Abstract
Although genetic variations in cell-cycle control genes have been previously linked to cancer risk, no study has specifically evaluated the role of these gene variants in endometrial carcinogenesis. Using data from the Shanghai Endometrial Cancer Study, a population-based case-control study with 1,199 cases and 1,212 age-matched controls (1997–2003), the authors carried out a systematic evaluation of the association of cell-cycle control genes with endometrial cancer risk. Sixty-five tagging or potentially functional single nucleotide polymorphisms in the CCNB1, CCND1, CCNE1, CDK2, CDK4, CDK6, CDKN1A, CDKN1B, and CDKN2A genes were genotyped and evaluated. Three single nucleotide polymorphisms in the CDKN1B gene (rs11055027, rs3759216, and rs34330) were related to endometrial cancer risk, although only the association with rs34330 remained statistically significant after adjustment for multiple comparisons. The odds ratios for rs34330 were 1.33 (95% confidence interval (CI): 1.06, 1.66) and 1.51 (95% CI: 1.16, 1.94) for the CT and TT genotypes, respectively, compared with the CC genotype. In vitro luciferase reporter assays showed that the minor allele (A) in rs3759216, which was associated with decreased endometrial cancer risk (odds ratio = 0.73, 95% CI: 0.56, 0.94) without adjustment for multiple comparisons, significantly increased promoter activity. These findings suggest that polymorphisms of the CDKN1B gene may play a role in endometrial carcinogenesis.
Keywords: cell cycle; China; endometrial neoplasms; polymorphism, genetic; polymorphism, single nucleotide; women
Carcinogenesis in humans is believed to result from uncontrolled cell proliferation. Cell-cycle checkpoints are one of the primary defense mechanisms against mutagenic exposure. These checkpoints are regulatory pathways that control the order and timing of cell-cycle transitions to ensure DNA replication and chromosome segregation (1). The key point in cell-cycle regulation is the transition of the restriction point late in the G1 phase, which is crucial in determining the cell's destiny—division, differentiation, senescence, or apoptosis. It is believed that once the restriction point has been overcome, cell-cycle progression occurs almost automatically. Cells may be arrested at the restriction point, temporarily halting the cell cycle and allowing DNA repair to be completed. The loss of this checkpoint and the perturbation of cell-cycle control not only may disrupt the balance between normal growth and terminal differentiation but may also be accompanied by genomic instability, which may facilitate the development of cancer, as evidenced by the frequent inactivation of cell-cycle control genes, including those for p53, p16, p27, and retinoblastoma protein, in various cancers (2). In mammalian cells, cell division is controlled by the activity of cyclin-dependent kinases (CDKs) and their essential activating coenzymes, the CDK inhibitors (3–5), which may be influenced by genetic variations in the corresponding genes.
Cyclin D1, encoded by the CCND1 gene, plays an important role in the progress of the cell cycle. It associates with CDKs to phosphorylate the retinoblastoma protein during the G1 phase (6). Cyclin E, dissimilar to the mitrogen-dependent cyclin D, is a mitrogen-independent activator of CDK and a critical regulator of G1/S transition (7). The CDK inhibitors are key regulators of the G1/S checkpoint; their concerted action prevents cells from undergoing subsequent division in response to oncogenic signaling or DNA damage (8). Among them, CDKN1B belongs to the Cip/Kip family and functions as a cell-cycle checkpoint at G1/S by encoding the p27 protein. The p27 protein is a putative tumor suppressor that inhibits phosphorylation of retinoblastoma protein to regulate G1 cyclin-CDK complexes (9, 10). In knockout mouse models, p27 deficiency has led to gigantism (11, 12), indicating the importance of this gene in tumorigenesis.
Anomalies in cell-cycle control genes have frequently been observed in many human malignancies. CCNB1, CCND1, and CCNE1 have distinct expressions in different breast cancer subtypes and invasive ovarian cancer (13, 14). CDK2, CDK4, CDK6, CDKN1A, CDKN1B, and CDKN2A are found to be associated with breast cancer (15, 16), lung cancer (17, 18), and ovarian, pancreatic, and prostate cancers (19–22). Together, these data suggest an important role for CDKs, cyclins, and CDK inhibitors in cancer development. However, to the best of our knowledge, no study has evaluated the effect of genetic variants in those cell-cycle control genes on susceptibility to endometrial cancer. In this study, we examined associations of polymorphisms in 9 cell-cycle control genes with endometrial cancer risk using data from the Shanghai Endometrial Cancer Study, a large population-based case-control study conducted in urban Shanghai, China.
MATERIALS AND METHODS
Study participants
Details on the Shanghai Endometrial Cancer Study have been published elsewhere (23). Briefly, of 1,449 newly diagnosed endometrial cancer patients aged 30–69 years who were identified between January 1997 and December 2003 through the population-based Shanghai Cancer Registry, 1,199 (82.7%) participated in the study. Controls were randomly selected from permanent female residents of urban Shanghai and were frequency-matched to cancer cases by age (5-year interval) at a 1:1 ratio. The random selection was performed by staff of the Shanghai Resident Registry. Women with a prior history of any cancer or hysterectomy were ineligible for the study. Of the 1,629 eligible controls contacted, 1,212 (74.4%) participated in the study. The study protocols were approved by the institutional review boards of all institutions involved in the study, and all participants provided written informed consent before participating in the study.
Detailed information on demographic factors, menstrual and reproductive history, hormone use, prior disease history, physical activity, tobacco and alcohol use, diet, weight history, and family history of cancer was collected for all participants via an in-person interview. Body weight, height, and waist and hip circumferences were measured according to a standardized protocol at the time of interview. Menopause was defined as cessation of menstruation for at least 12 months before the reference date (diagnosis date for cases and interview date for controls), excluding lapses caused by pregnancy or breastfeeding. Body mass index (weight in kilograms/squared height in meters) and waist-to-hip circumference ratio were calculated using measured anthropometric data.
Of the study participants who completed an in-person interview, 850 cases and 853 controls donated a blood sample and 280 cases and 274 controls provided a buccal-cell sample (187 cases and 186 controls provided samples using a mouthwash method, and 93 cases and 88 controls provided samples using a buccal swab method). Because of the very low DNA yield of the buccal swab method, we did not include buccal swab DNA samples in the genotyping. In addition, DNA samples from 19 controls who donated a blood sample were used up in other studies. Thus, DNA samples from 1,037 cases (86.5%; 850 blood and 187 buccal-cell) and 1,020 controls (84.2%; 834 blood and 186 buccal-cell) were included in this genotyping study. Genotyping data for cell-cycle control genes were obtained from 1,028 cases and 1,003 controls, with success rates of 99.1% and 99.6%, respectively.
SNP selection, identification, and genotyping
We selected haplotype-tagging single nucleotide polymorphisms (SNPs) by searching Han Chinese data from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) using the Tagger program (24). The following criteria were used to identify tagging SNPs: 1) the SNPs were located in the cell-cycle control gene or within the 5-kilobase region flanking the gene, 2) the SNPs had a minor allele frequency greater than or equal to 0.05, and 3) the other known unselected SNPs could be captured by one of the tagging SNPs with a linkage disequilibrium of r2 ≥ 0.90. Known functional or potentially functional SNPs were forced into the haplotype-tagging SNP selection process. SNP selection was completed in December 2005. Of the 67 selected SNPs, the design of the assay for 2 SNPs (rs2282992 in the CDK6 gene and rs1801270 in the CDKN1A gene) failed, which resulted in 65 SNPs being genotyped for this study (genotyped SNPs are listed in Web Table 1, which is posted on the Journal’s Web site (http://aje.oxfordjournals.org/)).
Genomic DNA was extracted from buffy coat fractions or buccal cells using the QIAamp DNA minikit (Qiagen Inc., Valencia, California) following the manufacture's protocol. The SNPs were genotyped using the Affymetrix MegAllele Targeted Genotyping System (Affymetrix, Inc., Santa Clara, California) with the molecular inversion probe method (25) as part of a large-scale genotyping effort that included 1,737 SNPs. Genotyping was conducted at the Vanderbilt Microarray Shared Resource (Vanderbilt University, Nashville, Tennessee) following the Affymetrix protocol. As a quality control procedure, we included 39 blinded duplicated quality control samples and 12 HapMap DNA samples in the genotyping. The average consistency rate for these samples was 99.6%. The genotyping of SNPs in cell-cycle control genes was highly successful, with call rates of 99.5%–100% (median, 99.95%). Finally, the laboratory staff was blind to the case-control status and identity of all samples.
In vitro functional experiments
To evaluate the function of SNPs located in the 5′ flanking region of the CDKN1B gene, we conducted a series of experiments with CDKN1B luciferase reporter constructs. First, a 3.6-kilobase fragment, spanning the 3,600 base pairs upstream of the ATG translation start codon and including rs11055027, rs3759216, and rs34330, was generated by polymerase chain reaction with the forward primer 5′- GTGAGCTTAGAAGAATGGTGGAGTTGAGTG -3′ and the backward primer 5′- CTATAAGCTTCTGCACGACCGCCTCTCTC -3′. Template DNA from persons known to have either a major allele or a minor allele for rs11055027, rs3759216, and rs34330 was used. The resulting fragments were cloned into a promoter-less luciferase reporter vector, pGL4.20 (Promega Corporation, Madison, Wisconsin). All DNA constructs were verified through sequencing. Transfection was performed with the use of FuGene 6 Transfection Reagent (Roche Diagnostics GmbH, Mannheim, Germany) in triplicate for each construct. Human 293 cells (2 × 105 each) were seeded in 24-well plates and cotransfected with pGL4.73 (a Renilla expressing vector serving as a reference for transfection efficiency) and the luciferase reporter constructs with different combinations of alleles. Thirty-six to 48 hours later, cells were lysed with passive lysis buffer, and luminescence (relative light units) was measured using the Dual-Luciferase Assay System (Promega Corporation). Relative luciferase activity was measured as the ratio of firefly luciferase activity to Renilla luciferase activity, and results from 5 independent experiments were averaged.
Statistical analyses
Chi-squared and t tests were used to evaluate case-control differences in the distributions and mean values of risk factors and genotypes of the cell-cycle control genes and deviations of genotype frequencies in controls from those expected under Hardy-Weinberg equilibrium. Unconditional multivariable logistic regression models were applied to estimate odds ratios and 95% confidence intervals for measurement of the association between endometrial cancer risk and cell-cycle control genes, adjusting for matching variables and known risk factors for endometrial cancer throughout. Known risk factors included age at interview, educational level, income level, menopausal status, age at menarche, number of livebirths, alcohol consumption (ever), and body mass index. Categorized variables were treated as dummy variables in the model. All tests for trend were performed by entering categorical variables as continuous parameters in the models. P values for trend tests on 65 SNPs in additive models were adjusted for multiple comparisons using an approach proposed by Conneely et al. (26) to reduce false-positive rates. SNPs with P < 0.05 in the additive model with no adjustment for multiple comparisons are presented here.
To evaluate whether multiple SNPs in the same gene may have an additive effect on endometrial cancer risk, we estimated the combined effect of these SNPs. We treated alleles with an odds ratio greater than or equal to 1 at each locus as the risk allele (the reference allele is the risk allele if the odds ratio is less than 1) and tallied the total number of risk alleles for each participant to calculate a genetic risk score (GRS). Linkage disequilibrium between polymorphisms was assessed using HaploView software, version 4.1 (27), and haplotype blocks were defined using a method reported by Gabriel et al. (28). The repeated-measures analysis of variance test was used to evaluate the relative value of luciferase activity for each SNP. SAS, version 9.2 (SAS Institute, Inc., Cary, North Carolina) was used to test associations between SNPs and endometrial cancer. All statistical tests were based on 2-tailed probability.
RESULTS
Table 1 presents selected demographic and risk factor characteristics of participants genotyped for polymorphisms of the cell-cycle control genes in this study. The 1,028 cases and 1,003 controls were similar with respect to age, educational status, income level, and cigarette smoking habits. Compared with controls, cases were more likely to have been younger at menarche, were less likely to be postmenopausal, had had fewer livebirths, and had a higher body mass index. There were no appreciable differences in the distribution of demographic or risk factors between the entire study population (data not shown) and persons with genotyping data.
Table 1.
Demographic Characteristics and Selected Risk Factors for Endometrial Cancer Cases and Controls, Shanghai Endometrial Cancer Study, Shanghai, China, 1997–2003
| Participant Characteristic | Cases (n = 1,028) |
Controls (n = 1,003) |
P Value | ||
| Mean (SD) | % | Mean (SD) | % | ||
| Age, years | 54.78 (8.51) | 54.88 (8.49) | 0.80 | ||
| Age at menarche, years | 14.47 (1.73) | 14.77 (1.81) | <0.001 | ||
| No. of livebirths | 1.67 (1.18) | 1.76 (1.14) | 0.07 | ||
| Body mass indexa | 25.75 (4.13) | 23.78 (3.48) | <0.001 | ||
| Education | |||||
| Primary school or below | 21.7 | 22.0 | |||
| Middle school | 38.2 | 37.9 | |||
| High school or above | 40.1 | 40.1 | 0.98 | ||
| Family income, RMB/year | |||||
| <10,000 | 12.2 | 10.8 | |||
| 10,001–19,999 | 41.6 | 39.7 | |||
| ≥20,000 | 46.2 | 49.5 | 0.30 | ||
| Occupation, % | |||||
| Professional | 29.1 | 25.4 | |||
| Clerical | 22.4 | 22.6 | |||
| Manual laborer | 48.5 | 51.9 | 0.15 | ||
| Ever smoking (yes) | 3.1 | 3.6 | 0.55 | ||
| Ever consuming alcohol (yes) | 3.1 | 5.3 | 0.01 | ||
| Postmenopausal | 56.8 | 61.5 | 0.03 | ||
Abbreviations: RMB, renminbi; SD, standard deviation.
Weight (kg)/height (m)2.
A total of 65 SNPs in 9 cell-cycle control genes—3 cyclins (CCNB1, CCND1, and CCNE1), 3 CDKs (CDK2, CDK4, and CDK6), and 3 CDK inhibitors (CDKN1A, CDKN1B, and CDKN2A)—were included in the study. Their associations with endometrial cancer risk are summarized in Web Table 1. The genotype distributions were consistent with Hardy-Weinberg equilibrium (P > 0.05) among controls for most SNPs, with the exception of rs649392, rs2237572, rs2282981, rs3829963, rs12528248, and rs12191972.
Of the 65 SNPs under study, 7 SNPs were associated with endometrial cancer risk at P < 0.05 (Table 2) before adjustment for multiple comparisons. These included 1 (rs2069433) of 3 SNPs in the CCNB1 gene, 2 (rs2282978 and rs2282979) of 32 SNPs in the CDK6 gene, and 4 (rs11055027, rs3759216, rs34330, and rs34322) of 9 SNPs in the CDKN1B gene. After multiple comparison adjustment, only the minor allele of rs34330 in the CDKN1B gene was significantly associated with endometrial cancer risk compared with its major homozygote, and the other 3 SNPs (rs2069433, rs11055027, and rs3759216) had associations of marginal statistical significance. There was no evidence for any association at a significance level of P ≤ 0.05 for the remaining 6 genes studied (CCND1, CCNE1, CDK2, CDK4, CDKN1A, and CDKN2A).
Table 2.
Genotype-Specific Risks of Endometrial Cancer for Selected Single Nucleotide Polymorphismsa, Shanghai Endometrial Cancer Study, Shanghai, China, 1997–2003
| Gene | Single Nucleotide Polymorphism | Minor/Major Alleles | Region | Minor Allele Frequency | PHWE | Endometrial Cancer Risk |
|||||
| Heterozygosity (AB) |
Minor-Allele Homozygosity (BB) |
Additive Ptrend | Adjusted P Valueb | ||||||||
| ORc | 95% CI | ORc | 95% CI | ||||||||
| CCNB1 | rs2069433 | C/T | Intron | 7.28 | 1.00 | 0.79 | 0.60, 1.02 | 0.35 | 0.08, 1.46 | 0.030 | 0.081 |
| CDK6 | rs2282978 | C/T | Intron | 11.30 | 1.00 | 0.79 | 0.63, 1.00 | 0.75 | 0.34, 1.68 | 0.040 | 0.413 |
| CDK6 | rs2282979 | C/T | Intron | 10.81 | 0.89 | 0.80 | 0.64, 1.01 | 0.67 | 0.28, 1.57 | 0.042 | 0.424 |
| CDKN1B | rs11055027 | C/G | Promoter | 18.24 | 0.73 | 1.21 | 0.99, 1.48 | 1.89 | 1.12, 3.19 | 0.006 | 0.068 |
| CDKN1B | rs3759216 | A/G | Promoter | 43.66 | 0.38 | 0.76 | 0.62, 0.93 | 0.73 | 0.56, 0.94 | 0.008 | 0.084 |
| CDKN1B | rs34330 | T/C | 5′ untranslated | 49.75 | 0.18 | 1.33 | 1.06, 1.66 | 1.51 | 1.16, 1.94 | 0.002 | 0.025 |
| CDKN1B | rs34322 | C/T | 3′ flanking | 48.35 | 0.49 | 0.84 | 0.67, 1.04 | 0.73 | 0.57, 0.94 | 0.015 | 0.142 |
Abbreviations: CI, confidence interval; HWE, Hardy-Weinberg equilibrium; OR, odds ratio.
Single nucleotide polymorphisms were selected on the basis of Ptrend < 0.05 and PHWE > 0.05.
Adjusted P values are presented because of multiple correlated tests (26).
Adjusted for age at interview, body mass index, menopausal status, education, income, alcohol consumption, age at menarche, number of livebirths, and occupation. Major-allele homozygosity (AA) was the reference category.
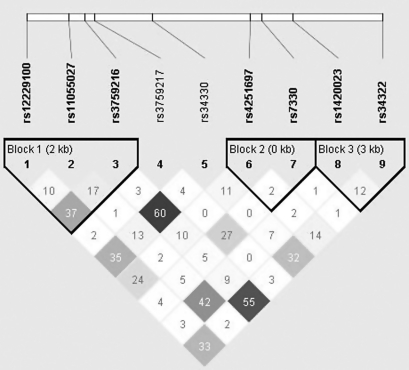
Figure 1 depicts the linkage disequilibrium structure of the CDKN1B gene with 3 blocks. The linkage disequilibrium plot of the 9 SNPs revealed low/moderate pairwise r2 values, and the highest r2 value (r2 = 0.60) was seen for rs3759216 and rs34330, indicating that linkage disequilibrium among these SNPs is not strong. Additive effects of multiple SNPs in the same gene were evaluated (Table 3). For the 5 CDKN1B SNPs with P < 0.10 (Web Table 1), the GRS for each participant ranged from 0 to 10. We categorized participants into 6 groups and used participants with a GRS of 0 as the reference group. The odds ratios for participants with GRS < 5 did not differ significantly from the reference group. However, the odds ratios for participants with GRS ≥ 5 exhibited a significantly increased risk of endometrial cancer. A woman with a GRS of 9–10 had a 2.5-fold increased risk of endometrial cancer (P = 0.02). We found no significant associations for either CCNB1 or CDK6.
Figure 1.
Pairwise r2 values for the relations between single nucleotide polymorphisms (SNPs) in the cyclin-dependent kinase inhibitor 1B gene (CDKN1B). Numbers shown in diamonds are the r2 values × 100 (10 means 0.10, 1 means 0.01). The block was defined using the method of confidence intervals (28), and the boldface SNPs are selected in blocks (blocks 1, 2, and 3). kb, kilobases.
Table 3.
Association Between Number of Risk Alleles in the CCNB1, CDK6, and CDKN1B Genes and Risk of Endometrial Cancer, Shanghai Endometrial Cancer Study, Shanghai, China, 1997–2003
| Gene (SNPa) and No. of Risk Alleles | No. of Cases | No. of Controls | Odds Ratiob | 95% Confidence Interval | P Value |
| CCNB1 (rs350104 and rs2069433) | |||||
| 0–2 | 381 | 392 | 1.0 | ||
| 3 | 419 | 359 | 1.25 | 1.03, 1.52 | 0.02 |
| 4 | 97 | 98 | 1.17 | 0.85, 1.61 | 0.34 |
| P for trendc | 0.1603 | ||||
| CDK6 (rs4272, rs2282978, and rs2282983) | |||||
| 0 | 547 | 500 | 1.0 | ||
| 1 | 239 | 238 | 0.93 | 0.75, 1.16 | 0.54 |
| >1 | 240 | 260 | 1.09 | 0.73, 1.63 | 0.66 |
| P for trend | 0.0570 | ||||
| CDKN1B (rs11055027, rs3759216, rs34330, rs1420023, and rs34322) | |||||
| 0 | 135 | 160 | 1.0 | ||
| 1–2 | 121 | 121 | 1.24 | 0.87, 1.76 | 0.23 |
| 3–4 | 332 | 353 | 1.17 | 0.88, 1.55 | 0.27 |
| 5–6 | 283 | 245 | 1.51 | 1.12, 2.02 | <0.01 |
| 7–8 | 131 | 100 | 1.61 | 1.13, 2.30 | <0.01 |
| 9–10 | 20 | 12 | 2.49 | 1.14, 5.46 | 0.02 |
| P for trend | 0.0008 |
Abbreviation: SNP, single nucleotide polymorphism.
Selection of SNPs for the analysis was based on a low Ptrend value (P < 0.10) for that SNP in Web Table 1 (http://aje.oxfordjournals.org/).
Adjusted for age at interview, body mass index, menopausal status, education, income, alcohol consumption, age at menarche, number of livebirths, and occupation.
Tests for trend were performed by entering categorical scores (0–2 for CCNB1 and CDK6 and 0–5 for CDKN1B) into the models as continuous parameters.
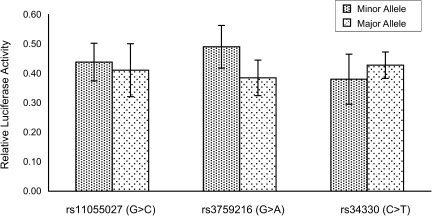
To evaluate whether SNPs in the CDKN1B gene (rs11055027, rs3759216, and rs34330) affect gene expression, we conducted an in vitro CDKN1B promoter/luciferase assay in human 293 cells. We generated several luciferase reporter constructs containing either the major allele or corresponding minor alleles in the promoter region of the CDKN1B gene. Human 293 cells were transiently transfected with these constructs. The luciferase assay results showed that the minor allele (A) of rs3759216 increased CDKN1B promoter activity by approximately 27% in comparison with the major allele (G) in the 293 cells (0.4906 vs. 0.3849; P <0.05), while no variant of rs34330 or rs11055027 had a significant effect on CDKN1B promoter activity in comparison with the major allele and minor constructs (Figure 2).
Figure 2.
Effects (allele-specific difference) of the single nucleotide polymorphisms rs11055027, rs3759216, and rs34330 on cyclin-dependent kinase inhibitor 1B gene (CDKN1B) promoter activity. The difference in relative luciferase activity between the A and G alleles was statistically significant (P < 0.05) for rs3759216 (G > A) Bars, standard deviation.
Since body mass index, menopausal status, years of menstruation, and oral contraceptive use are predominant risk factors for endometrial cancer and because these factors may influence the sex hormone milieu, we also evaluated the possible modifying effect of these factors. The inverse association between GRS for CDK6 and cancer risk appeared to be more evident among postmenopausal women and women who had not used oral contraceptives. In addition, the positive association between GRS for CDKN1B and cancer risk was stronger among postmenopausal women, women with more years of menstruation, women with a lower body mass index, and women who had not taken oral contraceptives. However, none of the tests for multiplicative interaction were statistically significant (data not shown).
DISCUSSION
In this population-based case-control study, we evaluated the association of 65 SNPs in 9 cell-cycle control genes with the risk of endometrial cancer. We found that 4 SNPs in the CDKN1B gene, 2 SNPs in the CDK6 gene, and 1 SNP in the CCNB1 gene were associated with the risk of endometrial cancer before adjustment for multiple comparisons. The association between the CDKN1B SNP rs34330 and endometrial cancer remained statistically significant even after adjustment for multiple comparisons. Our study found no evidence for an association between endometrial cancer and polymorphisms in the CCND1, CCNE1, CDK2, CDK4, CDKN1A, and CDKN2A genes.
Dysregulation of the cell cycle is a hallmark of many cancers, since control and timing of the cell cycle involves checkpoints and regulatory pathways that ensure the fidelity of DNA replication and chromosome segregation (1, 2, 29). These processes involve a large collection of key molecules, including the cyclins, CDKs, and CDK inhibitors. Therefore, genetic variations of those genes are excellent candidates for cancer susceptibility. Several studies have investigated the associations of genetic polymorphisms of the CDKN1B gene with cancer risk. The SNP rs34330 in CDKN1B has been previously related to risk of prostate cancer, particularly in men affected at ages less than 65 years (P = 0.0015) (20). The T allele of rs34330 has also been associated with increased risk of breast cancer in Chinese women (30) and United Kingdom women (31), but this association was not found among women in western Ireland (32). The variant-containing genotype of rs34330 (CT/TT) was significantly associated with increased risk of lung cancer (odds ratio = 1.27, 95% confidence interval: 1.01, 1.60) and was not associated with bladder cancer risk, compared with carriers of the homozygous wild type (18, 33). Few studies have investigated the association between rs3759216 in the CDKN1B gene and cancer. We found that rs3759216 was associated with reduced endometrial cancer risk. This was consistent with the finding in Spurdle et al.’s (34) study, in which this SNP was found to have a marginally significant association with breast cancer risk in carriers of BRCA1 and BRCA2 mutations.
The CCNB1 gene is a key regulator of the G2/M transition of the cell cycle through formation of a complex with cell division control protein 2 homolog, which may promote progression to mitosis and augment the cellular growth rate. Overexpression of CCNB1 has been reported in various tumor types and may be related to increased mitotic activity during malignant transformation (13, 35). In our study, we found that rs2069433 was related to a reduction in endometrial cancer risk. However, to our knowledge, there have been no investigations of associations of SNPs in the CCNB1 gene with endometrial cancer to date. Thus, the role of this SNP in the CCNB1 gene in endometrial tumorigenesis remains to be further studied. CDK6, as well as CDK4, has been shown to phosphorylate, and thus regulate, the activity of the tumor suppressor protein retinoblastoma protein. A previous study investigated 13 SNPs in the CDK6 gene and did not find associations between those SNPs and breast cancer risk in the British population (31). Two groups of investigators have reported that no association was found in their studies between CDK6 and lung and ovarian cancers, with the exception of rs8, which was associated with nonserous cancer, a histologic subtype of ovarian cancer (18, 36). Similar to the above studies, there were 32 SNPs in the CDK6 gene reviewed in our study, and none of them was associated with endometrial cancer risk after adjustment for multiple comparisons.
Although SNPs in other cell-cycle control genes, such as CCND1 (rs649392), CDK2 (rs2069414), CCNE1 (rs3218036), and CDKN2A (rs3731257), were found to be associated with ovarian cancer risk (21, 37) and although CDKN2A (rs3731239) was associated with breast cancer risk (31), we did not find any associations of these SNPs with endometrial cancer risk in our study.
We investigated the potential mechanisms underlying the associations of the CDKN1B SNPs with endometrial cancer risk through in vitro biochemical analyses. We found that the minor allele (A) of rs3759216 in the CDKN1B promoter region significantly increased luciferase activity, suggesting that rs3759216 may functionally up-regulate p27 protein expression in subpopulations carrying the A allele(s) of this SNP. This may suppress cell growth, thereby reducing risk of endometrial cancer, which is consistent with our finding that the A allele of rs3759216 was associated with reduced risk of endometrial cancer. Unlike the classic tumor suppressors that follow Knudson's “2-hit hypothesis,” homozygous loss or silencing of CDKN1B is extremely rare in human malignancies (38–39). The p27 protein, coded by CDKN1B, is an atypical tumor suppressor that regulates G1/S phase transition by binding to and regulating the activity of CDKs. The p27 protein has been linked to endometrial hyperplasia and/or carcinoma, which is consistent with its role in driving cell proliferation (40). A decrease in p27 protein level has been observed in colon, breast, lung, and other cancers (41–44). Our in vitro biochemical study data were consistent with the findings of these studies and suggest that rs3759216 may be a causal variant of endometrial cancer. These data need to be validated in other study populations. Studies are also needed to investigate the relation between the rs3759216 polymorphism and p27 expression.
Many association studies have presented inconsistent results for relations between rs34330 in the CDKN1B gene and the risks of breast, ovarian, lung, and prostate cancer (30–33). In a recent study, Landa et al. (45) found that rs34330 was a risk factor for developing the follicular variant of papillary thyroid carcinoma, a subtype of thyroid cancer, with an odds ratio of 2.12 (P = 0.023) only in a recessive model. In that study, the risk allele (T) led to a lower transcription rate in cells transfected with a luciferase reporter driven by the polymorphic p27 promoter (45). In our study, rs34330 had a highly significantly association with endometrial cancer risk. However, our in vitro biochemical study did not find that this SNP had a direct effect on promoter activity and suggested that rs34330 is unlikely to be the causal SNP, but rather a genetic marker associated with endometrial cancer via its linkage disequilibrium with rs3759216 or other unidentified functional variants.
This study had several strengths, including the population-based study design, the relatively high participation rate, the homogenous ethnic background, and the low frequency of hysterectomy in the study population. The application of the haplotype-tagging SNP approach in SNP selection made it possible to systematically evaluate the genetic markers of the above genes. In addition, we applied P value adjustment methods to control for multiple comparisons and in vitro assays to verify SNP-disease associations. Although, to our knowledge, our study was one of the largest epidemiologic studies of endometrial cancer carried out to date, the sample size was still not sufficiently large to investigate interactions or SNPs with a low minor allele frequency.
In conclusion, we found that 3 SNPs in CDKN1B were associated with endometrial cancer risk, and these associations remained statistically or marginally significant after adjustment for multiple comparisons. We found that 1 SNP, rs3759216, was related to increasing luciferase activity and may be a causal variant of endometrial cancer. Our study suggests that genetic variations in CDKN1B may contribute to endometrial carcinogenesis. Studies with high coverage of genetic variants in CDKN1B are needed to further validate our findings.
Supplementary Material
Acknowledgments
Author affiliations: Vanderbilt Epidemiology Center, Department of Medicine and Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, Tennessee (Hui Cai, Shimian Qu, Jirong Long, Qiuyin Cai, Wei Zheng, Xiao Ou Shu); and Department of Epidemiology, Cancer Institute of Shanghai Jiao Tong University, Shanghai Cancer Institute, Shanghai, China (Yong-Bing Xiang, Jing Gao).
This work was supported by US Public Health Service grant R01 CA92585 from the National Cancer Institute (US National Institutes of Health) and in part by grant P30 CA68485 from the Vanderbilt-Ingram Cancer Center.
The authors thank Dr. Fan Jin for her contributions to the implementation of the study in Shanghai, Regina Courtney and Dr. Shawn Levy for their contributions to the genotyping, and Bethanie Hull for her assistance in the preparation of the manuscript. The study would not have been possible without the support of the research staff of the Shanghai Endometrial Cancer Study.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Conflict of interest: none declared.
Glossary
Abbreviations
- CDK
cyclin-dependent kinase
- GRS
genetic risk score
- SNP
single nucleotide polymorphism
References
- 1.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274(5293):1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Morgan DO. Principles of CDK regulation. Nature. 1995;374(6518):131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 6.Moreno-Bueno G, Rodríguez-Perales S, Sánchez-Estévez C, et al. Cyclin D1 gene (CCND1) mutations in endometrial cancer. Oncogene. 2003;22(38):6115–6118. doi: 10.1038/sj.onc.1206868. [DOI] [PubMed] [Google Scholar]
- 7.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60(14):3689–3695. [PubMed] [Google Scholar]
- 8.Healy J, Bélanger H, Beaulieu P, et al. Promoter SNPs in G1/S checkpoint regulators and their impact on the susceptibility to childhood leukemia. Blood. 2007;109(2):683–692. doi: 10.1182/blood-2006-02-003236. [DOI] [PubMed] [Google Scholar]
- 9.Xiong Y, Hannon GJ, Zhang H, et al. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K, Lee MH, Erdjument-Bromage H, et al. Cloning of p27 Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78(1):59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama K, Ishida N, Shirane M, et al. Mice lacking p27 Kip1 display increased body size, multiorgan hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85(5):707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 12.Kiyokawa H, Kineman RD, Manova-Todorova KO, et al. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27 Kip1. Cell. 1996;85(5):721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal R, Gonzalez-Angulo AM, Myhre S, et al. Integrative analysis of cyclin protein levels identifies cyclin B1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res. 2009;15(11):3654–3662. doi: 10.1158/1078-0432.CCR-08-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbieri F, Cagnoli M, Ragni N, et al. Increased cyclin D1 expression is associated with features of malignancy and disease recurrence in ovarian tumors. Clin Cancer Res. 1999;5(7):1837–1842. [PubMed] [Google Scholar]
- 15.Debniak T, Cybulski C, Górski B, et al. CDKN2A-positive breast cancers in young women from Poland. Breast Cancer Res Treat. 2007;103(3):355–359. doi: 10.1007/s10549-006-9382-x. [DOI] [PubMed] [Google Scholar]
- 16.Mavaddat N, Dunning AM, Ponder BA, et al. Common genetic variation in candidate genes and susceptibility to subtypes of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(1):255–259. doi: 10.1158/1055-9965.EPI-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincenzi B, Schiavon G, Silletta M, et al. Cell cycle alterations and lung cancer. Histol Histopathol. 2006;21(4):423–435. doi: 10.14670/HH-21.423. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Spitz MR, Yang H, et al. Genetic variants in cell cycle control pathway confer susceptibility to lung cancer. Clin Cancer Res. 2007;13(19):5974–5981. doi: 10.1158/1078-0432.CCR-07-0113. [DOI] [PubMed] [Google Scholar]
- 19.Padua MB, Hansen PJ. Changes in expression of cell-cycle-related genes in PC-3 prostate cancer cells caused by ovine uterine serpin. J Cell Biochem. 2009;107(6):1182–1188. doi: 10.1002/jcb.22222. [DOI] [PubMed] [Google Scholar]
- 20.Chang BL, Zheng SL, Isaacs SD, et al. A polymorphism in the CDKN1B gene is associated with increased risk of hereditary prostate cancer. Cancer Res. 2004;64(6):1997–1999. doi: 10.1158/0008-5472.can-03-2340. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham JM, Vierkant RA, Sellers TA, et al. Cell cycle genes and ovarian cancer susceptibility: a tagSNP analysis. Br J Cancer. 2009;101(8):1461–1468. doi: 10.1038/sj.bjc.6605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heiferman MJ, Salabat MR, Ujiki MB, et al. Sansalvamide induces pancreatic cancer growth arrest through changes in the cell cycle. Anticancer Res. 2010;30(1):73–78. [PubMed] [Google Scholar]
- 23.Xu WH, Shrubsole MJ, Xiang YB, et al. Dietary folate intake, MTHFR genetic polymorphisms, and the risk of endometrial cancer among Chinese women. Cancer Epidemiol Biomarkers Prev. 2007;16(2):281–287. doi: 10.1158/1055-9965.EPI-06-0798. [DOI] [PubMed] [Google Scholar]
- 24.de Bakker PI, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 25.Hardenbol P, Yu F, Belmont J, et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15(2):269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conneely KN, Boehnke M. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet. 2007;81(6):1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 29.Nam EJ, Kim YT. Alteration of cell-cycle regulation in epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18(6):1169–1182. doi: 10.1111/j.1525-1438.2008.01191.x. [DOI] [PubMed] [Google Scholar]
- 30.Ma H, Jin G, Hu Z, et al. Variant genotypes of CDKN1A and CDKN1B are associated with an increased risk of breast cancer in Chinese women. Int J Cancer. 2006;119(9):2173–2178. doi: 10.1002/ijc.22094. [DOI] [PubMed] [Google Scholar]
- 31.Driver KE, Song H, Lesueur F, et al. Association of single-nucleotide polymorphisms in the cell cycle genes with breast cancer in the British population. Carcinogenesis. 2008;29(2):333–341. doi: 10.1093/carcin/bgm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mcinerney N, Colleran G, Rowan A, et al. Low penetrance breast cancer predisposition SNPs are site specific. Breast Cancer Res Treat. 2009;117(1):151–159. doi: 10.1007/s10549-008-0235-7. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Gu J, Grossman HB, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78(3):464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spurdle AB, Deans AJ, Duffy D, et al. No evidence that CDKN1B (p27) polymorphisms modify breast cancer risk in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2009;115(2):307–313. doi: 10.1007/s10549-008-0083-5. [DOI] [PubMed] [Google Scholar]
- 35.Koon N, Schneider-Stock R, Sarlomo-Rikala M, et al. Molecular targets for tumour progression in gastrointestinal stromal tumours. Gut. 2004;53(2):235–240. doi: 10.1136/gut.2003.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gayther SA, Song H, Ramus SJ, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67(7):3027–3035. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 37.Goode EL, Fridley BL, Vierkant RA, et al. Candidate gene analysis using imputed genotypes: cell cycle single-nucleotide polymorphisms and ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(3):935–944. doi: 10.1158/1055-9965.EPI-08-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiohara N, el-Deiry WS, Wada M, et al. Absence of WAF1 mutations in a variety of human malignancies. Blood. 1994;84(11):3781–3784. [PubMed] [Google Scholar]
- 39.Kawamata N, Morosetti R, Miller CW, et al. Molecular analysis of the cyclin-dependent kinase inhibitor gene p27/kip1 in human malignancies. Cancer Res. 1995;55(11):2266–2269. [PubMed] [Google Scholar]
- 40.Niklaus AL, Aubuchon M, Zapantis G, et al. Assessment of the proliferative status of epithelial cell types in the endometrium of young and menopausal transition women. Hum Reprod. 2007;22(6):1778–1788. doi: 10.1093/humrep/dem032. [DOI] [PubMed] [Google Scholar]
- 41.Lloyd RV, Erickson LA, Jin L, et al. P27kip1: a multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am J Pathol. 1999;154(2):313–323. doi: 10.1016/S0002-9440(10)65277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slingerland J, Pagano M. Regulation of the Cdk inhibitor p27 and its deregulation in cancer. J Cell Physiol. 2000;183(1):10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 43.Roy S, Kaur M, Agarwal C, et al. p21 and p27 induction by silibinin is essential for its cell cycle arrest effect in prostate carcinoma cells. Mol Cancer Ther. 2007;6(10):2696–2707. doi: 10.1158/1535-7163.MCT-07-0104. [DOI] [PubMed] [Google Scholar]
- 44.Caron D, Savard PE, Doillon CJ, et al. Protein tyrosine phosphatase inhibition induces anti-tumor activity: evidence of Cdk2/p27kip1 and Cdk2/SHP-1 complex formation in human ovarian cancer cells. Cancer Lett. 2008;262(2):265–275. doi: 10.1016/j.canlet.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 45.Landa I, Montero-Conde C, Malanga D, et al. Allelic variant at −79 (C>T) in CDKN1B (p27Kip1) confers an increased risk of thyroid cancer and alters mRNA levels. Endocr Relat Cancer. 2010;17(2):317–328. doi: 10.1677/ERC-09-0016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.