increased H3K4me2 could reflect decreased conversion of H3K4me2 to H3K4me3 in the absence of ASH2. Although the significance of ASH-2–dependent H3K4me2 in C. elegans embryos is currently unclear, a growing body of data suggests that different levels of H3K4 methylation may define distinct chromatin states (2, 42, 43).
Greer et al. (21) recently suggested that ASH-2, SET-2, and WDR-5 act in the same complex to influence H3K4 methylation in the germ-line and worm life span. However, the effect of loss of ASH-2 function on H3K4me2 or -me3 in the germ line was not directly investigated in that study. Our data suggest that although SET-2 is a major contributor to H3K4 methylation in that tissue, ASH-2 is likely to play a more complex role at least partly independent of SET-2.
SET-2 Is Required for Germ-Line Stability.
set-2 null mutants become progressively sterile and show evidence of chromosome fusion and defects in meiosis. SET-2 and/or H3K4 methylation could function to maintain meiotic fidelity through chromatin structure, as shown for mutants defective for H3K9me2 in the adult germ line (44). The accumulation of various forms of heritable cellular damage could also contribute to the mortal germ-line phenotype (45). A role in genome stability is suggested by the fact that set-2 mutants are more sensitive to UV-induced DNA damage, as recently reported for yeast Set1 (46). Alternatively, loss of SET-2/H3K4 methylation might affect the transcription of genes required for germ-line function. We have shown that, as in other species, SET-2 regulates H3K4 methylation of the 5′ region of genes. Loss of H3K4 methylation, however, was not associated with an obvious change in mRNA level for the three genes tested in this study. In yeast, no correlation was found between H3K4 methylation and gene expression (3), and additional factors are likely to recognize this mark to mediate downstream effects (2). However, we cannot exclude that SET-2 may have subtle effects on gene expression or influence the expression of genes only in specific tissues, including the germ line. A specific function for a SET1/MLL family protein in the germ line was recently reported for MLL2, which was found to be required in oocytes for bulk H3K4me3 and transcriptional silencing (47). In C. elegans, erasure of H3K4me has been shown to be important for resetting of germ-line transcriptional profiles (25). Our results suggest that establishing correct H3K4me may also be important for correct programming of germ-cell development. Altogether, our data highlight the importance of studying the functions of individual components of H3K4 HMTs in a variety of model organisms and developmental contexts. In light of the high degree of conservation in the composition and molecular mechanisms of H3K4 HMT complexes, the functions reported here are likely to be conserved in higher eukaryotes.
Materials and Methods
Strains and Alleles.
C. elegans strains were maintained and manipulated as described (48). The Bristol N2 strain was used as the wild type. Mutations used were as follows: LGII, ash-2(tm1905); and LGIII, set-2(ok952), set-2(bn129), wdr-5(ok1417), and glp-1(e2141ts).
Isolation of the bn129 Allele of set-2.
The bn129 allele was identified in a PCR screen of a deletion library generated according to the method of Koelle et al. http://medicine.yale.edu/labs/koelle/Site/Protocols.html. set-2(bn129) was backcrossed to wild type 10 times.
Quantitative Western Blot Analysis.
Staged worms were pelleted in M9 buffer and frozen at −80 °C. Pellets were recovered in TNET buffer [50 mM Tris·HCl (pH 8), 300 mM NaCl, 1 mM EDTA, 0,5% Triton X-100, 1 mM PMSF, and protease inhibitors] (Pierce) and sonicated. Homogenates were centrifuged and supernatants aliquoted and frozen at −80 °C. Total protein amount was quantified by the Bradford method. Serial dilutions of protein extracts were electrophoresed for Western blot analysis to verify linearity before quantification (examples are shown in Fig. S1). Blots were incubated with the following antibodies: anti-H3 (Abcam; 1791), 1:20,000; anti-H3K4me2 (Abcam; ab7766), 1:10,000; anti-H3K9me3 (Upstate; 07–442), 1:1,000; anti-H3K4me3 (Diagenode; cs-003-100), 1:3,000; and monoclonal anti-actin (MP Biomedicals; clone C4), 1:20,000 as internal controls of protein loading. Each analysis was repeated with at least four independently derived extracts. Quantification of scanned blots was carried out using Image J, and H3K4me signals were normalized to the level of histone H3 and actin.
Chromatin Immunoprecipitation.
ChIPs were performed as described in ref. 32 with some minor modifications. A detailed protocol is in SI Materials and Methods.
Quantitative RT-PCR.
cDNAs were synthesized using the iScript cDNA synthesis kit according to the manufacturer's protocol. qPCR reactions were performed using LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche) and the Light Cycler 1.5 Detection System (Roche).
Immunofluorescence.
Worms were dissected and permeabilized as described in ref. 20 and fixed in methanol for 2–3 min, followed by 1% paraformaldehyde for 5 min. Slides were blocked in 0.5% BSA in PBS with 0.25% Triton (PBSTB) for 30–60 min. Samples were incubated with primary antibody (diluted in PBSTB) overnight at 4 °C and secondary antibody at room temperature for 1–2 h. Samples were mounted in Fluoroshield plus DAPI (Sigma). Primary antibodies were anti-H3K4me3 (Abcam; ab8580, 1:20,000), anti-H3K4me2 (Abcam; ab7766, 1:20,000), and anti-PGL-1 (Developmental Studies Hybridoma Bank; 1:1,000). Similar results were obtained using H3K4me3 cs-003-100 from Diagenode and H3K4me2 308-34809 from Wako. Images were acquired using a Zeiss Axioplan2 imaging system with a Coolsnap HQ camera. Z-stacks were projected using ImageJ.
Characterization of the Mortal Germ-Line (Mrt) Phenotype.
Four independent lines were established from siblings of set-2(bn129) F1 freshly backcrossed homozygotes. For each line, five worms from each generation were picked to single plates with fresh Escherichia coli, in the presence of excess food. Average brood sizes were calculated by counting the total number of progeny per plate starting at the F3 at 20 °C and every generation starting at the F1 at 25 °C.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center and the National Bioresource Project for strains. F.P. and Y.X. were supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche, and the Association de la Recherche sur le Cancer. S.S. and S.D. were supported by National Institutes of Health Grant GM34059.
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019290108/-/DCSupplemental.
References
- 1.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller T, et al. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs SD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roguev A, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 7.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Goo YH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 11.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 14.Glaser S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 15.Heuser M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113:1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 16.Madan V, et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113:1444–1454. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood. 2009;113:1455–1463. doi: 10.1182/blood-2008-05-159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA. 2008;105:19229–19234. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher K, Southall SM, Wilson JR, Poulin GB. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev Biol. 2010;341:142–153. doi: 10.1016/j.ydbio.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Simonet T, Dulermo R, Schott S, Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol. 2007;312:367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 21.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamson AL, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltran S, Angulo M, Pignatelli M, Serras F, Corominas M. Functional dissection of the ash2 and ash1 transcriptomes provides insights into the transcriptional basis of wing phenotypes and reveals conserved protein interactions. Genome Biol. 2007;8:R67. doi: 10.1186/gb-2007-8-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics. 2001;159:1019–1029. doi: 10.1093/genetics/159.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Austin J, Kimble J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell. 1989;58:565–571. doi: 10.1016/0092-8674(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 28.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A novel non-set domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2010;286:3359–3369. doi: 10.1074/jbc.M110.174524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 31.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6(9):e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 34.Youds JL, O'Neil NJ, Rose AM. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics. 2006;173:697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanowitz JL. Genome integrity is regulated by the Caenorhabditis elegans Rad51D homolog rfs-1. Genetics. 2008;179:249–262. doi: 10.1534/genetics.107.076877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillers KJ, Villeneuve AM. Analysis of meiotic recombination in Caenorhabditis elegans. Methods Mol Biol. 2009;557:77–97. doi: 10.1007/978-1-59745-527-5_7. [DOI] [PubMed] [Google Scholar]
- 37.Cosgrove MS, Patel A. Mixed lineage leukemia: A structure-function perspective of the MLL1 protein. FEBS J. 2010;277:1832–1842. doi: 10.1111/j.1742-4658.2010.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 39.Dehé PM, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 40.Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roguev A, et al. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J Biol Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- 42.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orford K, et al. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessler JB, Andersen EC, Villeneuve AM. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 2010;6:e1000830. doi: 10.1371/journal.pgen.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smelick C, Ahmed S. Achieving immortality in the C. elegans germline. Ageing Res Rev. 2005;4:67–82. doi: 10.1016/j.arr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Faucher D, Wellinger RJ, et al. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8):e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreu-Vieyra CV, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 8(8):e1000453. doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.