Abstract
We developed a theoretical framework to prove the existence and quantify the Waddington landscape as well as chreode-biological paths for development and differentiation. The cells can have states with the higher probability ones giving the different cell types. Different cell types correspond to different basins of attractions of the probability landscape. We study how the cells develop from undifferentiated cells to differentiated cells from landscape perspectives. We quantified the Waddington landscape through construction of underlying probability landscape for cell development. We show the developmental process proceeds as moving from undifferentiated to the differentiated basins of attractions. The barrier height of the basins of attractions correlates with the escape time that determines the stability of cell types. We show that the developmental process can be quantitatively described and uncovered by the biological paths on the quantified Waddington landscape from undifferentiated to the differentiated cells. We found the dynamics of the developmental process is controlled by a combination of the gradient and curl force on the landscape. The biological paths often do not follow the steepest descent path on the landscape. The landscape framework also quantifies the possibility of reverse differentiation process such as cell reprogramming from differentiated cells back to the original stem cell. We show that the biological path of reverse differentiation is irreversible and different from the one for differentiation process. We found that the developmental process described by the underlying landscape and the associated biological paths is relatively stable and robust against the influences of environmental perturbations.
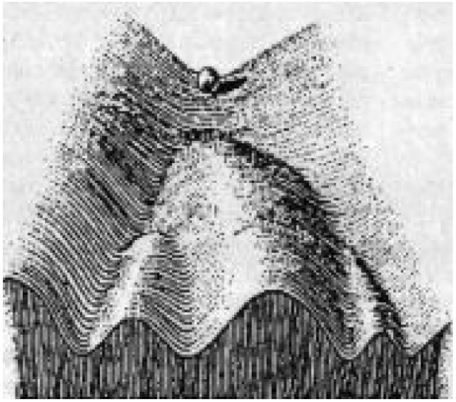
Cells are dynamical entities: They change their phenotype during development in an almost discontinuous manner, giving rise to discrete developmental stages (such progenitor and differentiated states) as well as discrete lineages and terminally differentiated types. This pattern of cell dynamics during development was already noted by C. Waddington in the 1940s, leading to his by now famous metaphor of the epigenetic landscape (1) (see Fig. 1) In this iconic picture a marble rolls down a surface (landscape), staying in valleys and seeking the lowest point. At watersheds, the valleys branch so that the marble takes one of two available paths. In Waddington’s picture, the ball represents a developing cell in an embryo and the landscape epitomizes some more abstract set of constraints, thus clearly heralding the notions of stability and instability in the modern sense of dynamics (1). Indeed, it has recently become clear that Waddington’s epigenetic landscape in principle represents the dynamics of a system of gene regulatory interactions that impose constraints to and drive cell development (2, 3), giving a metaphor for the qualitative understanding of developmental processes of cells. However, a detailed quantitative examination of how the dynamics of a gene regulatory circuit that governs binary cell fate decisions produces a generalized potential landscape that may recapitulate the epigenetic landscape has not been presented. In other words, it is not very clear on exactly what the Waddington landscape represents, how the qualitative picture of landscape can be quantified, and how the connection to the experiments can be made.
Fig. 1.
The original artistical picture of Waddington epigenetic landscape (1).
Here we develop a theoretical framework to show the existence of such a landscape as the formal representation of the dynamics of a gene circuit and quantify its detailed topography. We define Waddington’s chreodes—the biological paths (or trajectories) of development. Herein, the entity that changes, and hence is embodied by the marble, is the gene expression pattern of a cell that reflects the network state of the genes in a particular network. The state S(t) = (x1,x2,xi ..xN) thus reflects the vector consisting of the expression values (cellular concentration and activity of the products of gene i), x1, x2, in a gene regulatory network of N genes at a given time t. Because of the regulatory interactions not all states can be realized with equal ease and in a system with fluctuations, probabilities can be assigned to the states S. They can have a higher or lower probability of appearance that translates inversely into the elevation (potential) of the landscape (4, 5). The states with locally highest probability (lowest potential) represent attractor states of the gene regulatory network as a dynamical system, surrounded by their basins of attraction. Attractor states have been proposed to represent cell types (6).
The landscape metaphor has recently seen renewed interest with the arrival of cell reprogramming (5, 7, 8). If cell types are attractors, then reprogramming represents the transition between attractors. Thus the height of hills between two valleys, or the barrier heights between the attractors, can be correlated to the escape time from the basins, offering a quantitative measure for the nonlocal relative stability of the attractors or cell types. The landscape topography thus has a quantitative meaning.
Here we construct such a probability landscape quantitatively based on the underlying gene regulatory circuit for the biological process of a binary cell fate decision of a multipotent progenitor cell. Fate decision and subsequent lineage commitment to a particular fate is described by a path in this landscape from the attractor of the progenitor cell to that of either one of two differentiated cell types. These paths can be identified and quantified through counting the weights of all the possible paths and selecting the optimal paths with largest weights. We will do so with a path integral approach (9–11). The advantage of the path integral approach is that it addresses the fundamental issues of biological paths directly. Furthermore, the weights associated with the biological paths can also be determined. By varying the configurational states, we can also explore the corresponding probability landscape topography and investigate the association of escape time with the barrier height for global stability analysis of the cell types. Thus, landscape, paths, kinetics, and stability that are all of practical interest for determining reprogramming strategies, can be evaluated in one model.
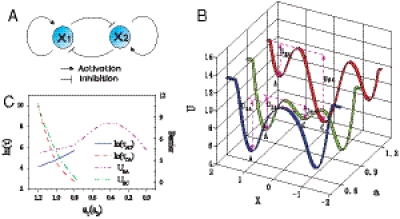
We will study an important example of cell developmental circuit (Fig. 2) (12) composed of a pair of self-activating and mutually inhibiting genes as a model, a gene regulatory motif that has been found in various tissues where a pluri/multipotent stem cells has to undergo a binary cell fate decision (8, 12). For instance, in the multipotent common myeloic progenitor cell (CMP), which faces the binary cell fate decision between the myeloid and the erythroid fate, the fate determining transcription factors (TF), PU.1, and GATA1, which promote the myeloid or the erythroid fates, respectively, form such a circuit. The relative expression levels x1 (PU.1) and x2 (GATA1) of these two reciprocal TFs can tilt the decision toward either lineage (7, 12).
Fig. 2.
(A) The illustration of cell fate decision module. (B) The landscape of the development at different stages or different paramenters. (C) Escape time under certain fluctuations versus the parameter a and the barrier height from the central undifferentiated state to differentiated state versus the parameter a. (a = a1 = a2, b1 = b2 = 1, k1 = k2 = 1, and S = 0.5, n = 4).
We quantitatively constructed the probability landscape for the GATA1-PU.1 type of circuit, showing its similarity and difference with Waddington’s epigenetic landscape, and quantified the developmental paths. We show that the cell development process proceeds from the basin of attraction of the undifferentiated state to that of the differentiated attractors. The heights of the barriers separating the basins of attractions correlate with the escape time that reflect the stability of cell types. Specifically, we demonstrate that the developmental process can be quantitatively described by the biological path on a quantified Waddington landscape and is governed by a combination of a gradient and a curl force on the landscape. The paths of differentiation do not follow the steepest descent path on the landscape (gradient) and the paths of the reverse differentiation process is different from the ones of the differentiation process indicating irreversibility.
Results and Discussions
Developmental Network and Cell Fate Decision Module.
A gene regulatory circuit that governs binary cell fate decision module is shown in Fig. 2A. It consists of mutual regulation of two opposing fate determining master TF X1 and X2. The module has been shown to control developmental cell fate decision and commitment in several instances of multipotent stem or progenitor cells that faces a binary fate decision, (i.e., GATA1 and PU.1) (8, 12). X1 and X2 are coexpressed in the multipotent undecided cell and committed to either one of the two alternative lineages is associated with either one factor dominating over the others, leads to expression patterns in a mutually exclusive manner (13). Importantly, in many cases the genes X1 and X2 also self-activate (positive autoregulate) themselves (Fig. 2A). The circuit can be described by the following minimal system equations (12): 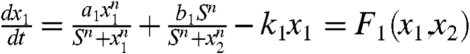 and
and 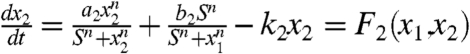 . In vector form, dx/dt = F(x) = [F1(x1,x2),F2(x1,x2)]. x1 and x2 represent the cellular expression or activation levels of the two lineage determining transcription factors X1 and X2, and a1, a2, b1, b2, k1, k2 are positive parameters that denote the strength of the following interactions or processes: The first expression represents, in the common formalization (12), a self-activation (of strength a1, a2) that obeys a sigmoidal transfer function, the second term represents mutual inhibition, given basal expression, (of strength b1, b2); and the last term is the first-order inactivation (degradation) of either factor with the rate k1, k2. For our purpose it suffice to consider the symmetric situation a = a1 = a2; b = b1 = b2; k = k1 = k2. The mutual regulations and self-activations are often separated and do not have to be simultaneously changed in a synchronized way in the experiments, therefore the underlying interactions described above follow an OR rather than AND logic. (Details in SI Text).
. In vector form, dx/dt = F(x) = [F1(x1,x2),F2(x1,x2)]. x1 and x2 represent the cellular expression or activation levels of the two lineage determining transcription factors X1 and X2, and a1, a2, b1, b2, k1, k2 are positive parameters that denote the strength of the following interactions or processes: The first expression represents, in the common formalization (12), a self-activation (of strength a1, a2) that obeys a sigmoidal transfer function, the second term represents mutual inhibition, given basal expression, (of strength b1, b2); and the last term is the first-order inactivation (degradation) of either factor with the rate k1, k2. For our purpose it suffice to consider the symmetric situation a = a1 = a2; b = b1 = b2; k = k1 = k2. The mutual regulations and self-activations are often separated and do not have to be simultaneously changed in a synchronized way in the experiments, therefore the underlying interactions described above follow an OR rather than AND logic. (Details in SI Text).
In this example, the self-activation strengths change due to the regulations on the transcription factors by other regulators such as Klf4 (14) during the developmental process. As we will see that parameter a of self-activation strength gives a sense of measure and direction of the development.
Landscape at a Given Stage of Development: Cross-Section of Waddington Landscape.
Based on the above dynamics of the cell fate decision, we quantitatively mapped out landscape of the development at different stages in Fig. 2B. In the resulting landscape representation, the vertical axis represents the potential landscape U (inversely related to the probability landscape P: U = - ln P) (5). The horizontal axis represents the expression level of either x1 or x2 (due to the symmetry between x1 and x2 as well as for easy visualization we only show one of them).
We see that the decision making circuit often has three basins of attractions. The center attractor C represents the undifferentiated undecided multipotent stem cell state with balanced expression of the two opposing fate determining transcription factors (13). The side attractors A and B represent the the differentiated states with mutually excluding expressions of x1 and x2 (12). In this view the three attractors become evident as potential wells.
When the self-activation is strong (characterized by large value of a), the central basin of attraction is deep and therefore the cell is more preferred to stay there, corresponding to undifferentiated conditions. As self-activation is weaker, the basins of attractions on the side become deeper, thus the differentiated states are preferred. At the extreme value of a where self-activation is weak, the central basins of attraction becomes hilltop and therefore unstable, only the differentiated states are preferred. Thus destabilization of the central progenitor attractor as the self-activation parameter a is gradually decreased (Fig. 2B) (12) represents one possible mechanism for the development undifferentiated cell C committed to either differentiated cell A or B.
Another possible way to formalize the fate commitment is to assume there is stochastic fluctuation-driven transition from the central undifferentiated attractor into either of the side differentiated attractors. The fluctuations can be intrinsic from small copy numbers of molecules or extrinsic from the environments. The strength of fluctuations is quantified by diffusion coefficient in method section. Experimental evidences support the role of both a destabilization of the progenitor undifferentiated attractor (12), as well as gene expression fluctuation-induced state transitions (15).
Fig. 2C shows the barrier height as a measure of the landscape topography) for the transition from the central undifferentiated state to the differentiated state (green) and the escape time (red) from the former state under certain fluctuations versus the parameter a mimicking the differentiation process. We can see that the escape time correlates with the barrier height. The barrier height decreases (increases) as parameter a decreases (increases) during development, and the escape time becomes faster (slower). This guarantees the stability of the developmental process to the differentiated states (when parameter a goes down) or stability of undifferentiated state (when parameter a goes up) (5).
As one can see, the chance for differentiation at initial stage of development when a is large is nonzero but small (long escape time). As development proceeds (a decreases) the chance for differentiation becomes larger and reaches the largest when the undifferentiated state becomes unstable. We see both induction for instability of undifferentiated state through the change of self-activation and fluctuations in action for the developmental processes in our picture.
Pathways at a Given Stage of Development.
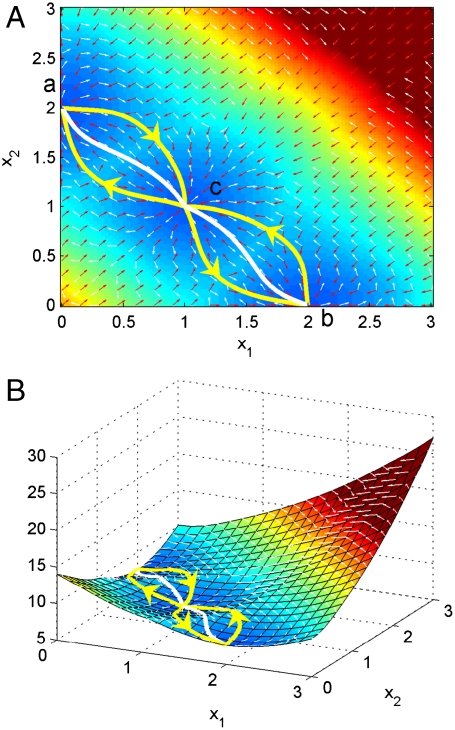
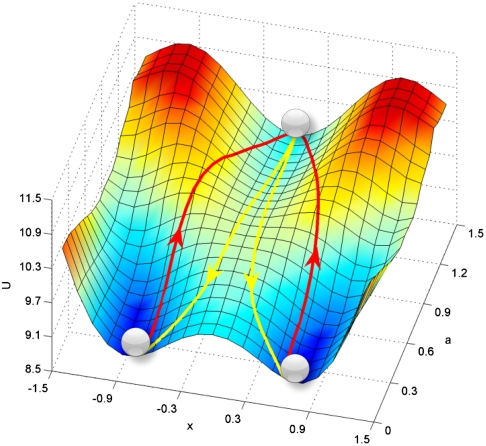
From our path integral approach, we can uncover quantitatively the developmental paths from the undifferentiated state to the differentiated state at certain stage of development (at certain value of self-activation parameter a). Fig. 3 shows the transcription factor expression level X1/X2 state space as two dimensional contour and three dimensional landscape and the paths from theoretical reversed multipotent undifferentiated state to differentiated state for development (and the paths from differentiated state to undifferentiate state) at certain developmental stage characterized by self-activation strength a.
Fig. 3.
(A) Two dimensional illustration of dominant kinetic path and flux between three basins of attraction in gene network. (B) Three dimensional illustration of dominant kinetic path and flux between three basins of attraction in gene network.
Fig. 3 A and B show that effective developmental paths (from central basin of the undifferentiated state to side basin of the differentiated state and vice versa) and the undifferentiated ones do not follow the gradient paths, that is the path determined by following the steepest directions of U. As we pointed out earlier, (for details see method and supporting information), the dynamics or paths of such developmental circuit is determined by both the force from the gradient of the landscape and an additional term representing the curl flux (4). The additional dynamical driving force emanating from the curl flux causes the paths to deviate from the naively expected steepest descent path computed from the gradient of U. This quantitative picture of paths is different from what would follow from Waddington’s picture where the developmental paths, symbolized by the rolling down of a marble, follow the gradient of the underlying landscape. By contrast, the real developmental paths do not simply go down the gradient but also are driven by a curl force leading to spiraling movements that can be quantitatively tested in experiments.
Furthermore, the forward developmental paths and backward retrodifferentiation paths are not identical. In other words, the developmental pathways are irreversible. The differentiation and the reverse process of retrodifferentiation follow different routes. This is unexpected from the original Waddington picture. The irreversibility of the developmental pathways is very fundamental and provides a unique prediction to test for developmental biology.
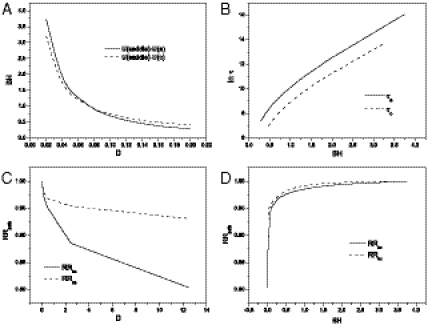
In Fig. 4 A and B, we show how fluctuations (defined in the method section as the strength of the autocorrelation function of protein concentrations quantitatively measured by the diffusion coefficient D) influence the (apparent) population barrier heights and the escape time for development. We see when the fluctuations increase, the population barrier height decreases, the escape time is faster. Nonzero but small fluctuations can help to accelerate the developmental processes. Large fluctuations can be damaging because the resulting population landscape becomes flat and therefore no essential preference or distinction among differentiated and undifferentiated state (equal probability) as well as other states. Therefore the landscape for development is stable against certain fluctuations.
Fig. 4.
(A) Barrier heights BH from side (center) to center (side) basin with solid line (dotted line) versus fluctuations via diffusion coefficients, D. (B) Logarithm of escape time τ from center (side minimum) versus barrier height, BH. (C) RRpath from from center (side) to side (center) versus D. RRpath represents the ratio of the weights between the path at fluctuation strength D compared with the low fluctuation strength D0 = 0.01. (D) RRpath from versus the barrier height BH.
In Fig. 4 C and D, we show how the fluctuations influence the paths. We can see that the consistency from the original paths for development decreases as fluctuations increase. For small fluctuations, the paths do not deviate much from the original ones. When fluctuations further increase, the barrier decreases, increasing the chances for different paths to go from undifferentiated state to the differentiated state. Therefore, the paths deviate more from the original paths when the fluctuations are larger. The paths for development is stable and robust against certain fluctuations.
Quantifying the Waddington Landscape and Paths of Development.
From experimental evidences (5, 12), both mechanisms are in action for development: the instructive change of landscape via decrease of a and the stochastic state transitions in cell differentiation. To cover the whole picture of the developmental process, one needs to explore the dynamics of different developmental stages (from the stage where undifferentiated state is preferred to the stage where the differentiated states are preferred). Because in our model the different stages of development are characterized by a single parameter, the self-regulation strengths. We use that as our reaction coordinate for development, and the other coordinate (cross-section coordinate connecting undifferentiated to differentiated state, as in Fig. 3) in which the relationship between the undifferentiated state to differentiated state can be displayed and the expression level changes of the transcription factors can be monitored. The third and vertical axis is the height of the potential landscape (see Fig. 5).
Fig. 5.
The quantified Waddington developmental landscape and pathways (a = a1 = a2, b1 = b2 = 1, k1 = k2 = 1, S = 0.5, n = 4, and λ = 0.01).
We have discussed the dynamics at certain stage of development in the previous section. The full landscape and dynamical paths for the development requires the specification of the dynamics of self-regulation parameter a. We assume the self-activation strengths decrease in the developmental process due to the influences of the other regulators in a self-degraded fashion. Then the dynamical equations for the developmental process is controlled by the following equations: 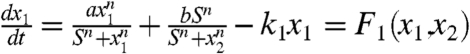 ; and
; and 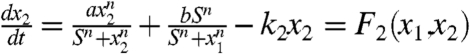 ; and
; and 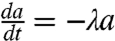 . Here λ is the self-degradation rate. We assume self-activation strength a representing the developmental process changes relatively slowly compared to the dynamics of the x1 and x2 (For full time scale dynamics, see discussions in SI Text). In this way, in each developmental evolution stage, the system has time to relax to steady state among x1 and x2.
. Here λ is the self-degradation rate. We assume self-activation strength a representing the developmental process changes relatively slowly compared to the dynamics of the x1 and x2 (For full time scale dynamics, see discussions in SI Text). In this way, in each developmental evolution stage, the system has time to relax to steady state among x1 and x2.
Based on the above developmental network circuit, we quantitatively obtain our probability landscape and associated paths for the developmental processes. This can be seen as the quantification of the Waddington landscape and associated chreodes.
Fig. 5 shows the developmental landscape and paths. The developmental reaction coordinate is parameter a. At large a, the undifferentiated basin of attraction dominates and stable as shown in the cross-section coordinate X linking side minimum-central minimum-side minimum representing the gene expression level. As the developmental process progresses (parameter a decreases), the undifferentiated state becomes less and less stable, the differentiated state becomes more and more stable forming basins of attractions. The qualitative similarities between our quantified landscape/paths in Fig. 5 and Waddington’s landscape/paths in Fig. 1 are obvious. So the basic picture and features conjectured by Waddington for development and differentiation do exist and can now be quantified based on the underlying gene regulatory circuit. The quantitative description of the landscape and paths now allow for predictions. The Waddington landscape is no longer a metaphor. It is physical and quantifiable by the underlying probability landscape.
However, we need to point out to the differences between the quantified Waddington landscape/paths in Fig. 5 and one suggested by Waddington’s marble shown in Fig. 1. Waddington describes the developmental processes as the downhill rolling of the marble. The undifferentiated state is unstable, which triggers the differentiation process. Whereas it is true that in vivo during embryonic development the stem cells are not stable (except when taken into culture and provided with the appropriate factors), in the adult the multipotent undifferentiated cells are stable. In contrast to Waddington, our model allows for temporal stabilization of the undecided stem or progenitor state. This is not captured by the original Waddington picture of landscape. Our quantified landscape covers all stages of development process. Before or near the very early developmental stage, the undifferentiated state can still be stable but has a small but finite chance to climb up (induced from fluctuations) from the basin of attraction for escaping to the differentiated states. This process can happen even before the undifferentiated state become unstable.
The cell fate decision making process for development is also different in our picture compared with the Waddington picture. Whereas Waddington believes that the cell fate decision happens at the hill top, in our potential landscape we see that even before climbing up (induced by fluctuations) to the barrier top, the developmental paths (yellow lines) have already started to depart or bifurcate from each other. The bifurcation starts from the undifferentiated state even when it is still stable (down in the basin of attraction). The random directions of climbing out of the undifferentiated basin can bias to the choice of the different cell fates later on. This is the second quantitative difference between our quantified Waddington landscape (Fig. 5) and the original Waddington picture (Fig. 1).
We also quantified the dominant course of developmental paths from undifferentiated states (central basins of attraction) to the differentiated states (side basins of attractions) shown in yellow lines (Fig. 5). The developmental paths start from the undifferentiated states, climb up from the basin to reach the top, and then gradually bifurcate to two distinct pathways along the landscape valley to the differentiated states. As shown in Fig. 5, the developmental paths clearly do not follow gradient paths that the gravity driven metaphor of Waddingon would predict. This is the third quantitative deviation of our pathways (Fig. 5) from the Waddington paths or chreodes (Fig. 1). This is due to the fact that dynamics is controlled by both the force from landscape gradient and the force from the curl flux. The curl flux force makes the developmental path deviate from the steepest descent gradient path. This provides testable predictions.
Furthermore, there is a forth difference between our developmental paths (Fig. 5) and the Waddington paths (Fig. 1). The reverse paths in the original Waddington picture is supposed to be the same as the forward path. In other words, the retrodifferentiation process would follow exactly the same paths as differentiation except with opposite direction. By contrast, in our quantitative path picture, the paths are no longer reversible. The developmental paths (yellow lines) are clearly distinct from the retrodifferentiation paths (shown in red lines). This could be tested by studying the effective developmental path for instance in the iPS generation process. The study of paths on a potential landscape of development suggests a way to reverse the developmental process. We can increase the parameter a by increasing the self-activation strength of the transcription factors through the regulations or over expression of both transcription factors X1 and X2, as of the ongoing efforts in the stem cell research.
One should keep in mind that the quantified Waddington landscape topology depend on the structure of the underlying gene regulatory circuits. Accordingly, genetic mutations that change the network architecture through the wiring node, resulting rewiring (i.e., coupling strength b), change the resulting potential landscape as well as the paths for development. A slight distortion of the landscape may result in biasing paths to entire distinct, but independently robust valleys. This would straightforwardly explain homeotic mutations that Waddington noticed from studying drosophila. On the other hand, the landscape topology can also be altered by the links and connectivity strengths even when there is no missing nodes or genetic mutations. So through the epigenetics with environmental and local chemical reaction changes as Waddington envisioned, the network connection strengths can be altered, resulting the changes of the Waddington landscape and associated paths for development. Therefore both genetic changes (mutations on gene nodes) and epigenetic changes (link strengths between genes) are important and can alter the topology of landscape and paths for development.
Our study provides a framework for studying the global nature of the binary fate decision and commitment of a multipotent cell into one of two cell fates (8). The two fate options for CMP in hematopoietic system correspond to the erythroid fate and the myeloid fate attractor (side minimum). Measurement of PU.1 and GATA1 are confirming the predicted symmetric gene expression configuration in the progenitor state, which exhibits the coexpression of both fate determining factors at intermediate levels (center minimum) (12). Further experimental verifications and design can be studied by the use of two different GFPs for PU.1 and GATA1 and collecting the expression data. The joint histogram will directly give the information of the underlying landscape, confirming the tristable basins for cell decision making. Further experimental exploration can be carried out through the modulation of the transcription factor Klf4 for controlling the self-regulation strength and therefore the differentiation/undifferentiation process. The real time trajectories of expressions can provide us information on dynamics (jumps between basins) and kinetic paths, linking to the underlying landscape topography such as basins and barrier heights in between (through joint histograms). Previous experimental and theoretical studies (12) explored the natures of the cell fate decision systems using flow cytometry and dynamical system analysis at certain stages of development. In the present work, we focus on the underlying global landscape, transitions between the attractors and associated kinetic paths for the dynamical developmental process.
Methods and Models
Potential and Flux Landscape for Development.
We first evaluated the global dynamics of the developmental network motif so that we can quantitatively assign potentials to the attractor states and determine their distinct relative depth within the same frame of reference. The idea of a potential landscape describes how forces acting in a system relate to its global behavior. They are particularly useful for systems of interacting components, such as chemical reactions and protein dynamics, motion, and folding (16). However, these applications deal with equilibrium systems where the potential function is a priori knowable. For nonequilibrium systems, such as gene circuits that exhibit stable stationary states in higher (N > 1) dimensional state space, the intuition of some form of a potential is still warranted and widely used metaphorically (1); however, its functional form is not easy to obtain. In the gene circuit dx/dt = F(x), exemplified in Eq. [S2] of SI Text, the vector F(x) is the force that drives the system. However, F(x) cannot in general be written as a gradient of a potential U: F(x) ≠ -grad(U) for systems of more than one dimensions. For stochastic systems, the system is not determined by the local trajectories but by the probability distributions. The probability distribution follows master equations for discrete state space or diffusion equations for continuous case.
In continuous representation, the diffusion equation for probability evolution can be written in the form of the probability conservation: ∂P/∂t + ∇·J = 0 where J is the probability flux J = FP - D∇P. This means flux in and out of the system gives the increase or decrease of the local probability. Whereas in nonequilibrium systems at steady-state the divergence of the probability flux Jss vanishes ∇·J = 0, the flux itself need not vanish. When local flux is equal to zero, the detailed balance is preserved and the system is in equilibrium state. When local flux is not equal to zero, the detailed balance is broken and the system is in nonequilibrium state. We found (details in SI Text) (4, 5) that F = -D·∇U + Jss/Pss. We have decomposed the force driving the dynamics of the system into two terms, the gradient of the potential U where U is linked with the steady-state probability by U = - ln(Pss) and curl flux force linking the divergence-free steady-state (long time limit) probability flux Jss (velocity current) and the steady-state probability Pss (density). Divergence-free flux has no place to start or end. It is in this sense that the flux has a curl nature.
We treated x1 and x2 as independent variables. For the complete model of development, both concentration variables x1 and x2 characterizing the gene expression and self-regulation variable a are dynamical (as shown in Fig. 5). For simplicity of the treatment, we assume here the D is a constant independent of concentrations. Detailed discussions are in SI Text.
Developmental Pathways Through Path Integral.
The dynamics of the cellular network has often been studied by the chemical reaction rate equations of various local protein concentration species. However, both internal statistical fluctuations from finite number of molecules within the cell and external fluctuations from cellular environments can be significant (17). Therefore it is more appropriate to formulate the dynamics of the chemical rate equations in the noisy fluctuating environments. The dynamics therefore can be formulated as: 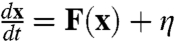 where x is the protein concentration vector, F(x) is the chemical rate flux vector. η is Gaussian noise term where its autocorrelation function is 〈η(x,t)η(x,0)〉 = 2Dδ(t). D is the diffusion coefficient. The noise term is related to the intensity of cellular fluctuations either from the environmental external fluctuations or intrinsic fluctuations (under large number expansions, the process follows Brownian dynamics and diffusion coefficient is often concentration dependent).
where x is the protein concentration vector, F(x) is the chemical rate flux vector. η is Gaussian noise term where its autocorrelation function is 〈η(x,t)η(x,0)〉 = 2Dδ(t). D is the diffusion coefficient. The noise term is related to the intensity of cellular fluctuations either from the environmental external fluctuations or intrinsic fluctuations (under large number expansions, the process follows Brownian dynamics and diffusion coefficient is often concentration dependent).
We can now formulate the dynamics for the probability of starting from initial configuration xinitial at t = 0 and end at the final configuration of xfinal at time t, with the Onsager–Machlup functional (11) as 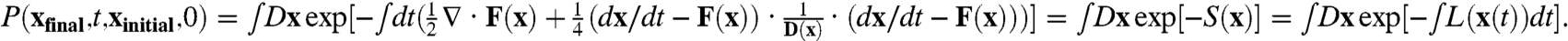
The integral over Dx represents the sum over all possible paths connecting xinitial at time t = 0 to xfinal at time t. D(x) is the diffusion coefficient matrix tensor. The second term of the exponent represents the weight contribution from specific trajectory path due to the underlying Gaussian noise. The first term of the exponent represents the contribution due to the variable transformation from the Gaussian noise η to the trajectory path x (Jacobian). The exponential factor gives the weight of each path. So the probability of network dynamics from initial configurations xinitial to the final state xfinal is equal to the sum of all possible paths with different weights. The S(x) is the action and L(x(t)) is the Lagrangian or the weight for each path (Fig. 2).
Notice that not all the paths give the same contribution. We can approximate the path integrals with a set of dominant paths. Because each path is exponentially weighted, the other subleading path contributions are often small and can be ignored. One can easily use this observation to find the paths with the optimal weights. We identify the optimal paths as biological paths or developmental pathways in our case.
Once the paths are known, we can substitute back to the path integral formulation to calculate the probability evolution in time. We can obtain the rate or speed of kinetics from one state to another. One can also use the long time limit to infer the weights of states and therefore map out the landscape. Details are given in SI Text.
We further notice that if the force F is a gradient, then the term F·dx in the weight functional above is a constant depending only on ending points. When force F is not purely a gradient (nonequilibrium with no detailed balance), then the curl flux component of the force leads to path dependent weights ∫F·dx. It will create a topological winding (nonzero) contribution to the weights of the paths going back to itself (∮F·dx ≠ 0). This will result the deviation of the pathways from the pure gradient paths and furthermore the forward path and backward path will not be the same (∫forwardF·dx ≠ -∫backwardF·dx) and therefore the corresponding developmental pathways are irreversible. Details are shown in SI Text. We point out that curl flux as a result of detailed balance breaking of nonequilibrium system is the origin of optimal kinetic paths deviating from the steepest descent one, which was not previously known (9, 10).
Conclusion
We developed a theoretical framework to quantify the Waddington landscape and biological paths for development and differentiation. We quantified the Waddington landscape through the construction of the underlying probability landscape for the cell development. We show the developmental process proceeds as moving from undifferentiated basin of attraction to the differentiated attractors. The barrier heights between the basins of attractions correlate with the escape time that determines the stability of cell types.
We show that the developmental process can be quantitatively described and uncovered by the biological paths on the quantified Waddington landscape and the dynamics of the developmental process is controlled by a combination of the gradient and curl force on the landscape. The biological paths do not follow the normally expected steepest descent path on the landscape. We show that the biological paths of the reverse differentiation process or reprogramming are irreversible and different from the ones of the differentiation process.
We found that the developmental process described by the underlying landscape and the associated biological paths is stable and robust against the influences of environmental perturbations.
Supplementary Material
Acknowledgments.
We thank Dr. Sui Huang for help discussions and comments. We thank Virtual Cell (supported by National Institutes of Health Grant P41RR013186 from the National Center For Research Resources) for software support. We acknowledge support from the National Science Foundation and National Natural Science Foundation of China Grants 20735003 and 90713022, and 973 Project Grants 2009CB930100 and 2010CB933600.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017017108/-/DCSupplemental.
References
- 1.Waddington CH. The Strategy of the Genes. London: Allen &Unwin; 1957. [Google Scholar]
- 2.Huang S. Reprogramming cell fates: reconciling rarity with robustness. Bioessays. 2009;31:546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 3.MacArthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Xu L, Wang EK. Potential landscape and flux framework of nonequilibrium networks: Robustness, dissipation, and coherence of biochemical oscillations. Proc Natl Acad Sci USA. 2008;105:12271–12276. doi: 10.1073/pnas.0800579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Xu L, Wang EK, Huang S. The potential landscape of genetic circuits imposes the arrow of time in stem cell differentiation. Biophys J. 2010;99:29–39. doi: 10.1016/j.bpj.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 7.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–590. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 8.Zhou JX, Huang S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends Genet. 2010;27:55–62. doi: 10.1016/j.tig.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Weinan E, Vanden-Eijnden E. Transition-oath theory and path-finding algorithms for the study of rare events. Annu Rev Phys Chem. 2010;61:391–420. doi: 10.1146/annurev.physchem.040808.090412. [DOI] [PubMed] [Google Scholar]
- 10.Roma DM, O’Flanagan RA, Ruckenstein AE, Sengupta AM, Mukhopadhyay R. Optimal path to epigentic switching. Phys Rev E. 2005;71(011902):1–5. doi: 10.1103/PhysRevE.71.011902. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Zhang K, Wang EK. Kinetic paths, time scale, and underlying landscapes: A path integral framework to study global natures of nonequilibrium systems and networks. J Chem Phys. 2010;133(125103):1–13. doi: 10.1063/1.3478547. [DOI] [PubMed] [Google Scholar]
- 12.Huang S, Guo YP, May G, Enver T. Bifurcation dynamics of cell fate decision in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 13.Hu M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 15.Kalmar T, et al. Regulated fluctuations in Nanog expression mediate cell fate decisions in ES and EC cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolynes PG, Onuchic JNO, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1622. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 17.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.