Abstract
Background and Aims
Current understanding of stomatal development in Arabidopsis thaliana is based on mutations producing aberrant, often lethal phenotypes. The aim was to discover if naturally occurring viable phenotypes would be useful for studying stomatal development in a species that enables further molecular analysis.
Methods
Natural variation in stomatal abundance of A. thaliana was explored in two collections comprising 62 wild accessions by surveying adaxial epidermal cell-type proportion (stomatal index) and density (stomatal and pavement cell density) traits in cotyledons and first leaves. Organ size variation was studied in a subset of accessions. For all traits, maternal effects derived from different laboratory environments were evaluated. In four selected accessions, distinct stomatal initiation processes were quantitatively analysed.
Key Results and Conclusions
Substantial genetic variation was found for all six stomatal abundance-related traits, which were weakly or not affected by laboratory maternal environments. Correlation analyses revealed overall relationships among all traits. Within each organ, stomatal density highly correlated with the other traits, suggesting common genetic bases. Each trait correlated between organs, supporting supra-organ control of stomatal abundance. Clustering analyses identified accessions with uncommon phenotypic patterns, suggesting differences among genetic programmes controlling the various traits. Variation was also found in organ size, which negatively correlated with cell densities in both organs and with stomatal index in the cotyledon. Relative proportions of primary and satellite lineages varied among the accessions analysed, indicating that distinct developmental components contribute to natural diversity in stomatal abundance. Accessions with similar stomatal indices showed different lineage class ratios, revealing hidden developmental phenotypes and showing that genetic determinants of primary and satellite lineage initiation combine in several ways. This first systematic, comprehensive natural variation survey for stomatal abundance in A. thaliana reveals cryptic developmental genetic variation, and provides relevant relationships amongst stomatal traits and extreme or uncommon accessions as resources for the genetic dissection of stomatal development.
Keywords: Natural variation, Arabidopsis thaliana, stomatal abundance, pavement cell, stomatal lineage, satellite stomatal lineage, epidermis, development
INTRODUCTION
The potential surface available for regulated gas exchange between plants and the atmosphere is set by stomatal number and distribution in the aerial epidermis. In Arabidopsis thaliana, stomata differentiate gradually during organ development, through a series of stereotyped yet flexible cell division and fate acquisition events. Stomatal abundance in different plant surfaces and environments is regulated, resulting in variable stomatal numbers and distribution patterns in mature organs (Bergmann and Sack, 2007; Casson and Hetherington, 2010, and references therein). This suggests that overlapping, partly redundant developmental pathways involving many genes must operate to produce a diversity of stomatal patterns and numbers while guaranteeing their functionality. Dissecting such gene circuits has only just begun, and several positive and negative regulators of stomata differentiation have been identified genetically and molecularly (reviewed by Bergmann and Sack, 2007; Nadeau, 2009; Dong and Bergmann, 2010).
The first recognizable stomata developmental event in A. thaliana Col-0 (see Fig. S1 in Supplementary Data, available online) is the asymmetric division of a protodermal cell [meristemoid mother cell (MMC)], termed entry division, that initiates a stomatal cell lineage; the smaller product, the meristemoid (M), undergoes up to three sequential asymmetric amplifying divisions, oriented in an inward spiral that places the M in the centre of a recognizable structure made by the larger division products (Bergmann and Sack, 2007). While the central M differentiates into a guard mother cell, which divides symmetrically to make a stoma, each of the larger cells can differentiate into a pavement cell or can become a MMC, experience an asymmetric entry division and start a satellite stomatal lineage (Bergmann and Sack, 2007). This is termed a spacing division because it puts at least one non-stomatal cell between the primary (also termed planet; Lucas et al., 2006) and the satellite stomata. Spacing divisions must involve cell–cell signalling events that hinder the development of stomata in contact, while amplifying divisions may also rely on unequal distribution of stomatal fate determinants and, thus, be regarded as executing a lineage-based programme. Therefore, stomatal development is an iterative process involving cell lineage and cell interaction-based processes.
The genetic and molecular dissection of stomatal development is mostly based on severe, aberrant phenotypes produced by induced (often loss-of-function) mutations. Such alleles, which impede stomata formation or produce stomatal clustering, have identified a large suite of stomata developmental regulators. Positive regulators are needed for stomata lineage initiation and development, while negative regulators are determinant for enforcing correct spacing (reviewed by Dong and Bergmann, 2010; Rowe and Bergmann, 2010). A few studies found some genes whose loss-of-function influence stomatal abundance without altering normal stomata development (Zhang et al., 2008; Dong and Bergmann, 2010, and references therein), but little is known about how diversity in functional, non-aberrant stomatal patterns could be set or how primary and satellite lineages contribute to final stomatal abundance.
The quantitative traits currently used to score stomatal abundance are stomatal index (SI), which measures the proportion of epidermal cells that are stomata, and stomatal density (SD) or number of stomata per area unit. SI and SD are the result of cell division patterns and of cell differentiation and expansion during organ growth (Geisler et al., 1998; Geisler and Sack, 2002). SD depends on stomatal number and on the size and number of non-stomatal epidermal cells (mostly pavement cells), while SI depends solely on cell-type proportion, regardless of cell size, and therefore both traits provide complementary information on final stomatal abundance and pattern. In developmental terms, the more widely examined organs for SI and SD are the cotyledon and first true leaf. Though both contribute little to total transpiration and photosynthesis in the adult plant, they are crucial in the first stages of plant development, when transpiration is needed for cell expansion-mediated plant growth while seedlings still have a shallow root system for soil water uptake. Arabidopsis thaliana seed resources are mostly depleted 3 days after germination (Penfield et al., 2005; Graham, 2008), and seedlings rely on photosynthesis as the sole source of nutrients and energy for cell division and dry matter increase. Therefore, in early postembryonic development, plant survival also depends on the compromise between photosynthesis and transpiration, and seedling stomatal abundance and distribution are probably under selective pressure in natural environments.
Naturally occurring variation among wild genotypes is an alternative genetic resource for studying stomatal development. Although many genes involved in stomatal development are remarkably conserved during plant evolution (Peterson et al., 2010), natural selection under diverse conditions may have resulted in natural variants that incorporate adaptive responses of stomata developmental gene networks to environmental cues. Indeed, natural variants in poplar and rice are being used to address the genetic basis of stomatal abundance in these taxa (Ferris et al., 2002; Laza et al., 2009). In A. thaliana, natural variation of complex biological processes has been successfully studied at the molecular level, and natural alleles with a broad range of quantitative effects have been isolated (Koornneef et al., 2004; Alonso-Blanco et al., 2009; Lefebvre et al., 2009). Variation in the stomatal density response to CO2 doubling has also been reported for a number of A. thaliana accessions (Woodward et al., 2002). However, a detailed record of natural stomatal-related trait variants with traceable accessions is not yet available in this model species.
MATERIALS AND METHODS
Plant material and growth conditions
The Arabidopsis thaliana wild genotypes studied (see Table S1 in Supplementary Data, available online) included 47 accessions of the Versailles 48 nested Core Collection (McKhann et al., 2004) and 18 accessions analysed by Clark et al. (2007) for sequence polymorphisms. Ler was excluded from the original Clark's collection because it carries non-natural alleles derived from a fast-neutron mutagenesis (Rédei, 1962). These collections were built to maximize global genetic diversity in A. thaliana. Col-0 (N1092) was used as a laboratory reference accession for both collections. Three accessions were common to the two collections; thence, this study includes 62 different wild accessions plus Col-0. The INRA set seeds were obtained from the National Institute of Versailles's Agronomic Research (INRA, France), and the remaining accessions were provided by the Notthingham A. thaliana Stock Centre (NASC). Seeds were stratified at 4 °C in darkness for 3 d, then sown on pots containing a 2 : 1 : 1 mixture of soil (Prohumin, Klasmann–Deilmann 50–50), perlite and vermiculite, and grown in controlled chambers (Conviron MTR30) under a 16-h photoperiod at 21 ± 1 °C, 60–70 % relative humidity and 150 ± 20 µmol m−2 s−1 irradiance. Data were obtained from four or five plants for each accession (INRA and Clark sets, respectively), simultaneously grown in two random blocks. Accessions of INRA and Clark sets were grown separately.
Trait scoring
The mature adaxial epidermis of cotyledons and first leaves of accessions were scored for stomata and pavement cell abundance. The adaxial epidermis was selected because it has simpler cellular patterns and fewer cell numbers than the abaxial epidermis. Cotyledons and first leaves of the same individuals were analysed. To ensure that organ growth was completed, cotyledons and first leaves were harvested 2 and 7 d after bolting, respectively. Sixteen accessions (see Table S1 in Supplementary Data) did not flower in the present conditions, and their organs were collected 1 week later than those of the latest bolting accession in the assay. Surface replicas were obtained with dental resin (Geisler et al., 2000) and micrographed with a Leica® IRB microscope equipped with a Leica® DC300F camera. Images were digitally processed with Adobe Photoshop CS3 (Adobe Systems Inc.). Epidermal cell counts of each individual were an average from two 0·327-mm2 areas centred along the organ apical-to-basal and the median-to-margin axes. Cells having at least 25 % of their surface inside the sampling area were scored. In leaves, this area excluded trichomes and their surrounding socket cells. Stomatal and pavement cell densities (SD and PD) were calculated as number of stomata or pavement cells per area unit (cell number mm−2), respectively, and stomatal index (SI) as percentage of epidermal cells that were stomata.
Organ areas were measured in the same individuals scored for epidermal traits. Organ replicas were photographed with a Leica® Stereomicroscope equipped with a Leica® -DC300F camera and areas determined with Leica® IM50 image manager v. 4.0 software.
Environmental maternal effects assay
The maternal environment (environmental growth conditions of the mother plants of a seed progeny; ME) can have effects on offspring phenotypes (Donohue, 2009). Since ME are often non-uniform across different experiments, to find out if the traits under study were affected by differences in laboratory ME, tests were carried out. Several separate seed harvests were obtained for a number of accessions. Seven accessions representing the range of stomatal abundance variation in the Clark set (Bur-0, Col-0, Cvi-0, Nfa-8, Shakdara, Ts-1 and Van-0) were grown and selfed from the seed lot used for trait description, in two independent experiments (referred as ME1 and ME2) conducted in the same chamber at different times. Additionally, the four accessions present in both the Clark and the INRA collections (Bur-0, Col-0, Cvi-0 and Shakdara) were grown and selfed from the INRA collection seed stocks in a chamber different from that used for ME1 and ME2 (ME3). Both chambers were Conviron MTR30 and plants in the three ME were grown using the same environmental setting described above. Subsequently, seeds derived from a single plant for each accession and ME were simultaneously grown, and six individuals per accession and ME were scored for the various traits. Data were analysed in two sets: one set contained four accessions [genotypes (G)] in three maternal environments (4G × 3ME), and the other had seven accessions in two ME (7G × 2ME).
Analysis of primary and satellite stomatal lineages
Seeds harvested from plants grown simultaneously (i.e. under the same ME) were grown as described above. Developmental homogeneity was established by monitoring germination every 6 h. Radicle emergence set germination time point at 0, and ten synchronous seedlings per accession were collected 48 h later. Specimens were fixed in ethanol : acetic acid 9 : 1 (v/v), dehydrated through ethanol : water series, rehydrated, mounted in Hoyer's medium (Liu and Meinke, 1998), and inspected with differential interference contrast (DIC) (Eclipse 90i microscope and Nikon DXM1200C camera). Epidermal cell types were recorded in two fields per cotyledon, avoiding the central zone and edges, and represented about 70 % of the organ surface. Cell types were identified accordingly to Zhao and Sack (1999) and Geisler and Sack (2002). Primary and satellite stomatal lineages were scored as previously defined (Berger and Altmann, 2000; Kutter et al., 2007; Zhang et al., 2008). No distinction was made between secondary and higher-order satellite lineages. Ten additional individuals per accession were grown to maturity and their stomatal index determined.
Statistical analysis
Cell density values were loge transformed, while cell index values were arcsin-root transformed to improve normality of distributions and homogeneity of variances. None of the outcomes and conclusions changed when using the original data, and therefore most descriptions are based on untransformed data to simplify interpretation. INRA and Clark sets are described separately because two-way ANOVA and t-test analyses showed that some trait values in the four accessions common to both sets (Bur-0, Col-0, Cvi-0 and Shakdara) differed significantly between assays, probably due to slight environmental differences resulting from their independent growth. For each trait, the amount of variation was calculated by the coefficient of variation (CV = standard deviation/average) and by the fold change (maximum accession value/minimum accession value). Broad-sense heritability (h2) was calculated as:
where VG (genetic variance) is the among-genotype (accession) variance component, and VE (environmental variance) is the residual error variance component estimated by restricted maximum likelihood (REML) analysis (Lynch and Walsh, 1998). Genetic correlations between traits were estimated by Pearson's and Spearman's tests using accession mean trait values (family mean of the selfing offspring, which carry practically the same genotype; Lynch and Walsh, 1998). Similar relationships and conclusions were achieved from both analyses (unless indicated) and, hence, only Pearson coefficients are reported. Fisher's z-test (Zar, 1996) was used to identify correlations that differed significantly between the two accessions sets. Differences between mean stomatal lineage indices (arcsin-root transformed) of selected accessions were tested by Student's t-tests. Environmental maternal effects on traits were evaluated by a two-factorial analysis of variance (ANOVA), with genotype (accession) and maternal environment as fixed factors. All statistical analyses were performed with the SPSS 11·0 package (SPSS Inc., Chicago, IL, USA). To identify and classify phenotypic diversity within the accession sets, hierarchical cluster analysis was carried out with MeV v4.2 (TM4 Microarray Software Suite, Boston, MA, USA) using standardized data to perform an average linkage clustering based on uncentred Pearson's correlation as the distance metric.
RESULTS
Natural variation in stomatal abundance
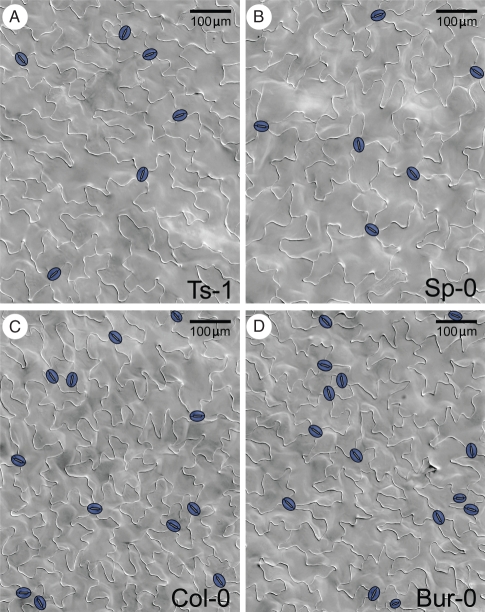
Stomatal abundance traits in 62 wild genotypes of A. thaliana, grouped in two sets (namely, the INRA and the Clark sets), which were grown independently and therefore described separately, were analysed (see Materials and methods, and Table S1 in Supplementary Data, available online). Col-0 was included in both sets as a reference genotype. Accessions were examined for six traits: stomatal index, stomatal density and pavement cell density in cotyledons (C) and first leaves (L) in the adaxial epidermis (namely SIC, SDC, PDC SIL, SDL and PDL). Pavement cell density was included to address its impact on stomatal density variation. Representative sampled epidermes for accessions with distinct stomatal abundance are shown in Fig. 1. No aberrant phenotypes were found in any of the natural accessions studied (not shown).
Fig. 1.
Representative epidermes of A. thaliana accessions with distinct stomatal abundance. DIC micrographs of mature adaxial cotyledon epidermis (21 dpg) are shown for four accessions. Stomatal index and density values are low for Ts-1 (A) and Sp-0 (B), and high for Col-0 (C) and Bur-0 (D). Stomata have been coloured in blue. Scale bars = 100 µm.
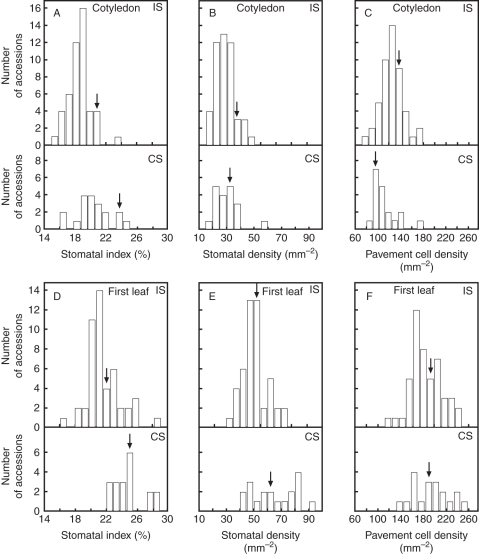
All traits displayed considerable and continuous variation among the genotypes scored in each set (Fig. 2, and Table S2 in Supplementary Data). Stomatal indices showed the lowest variation both in cotyledons and first leaves (1·3- to 1·7-fold). In cotyledons, SI varied 1·5-fold, ranging from the lowest values in Can-0 and Ts-1 to the highest values in Bur-0 and Van-0 in the INRA and Clark sets, respectively (see Table S1 in Supplementary Data). Bur-0, represented in both sets, also had the second maximum SIC within the Clark group. Remarkably, Col-0 cotyledons, commonly used to study stomatal development, showed one of the highest stomatal index values, ranking right after Bur-0 in the two data sets. In leaves, minimum stomatal index values occurred for Can-0 and Nfa-8, while maximums corresponded to Ishikawa (followed by Bur-0) and Rrs-7 (followed by Bur-0 and Van-0) in the INRA and Clark sets, respectively.
Fig. 2.
Frequency distributions of stomatal and pavement cell-abundance traits. Each panel shows a histogram for the 48 accessions of the NRA set (IS; upper part) and the 19 accessions of the Clark set (CS; lower part). Cotyledons and first leaves were scored for stomatal index [SIC (A) and SIL (D), respectively], stomatal density [SDC (B) and SDL (E), respectively] and pavement cell density [PDC (C) and PDL (F), respectively]. Col-0 mean values are indicated by a black arrow.
The largest variation was observed in stomatal densities, which varied between 2·1- and 3·5-fold (see Table S2 in Supplementary Data). Sakata and Got-7 had the lowest SD values in cotyledons, and Can-0 and Tsu-1 in leaves (INRA and Clark sets, respectively). The highest SD corresponded to Jm-0 and Van-0 cotyledons, and Bur-0 and Van-0 leaves (INRA and Clark sets, respectively).
Pavement cell density showed an intermediate amount of variation in both organs (ranging between 1·9- and 2·3-fold). Accessions setting the upper and lower limits of PDC were the same that flanked the SDC variation range (Sakata and Got-7 had the lowest PDC, and Jm-0 and Van-0 the highest one). In leaves, Ishikawa and Tsu-1 showed the lowest PDL, while Jm-0 and Bor-4 had the highest PDL values (INRA and Clark sets, respectively).
Mean, minimum and maximum values for all traits were always higher in leaves than in cotyledons (Table S2 in Supplementary Data), suggesting a supra-organ regulation of stomatal abundance and pavement cell density, likely related to heteroblasty (Tsukaya et al., 2000). Ratios between equivalent traits in leaves and cotyledons were calculated for each accession (Table S1 in Supplementary Data). Few exceptions to the general behaviour were found, ratios varying from 1·3 to 3·2 for SD, from 1·1 to 2·3 for PD and from 1·0 to 1·6 for SI. Remarkably, Col-0 is one of six accessions with nearly identical SI in leaves and cotyledons.
The six traits showed high to moderate broad-sense heritabilities (h2), which ranged from 0·33 to 0·71 (Table S2 in Supplementary Data). Overall, stomatal and pavement cell densities had similar heritability in cotyledons and leaves, with higher h2 values than for stomatal index. Therefore, our data show that there is substantial genetic variation among accessions for all six traits.
Genetic correlations among stomatal abundance traits
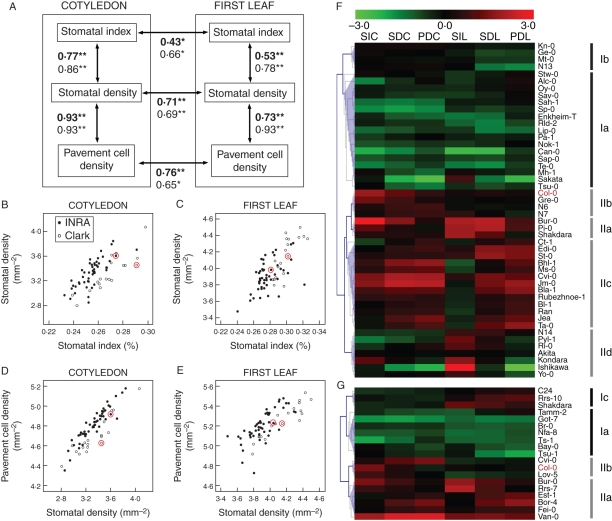
To test for co-ordinated regulation of stomatal abundance in cotyledons and leaves and for relationships among epidermal traits within each organ, correlation coefficients between all pair combinations of the six traits were estimated (Fig. 3A–E, and Table S3 in Supplementary Data). Stomatal and pavement cell densities showed the strongest correlations, both in cotyledons and leaves (r = 0·73–0·93; P < 10−8), indicating that variation in pavement cell size is a major cause for SD variation among accessions. In addition, SI and SD positively correlated in both organs, with larger correlations in cotyledons (cotyledons: r = 0·86, P < 10−5; leaves: r = 0·78, P < 10−4). The relationship between SI and PD differed between organs; in cotyledons it was moderate in the two accession sets (r = 0·49–0·61; P ≤ 5 × 10−3), and in leaves it was weak in the Clark set (r = 0·5, P ≤ 3 × 10−2) or absent in the INRA set. In addition, correlations between cotyledon and leaf values for SI, SD and PD traits were moderately positive (Fig. 3A, and Table S3 in Supplementary Data).
Fig. 3.
Correlation analysis among epidermal cell-abundance traits. (A) Diagram showing selected correlations between traits in cotyledons and first leaves. Double-headed arrows connect significantly related traits. Pearson's coefficient and significance level (**, P < 0·001; *, P < 0·01) are indicated for the INRA set (bold) and the Clark set (normal font). (B–E) Scatter plots showing the relationship between stomatal density and stomatal index or pavement cell density, in cotyledon (B, D) and first leaf (C, E). Closed circles and open circles correspond to INRA and Clark accessions, respectively. The Col-0 positions are indicated with red circles. (F, G) Heat-map visualization of a hierarchical cluster analysis of the cell-type abundance phenotypes at INRA (F) and Clark (G) accession sets. Each row represents one accession and each column represents one trait. Red and green rectangles indicate, for each trait and accession, mean values higher or lower than the set average; colour intensity scales are given in the top of the panel. Group designation is shown on the right and the corresponding nodes are highlighted in blue on the left (see text for details). Col-0 is highlighted in red.
To identify common and distinct phenotypic patterns underlying the observed correlations between trait pairs, clustering analyses were carried out using an uncentred Pearson correlation with average linkage metric (Fig. 3F, G). Dendrograms show, in a heat-map format, the standardized trait values (z-scores) for each accession, represented as a rank of divergence for the trait mean in the set. In both collections, accessions were assigned to two main clusters: one with prevalently low-to-medium values (cluster type I) and another containing accessions with mostly high-to-medium values (cluster type II). Both cluster types included a group with all trait values ranking similarly, either low-moderate (type Ia; 36 % of accessions) or high-moderate (type IIa; 13 % of accessions). These two main clusters also included groups with trait values differing among organs, either standing higher (Ib and IIb; 6 % and 10 %, respectively) or lower (Ic; 4 %) in cotyledons than in leaves. In the INRA collection, cluster II included two additional groups with lower (IIc; 19 % of accessions) or higher (IId; 10 % of accessions) SI values than other traits. Therefore, about 49 % of accessions showed a phenotypic pattern with strong relationships among all trait values (Ia and IIa), in agreement with the correlations found among traits (Fig. 3A). The remaining groups contained phenotypes that deviated from these general correlations and showed, for instance, leaf or cotyledon stomatal index values differing from cell density values (see Discussion). Thus, clustering analyses revealed natural prevalent phenotypic patterns, and also accessions with distinct uncommon phenotypes.
Relationship between organ size and stomatal abundance
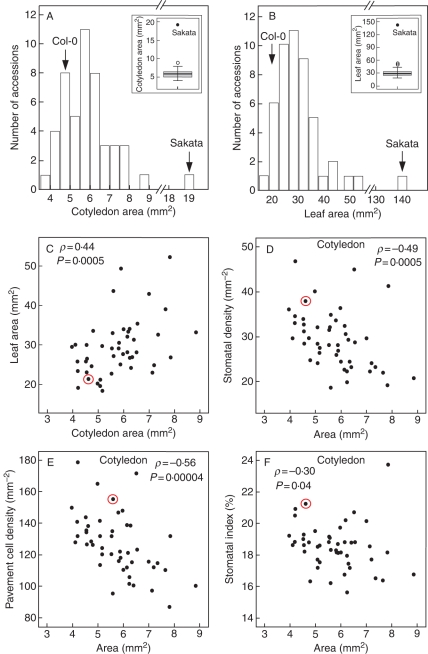
Organ size regulation is expected to integrate mechanisms controlling stomatal lineage initiation and proliferation. To explore relationships between organ size and stomatal abundance, cotyledon and first leaf areas were measured in the INRA set. Correlations between cell and organ traits were estimated (see Fig. 4, and Tables S1, S4 and S5 in Supplementary Data). Substantial variation was found for cotyledon and leaf area (Fig. 4A, B). Notably, Sakata displayed extremely large organs, appearing as an outlier of the observed continuous distribution. Hence, organ area data are presented with and without Sakata (Fig. 4C–F and Tables S4 and S5 in Supplementary Data). Broad-sense heritability estimates were high in both cases (ranging from 0·59 to 0·76; Table S4 in Supplementary Data), indicating that there is substantial genetic variation for these traits among natural accessions.
Fig. 4.
Natural diversity in cotyledon and first leaf area and their correlations with epidermal cell-abundance traits. (A, B) Histograms depict the frequency of accession mean values for the cotyledon area (CA) and the first leaf area (LA) in the INRA set. Values of the reference Col-0 and the extreme Sakata accessions are indicated (note that area scale is discontinuous). Inset: box plots for CA and LA where the upper and lower limits of the boxes represent the third and first quartiles and the bars inside the boxes indicate the medians; open circles indicate outlying accessions (values 1·5–3 times the interquartile range) and closed circles highlight the extreme Sakata value (above 3 times the interquartile range). (C–F) Scatter plots depicting relationships between (C) cotyledon and first leaf areas, and between cotyledon area and (D) adaxial stomatal density (SDC), (E) pavement cell density (PDC) and (F) stomatal index (SIC). Col-0 is highlighted with circles. Inside each panel, the corresponding Spearman coefficient ρ and P-value are indicated. Sakata values were excluded in the scatter plots.
Relationships among cellular and organ size traits were analysed by Spearman correlation (Table S5 in Supplementary Data), since organ area data did not meet parametric test assumptions required for Pearson correlations. Cotyledon and first leaf areas were positively correlated (ρ = 0·48 and 0·44; P < 0·05; with and without Sakata, respectively). Both showed negative correlations with their respective SD and PD (ρ between –0·29 and –0·59; P < 0·05), particularly in cotyledons. Organ area and SI were negatively correlated in cotyledons (ρ = –0·3; P < 0·05), but not in leaves. Total pavement cell number for all accessions was estimated from their PD and organ area values (not shown). Cell number and organ area showed a strong positive correlation, which in leaves was higher than the relationship with any other trait (Table S5 in Supplementary Data), as previously reported (Granier et al., 2000; Cookson et al., 2005, 2007). Hence, in this sample of accessions, larger organs often had more and larger pavement cells (leading to lower stomatal densities) and vice versa, showing a relationship between organ and pavement cell number and size. All together, the present results show that variation in cell number and size contributes to genetic diversity in cotyledon and first leaf size. However, accessions with larger cotyledons tended to have a lower proportion of stomata (SI), suggesting a distinct genetic link between cotyledon size and stomatal lineage development.
Environmental maternal effects on epidermal cell-type abundance and organ size traits
Two complementary data sets were obtained to evaluate whether small differences in laboratory maternal environment (ME), expected to occur among separate plant growth experiments, would affect the traits surveyed in this study. One set assessed effects of three MEs on four accessions [genotypes (G)], while the other used two MEs on seven accessions (4G × 3ME and 7G × 2ME, respectively; see Materials and methods). Each ME refers to a separate experiment where plants of the various accessions were grown simultaneously from seed germination to seed production. Plants from all maternal sources were simultaneously grown and the traits scored. ME effects were estimated by two-factorial ANOVA, with genotype (G; accession) and ME as factors (Table S6 in Supplementary Data). A few significant effects of ME (P < 0·05) were only detected in the 4G × 3ME data set. MEs had no effect on stomatal abundance traits, except for a marginal influence on leaf SD (R2 = 2·9). Minor effects were also found on leaf PD (R2 = 7·09). However, MEs and genotype × ME interactions show significant effects on organ sizes, being larger in cotyledons (ME, R2 = 10·4; G × ME, R2 = 20·3) than in leaves (ME, R2 = 3·9; G × ME, R2 = 13·24).
Natural variation in stomata developmental pathways
To address if natural variants differ in the contribution of distinct developmental processes affecting stomatal index in mature organs, two quantitative processes were analysed: the proportion of primary lineages stemming from protodermal cells, and the proportion of satellite lineages that arise from non-stomatal cells in primary lineages (Fig. S1 in Supplementary Data). The analyses were restricted to cotyledons in four accessions with extreme stomatal index values (Fig. 3F, G): Ts-1 and Sp-0, with very low SI, and Col-0 and Bur-0 as accessions with the highest SI values in the two collections. Adaxial cotyledon epidermes were examined at two time points: 48 h post-germination (hpg), the shortest time amenable to analyses, and 21 d post-germination (dpg), when satellite lineages have appeared extensively. Lineage initiation was monitored by the primary lineage index (PLI) or proportion of primary stomata plus primary stoma precursors to total epidermal cells, and the satellite lineage index (SLI), or proportion of satellite stomata plus satellite stomata precursors (see Fig. 5 and Materials and methods). At 48 hpg (Table 1) the two high SI accessions Bur-0 and Col-0 had initiated a similar number of lineages (primary plus satellite), which was significantly higher than that of the low SI accessions Sp-0 and Ts-1. The total lineage index (TLI = PLI + SLI) at 48 hpg showed highly significant correlation with proportion of stomata (SI) in mature cotyledons at 21 dpg (TLI vs. SI: r = 0·99, P = 0·01), indicating that stomatal phenotypes observed at 48 hpg are predictive of mature organ phenotypes. In spite of this correlation, the low SI but not the high SI accessions showed a higher TLI at 48 h than SI at 21 dpg (Table 1). This could be due to an increased pavement cell production through extended stomatal lineage amplification divisions and/or to more pavement cell symmetric divisions.
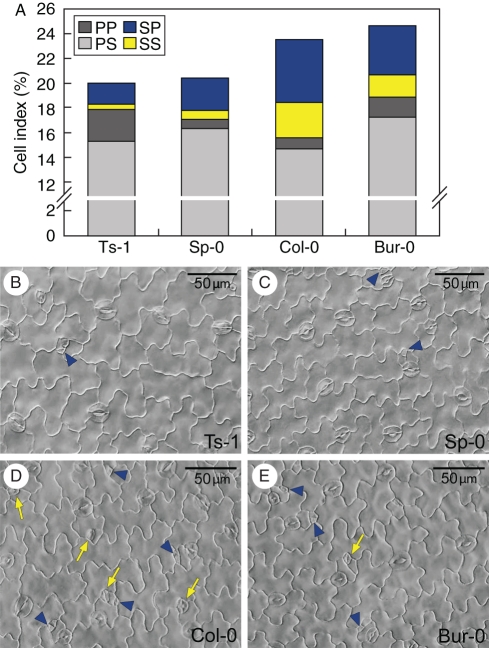
Fig. 5.
Diversity in distinct stomata developmental pathways in selected accessions. (A) Cell index on the adaxial epidermis of cotyledons 48 h after germination for primary stomata (PS) or precursors (PP) and satellite stomata (SS) or precursors (SP) in Ts-1, Sp-0, Col-0 and Bur-0. The data represent average values of ten individuals. Note that the cell index scale is discontinuous. (B–E) Representative DIC images of developing cotyledon epidermes in Ts-1 (B), Sp-0 (C), Col-0 (D) and Bur-0 (E). Satellite stomata are marked by yellow arrows and satellite precursors by blue arrowheads. Scale bars = 50 µm.
Table 1.
Differential contribution of primary and satellite lineage initiation to stomatal abundance in selected accessions
| 48 hpg |
21 dpg | ||||
|---|---|---|---|---|---|
| Accession | Primary lineages index | Satellite lineages index | Total lineages index | % Satellite lineages | Stomatal index |
| Ts-1 | 17·9 ± 0·4** | 2·1 ± 0·3*** | 20·0 ± 0·5*** | 10·2 ± 1·4*** | 15·8 ± 0·4*** |
| Sp-0 | 17·1 ± 0·3* | 3·3 ± 0·4*** | 20·4 ± 0·5*** | 16·1 ± 1·7*** | 16·2 ± 0·3*** |
| Col-0 | 15·6 ± 0·6 | 7·9 ± 0·7 | 23·4 ± 0·4 | 33·4 ± 3·0 | 22·3 ± 0·6 |
| Bur-0 | 18·8 ± 0·5*** | 5·8 ± 0·7 (*) | 24·6 ± 0·6 | 23·3 ± 2·5* | 23·3 ± 0·5 |
The index of stomatal primary lineages, satellite lineages and total lineages (primary + satellite lineages), and the percentage of satellite lineages over the total stomatal lineages were determined in the adaxial epidermis of cotyledons 48 hpg in Ts-1, Sp-0, Col-0 and Bur-0. The stomatal index at maturity (21 dpg) in the same assay is provided. Significance levels of t-tests with respect to Col-0 values are indicated. [***, P < 0·001; **, P < 0·01; *, P < 0·05; (*), P < 0·052].
Satellite lineage contribution to total stomatal lineages showed considerable variation among accessions, ranging from 10 % in Ts-1 to 33 % in Col-0 (Table 1). Although Bur-0 and Col-0 had different SLIs, they had a similar stomatal precursors index, which presented larger individual variability (as expected for a parameter very sensitive to small variations in temporal development; Fig. 5A). Regarding primary lineage index, Bur-0 and Col-0 showed the largest difference, while Bur-0 and Ts-1 displayed similar PLI values (P > 0·05). Therefore, two accessions with similar stomatal index showed a different primary lineage index (Col-0 and Bur-0), while accessions with a different stomatal index presented an equivalent primary lineage relative abundance (Bur-0 and Ts-1). Thus, primary and satellite lineage initiation frequencies can combine in different ways: low production of primary stomata with high production of satellite lineages (Col-0); high production of primary stomata and low satellite initiation (Ts-0); or moderate (Sp-0) or high (Bur-0) production of both primary and satellite lineages. These results reveal substantial natural variation in the initiation of both primary and satellite stomatal lineages, suggesting that the two processes are partially under independent genetic control.
DISCUSSION
Wild genotypes of A. thaliana show substantial genetic variation in epidermal cell-type abundance
Analyses of quantitative traits related to cotyledon and first leaf stomatal abundance in 62 wild A. thaliana genotypes, evaluated in two sets, have revealed considerable natural genetic variation. Stomatal density is the most variable trait, while stomatal index showed the least variation. SI relates to cell division and differentiation, and its variation may result from relatively narrow limits that ensure a functional epidermis co-ordinated with the underlying mesophyll. The moderate pavement cell density variation found in the present study would mostly arise from cell-size phenotypes. Combinations of low SI and moderate PD variations most likely account for the higher SD diversity.
The genetic variation found in this work suggests that phenotypes of the genotypes examined are not deleterious under natural environments. In agreement, aberrant stomatal phenotypes were not found, indicating that incorrect patterns reduce plant fitness. To determine if the genetic variation observed may be involved in environmental adaptation, correlations were inspected among stomatal traits and geographic and historical climate data from the accession collection sites. Marginal positive correlations were detected between leaf SI and mean monthly precipitation from October to April (Spearman ρ = 0·29–0·35; P = 0·014–0·044), which suggests adaptive population differentiation to water availability. However, the broad geographic distribution of the accessions studied and imprecise information on their original habitats may influence these findings. In addition, given the high plasticity of the traits studied (Casson and Gray, 2008; Lampard, 2010, and references therein), phenotypes may differ between laboratory conditions and natural environments, fading a putative adaptive basis in the underlying genetic variation.
Maternal environments may influence offspring traits, especially those expressed early in the life cycle (reviewed by Donohue, 2009). It was found that cotyledon and first leaf cell-abundance traits were marginally or not affected by differences in the ME occurring in laboratory experiments. In agreement, A. thaliana stomatal density showed no ME effect during 15 generations grown at different CO2 concentrations (Teng et al., 2009). As demonstrated for other species (Roach and Wulff, 1987), ME had a moderate but significant effect on cotyledon size, although genetic differences accounted for most of the phenotypic variation. Minor ME effects were also detected on first leaf size. Therefore, the phenotypic differences found among accessions for cellular traits are mainly determined by genotypic variation and not by the maternal environments used.
Relationship between epidermal cell abundance and organ size traits
Correlation analyses uncovered an overall pattern of positive genetic relationships among stomatal and pavement cell-abundance traits within and between organs. The correlations we found most likely have a common genetic basis, because A. thaliana mutations in several stomata developmental genes simultaneously affect all or most traits examined here [notably GPA1 and ERECTA (Zhang et al., 2008; Nilson and Assmann, 2010; van Zanten et al., 2010); other genes are reviewed by Dong and Bergmann (2010)]. Nonetheless, such trait relationships may also originate from trait co-evolution (Armbruster and Schwaegerle, 1996).
Relationships between all traits for each organ suggest that shared genetic networks control cell-type proportion and density at the organ level, although they appear more closely related in cotyledons than in leaves. These results are consistent with the proposed mechanism co-ordinating cell proliferation and expansion at the organ level (Tsukaya, 2006; Fujikura et al., 2007, 2009; Tsukaya, 2008; Micol, 2009). Correlations were also found for each trait between organs. In agreement with these correlation patterns, cluster analyses identified two consistent groups of accessions where all six cell-abundance traits were either low (group Ia) or high (group IIa), indicating that similar genetic networks control cell-type abundance in both organs. These two groups account for nearly half of the accessions, suggesting a mechanism for supra-organ co-ordination of stomatal abundance in A. thaliana. Interestingly, some accessions deviated from the prevalent trait correlations. Clustering analysis singled out groups with uncommon phenotypic patterns of opposite SI and cell densities values (e.g. Ishikawa and Jm-0) or having weak or absent relationships for all traits between organs (e.g. C24 or Shakdara). These accessions that deviate from the general tendencies suggest that the genetic programmes controlling the various traits and their co-ordination also involve unshared factors.
Cotyledon and leaf areas, which also showed significant genetic variation, negatively correlated with most cell-abundance traits in the 48 accessions examined (INRA set). In most accessions, large organs are built not only by more cells but also by larger ones (as inferred by their low pavement cell densities) and have lower stomatal densities; conversely, high stomatal and pavement cell densities (small cells) and lower cell numbers are the norm in small organs. In cotyledon, stomatal index and organ area were also negatively correlated. Overall, the present results show that wild accessions comply with the above-mentioned theories of co-ordinated cell behaviour in leaf growth (reviewed by Tsukaya, 2008), and suggest that, at least in cotyledons, stomatal lineage development is partially linked to an organ level growth control.
Some accessions carry allele combinations determining extremely low or high stomata proportions and cell densities, like Ts-1, Sp-0, Van-0 or Bur-0. The highest SD was recorded for Van-0, an erecta mutant (van Zanten et al., 2010). Loss-of-function ERECTA mutations in Col and Ler increase SD (Shpak et al., 2005) and total epidermal cell densities (Tisné et al., 2008) but do not affect SI in adult leaves (Masle et al., 2005), or lower it in the abaxial cotyledon epidermes (Shpak et al., 2005). It was found that Van-0 also has a very high pavement cell density and SI, suggesting differences in erecta phenotypes between epidermes or the presence of natural erecta modifiers in Van-0. Thus, the present analyses discriminated a loss-of-function allele of a gene involved in epidermal cell density determination, further revealing additional features associated with this gene.
The wide natural genetic variation in stomatal abundance traits that are described here can be readily dissected by QTL mapping through available recombinant inbred line populations (Simon et al., 2008). Its combination with the complete genome sequences of a fast growing number of A. thaliana accessions (Weigel and Mott, 2009) will provide essential information on the mechanisms involved in this developmental process. Given the impact of stomatal abundance on transpiration and photosynthesis, these studies might provide new alleles of agronomic interest for genetic improvement of crop performance.
Stomatal index encompasses cryptic genetic variation in distinct developmental processes
Primary and satellite stomatal pathways have been extensively described (Bergmann and Sack, 2007; see Fig. S1 in Supplementary Data). However, mechanisms regulating their proportions are poorly understood. The present analysis has found genetic variation in the relative frequency of primary and satellite lineages. A similar stomatal index could also be achieved with different relative frequencies. Therefore, primary to satellite stomata proportion is a distinct trait that reveals different phenotypes hidden under the same SI. This range of previously unknown genetic diversity suggests an even higher complexity of genetic factors involved in stomatal abundance control.
Primary stomata abundance directly relates to entry divisions, while satellite stomata abundance will be affected directly by spacing divisions, and indirectly by amplifying and entry divisions (Bergmann and Sack, 2007). Several genes are known to control these three asymmetric divisions (Dong and Bergmann, 2010), but very few seem to affect satellite lineages specifically. One is AGL16, whose microRNA-mediated regulation limits satellite lineage production with no apparent effect on primary stomata (Kutter et al., 2007). Also, two G-protein subunits, AGB1 and GPA1, function as mutually antagonistic modulators of satellite lineages (Zhang et al., 2008). GPA1 also controls epidermal cell size (Nilson and Assmann, 2010). The satellite stomata fraction may largely depend on the cell-proliferation time window in Col-0 adaxial cotyledon epidermes (Geisler and Sack, 2002). The earlier cell division arrest in adaxial versus abaxial epidermes concurs with lower satellite stomata production. Therefore, changes in the proliferation-window length may alter satellite stomata abundance. Division rate differences would have a similar influence, and both factors may also impact on higher-order satellization.
Primary and satellite stomatal pathways are assumed to contribute to the phenotypic plasticity of A. thaliana stomatal development in response to internal and environmental cues (Bergmann and Sack, 2007; Casson and Gray, 2008; Lampard, 2010), but whether each pathway displays specific responses remains to be established. Unravelling the molecular genetic basis of the observed variation would shed light on a basic developmental programme, and may also provide tools to understand plant productivity under different environments.
Conclusions
A systematic data collection is provided on cotyledon and first leaf traits related to stomatal abundance for 62 wild Arabidopsis thaliana accessions analysed in two sets. This survey reveals substantial genetic variation in SI, SD and PD in both organs, and shows that these traits are largely unaffected by the laboratory maternal environments used. SD shows the highest correlation with the other traits within each organ, strongly suggesting that these traits share genetic bases. Inter-organ correlations were also found, supporting the operation of supra-organ mechanisms for the control of cell-type abundance in the accessions surveyed. Clustering analyses showed that while half of the accessions had these strong relationships among all traits, accessions with uncommon phenotypic patterns, suggestive of differences among genetic programmes controlling the various traits, do also occur in nature. The 47 wild accessions surveyed had considerable variation in cotyledon and organ size, which were negatively correlated with cell densities. Notably, variation was also identified in two distinct processes during stomatal differentiation (primary and satellite lineage initiation), which can be hidden under stomatal abundance trait values, because accessions with a similar SI could have very different lineage class ratios. Thus, genetic determinants of primary and satellite lineage initiation can combine in several ways and produce a similar final stomatal abundance phenotype. In addition to revealing this cryptic diversity in a developmental process, this first systematic, comprehensive natural-variation survey for stomatal abundance in A. thaliana provides relevant relationships amongst stomatal traits at the organ and supra-organ levels. This survey also identifies accessions with extreme values or uncommon trait correlations, which are valuable tools for further understanding of stomatal development in natural germplasm.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported by the Consejería de Ciencia y Educación-JCCM (PAI07-0036-3278 to M.M.) and the Ministerio de Ciencia e Innovación (BIO2007-60276 to C.F. and M.M., CSD2007-00057 to C.F. and C.A.-B., and BIO2007-62632 to C.A.-B.). We thank C. Donaire for technical assistance. The authors thank Prof. Pamela Diggle and two anonymous reviewers for their helpful suggestions.
LITERATURE CITED
- Alonso-Blanco C, Aarts MGM, Bentsink L, et al. What has natural variation taught us about plant development, physiology, and adaptation? The Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster WS, Schwaegerle KE. Causes of covariation of phenotypic traits among populations. Journal of Evolutionary Biology. 1996;6:261–276. [Google Scholar]
- Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes and Development. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. Stomatal development. Annual Review of Plant Biology. 2007;58:163–181. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- Casson SA, Gray JE. Influence of environmental factors on stomatal development. New Phytologist. 2008;178:9–23. doi: 10.1111/j.1469-8137.2007.02351.x. [DOI] [PubMed] [Google Scholar]
- Casson SA, Hetherington AM. Environmental regulation of stomatal development. Current Opinion in Plant Biology. 2010;13:90–95. doi: 10.1016/j.pbi.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Cookson SJ, van Lijsebettens M, Granier C. Correlation between leaf growth variables suggests intrinsic and early controls of leaf size in Arabidopsis thaliana. Plant, Cell & Environment. 2005;28:1355–1366. [Google Scholar]
- Cookson SJ, Chenu K, Granier C. Day-length affects the dynamics of leaf expansion and cellular development in Arabidopsis thaliana partially through floral transition timing. Annals of Botany. 2007;99:703–711. doi: 10.1093/aob/mcm005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Bergmann DC. Stomatal patterning and Development. Current Topics in Development Biology. 2010;91:267–297. doi: 10.1016/S0070-2153(10)91009-0. [DOI] [PubMed] [Google Scholar]
- Donohue K. Completing the cycle: maternal effects as the missing link in plant life histories. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:1059–1074. doi: 10.1098/rstb.2008.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris R, Long L, Bunn SM, et al. Leaf stomatal and epidermal cell development: identification of putative quantitative trait loci in relation to elevated carbon dioxide concentration in poplar. Tree Physiology. 2002;22:633–640. doi: 10.1093/treephys/22.9.633. [DOI] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Tsukaya H. Dissection of enhanced cell expansion processes in leaves triggered by a defect in cell proliferation, with reference to roles of endoreduplication. Plant Cell Physiology. 2007;48:278–286. doi: 10.1093/pcp/pcm002. [DOI] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H. Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. The Plant Journal. 2009;59:499–508. doi: 10.1111/j.1365-313X.2009.03886.x. [DOI] [PubMed] [Google Scholar]
- Geisler M, Sack FD. Variable timing of developmental progression in the stomatal pathway in Arabidopsis cotyledons. New Phytologist. 2002;153:469–476. doi: 10.1046/j.0028-646X.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- Geisler M, Yang M, Sack FD. Divergent regulation of stomatal initiation and patterning in organ and suborgan regions of the Arabidopsis mutants too many mouths and four lips. Planta. 1998;205:522–530. doi: 10.1007/s004250050351. [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. The Plant Cell. 2000;12:2075–2086. doi: 10.1105/tpc.12.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. Seed storage oil mobilization. Annual Review of Plant Biology. 2008;59:115–142. doi: 10.1146/annurev.arplant.59.032607.092938. [DOI] [PubMed] [Google Scholar]
- Granier C, Turc O, Tardieu F. Co-ordination of cell division and tissue expansion in sunflower, tobacco, and pea leaves: dependence or independence of both processes? Journal of Plant Growth Regulation. 2000;19:45–54. doi: 10.1007/s003440000006. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Kutter C, Schob H, Stadler M, Meins F, Jr, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. The Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR. Plasticity in stomatal development: what role does MAPK signaling play? Plant Signaling and Behavior. 2010;5:576–579. doi: 10.4161/psb.11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laza M, Kondo M, Ideta O, Barlaan E, Imbe T. Quantitative trait loci for stomatal density and size in lowland rice. Euphytica. 2009;172:149–158. [Google Scholar]
- Lefebvre V, Kiani SP, Durand-Tardif M. A focus on natural variation for abiotic constraints response in the model species Arabidopsis thaliana. International Journal of Molecular Sciences. 2009;10:3547–3582. doi: 10.3390/ijms10083547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Meinke DW. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. The Plant Journal. 1998;16:21–31. doi: 10.1046/j.1365-313x.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- Lucas JR, Nadeau JA, Sack FD. Microtubule arrays and Arabidopsis stomatal development. Journal of Experimental Botany. 2006;57:71–79. doi: 10.1093/jxb/erj017. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- McKhann HI, Camilleri C, Bérard A, et al. Nested core collections maximizing genetic diversity in Arabidopsis thaliana. The Plant Journal. 2004;38:193–202. doi: 10.1111/j.1365-313X.2004.02034.x. [DOI] [PubMed] [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature. 2005;436:866–870. doi: 10.1038/nature03835. [DOI] [PubMed] [Google Scholar]
- Micol JL. Leaf development: time to turn over a new leaf? Current Opinion in Plant Biology. 2009;12:9–16. doi: 10.1016/j.pbi.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Nadeau JA. Stomatal development: new signals and fate determinants. Current Opinion in Plant Biology. 2009;12:29–35. doi: 10.1016/j.pbi.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson SE, Assmann SM. The α-subunit of the arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiology. 2010;152:2067–2077. doi: 10.1104/pp.109.148262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Graham S, Graham IA. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochemical Society Transactions. 2005;33:380–383. doi: 10.1042/BST0330380. [DOI] [PubMed] [Google Scholar]
- Peterson KM, Rychel AL, Torii KU. Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. The Plant Cell. 2010;22:296–306. doi: 10.1105/tpc.109.072777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei GP. Single locus heterosis. Zeitschrift für Vererbungsiehre. 1962;93:167–170. [Google Scholar]
- Roach DA, Wulff RD. Maternal effects in plants. Annual Review of Ecology and Systematics. 1987;18:209–235. [Google Scholar]
- Rowe MH, Bergmann DC. Complex signals for simple cells: the expanding ranks of signals and receptors guiding stomatal development. Current Opinion in Plant Biology. 2010;13:548–555. doi: 10.1016/j.pbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, et al. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics. 2008;178:2253–2264. doi: 10.1534/genetics.107.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng N, Jin B, Wang Q, et al. No detectable maternal effects of elevated CO2 on Arabidopsis thaliana over 15 generations. PLoS ONE. 2009;4:e6035. doi: 10.1371/journal.pone.0006035. doi:10.1371/journal.pone.0006035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisné S, Reymond M, Vile D, et al. Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis. Plant Physiology. 2008;148:1117–1127. doi: 10.1104/pp.108.124271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. Mechanism of leaf-shape determination. Annual Review of Plant Biology. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. Controlling size in multicellular organs: focus on the leaf. PLoS Biology. 2008;6:e174. doi: 10.1371/journal.pbio.0060174. doi:10.1371/journal.pbio.0060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Shoda K, Kim GT, Uchimiya H. Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta. 2000;210:536–542. doi: 10.1007/s004250050042. [DOI] [PubMed] [Google Scholar]
- Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biology. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. doi:10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward FI, Lake JA, Quick WP. Stomatal development and CO2: ecological consequences. New Phytologist. 2002;153:477–484. doi: 10.1046/j.0028-646X.2001.00338.x. [DOI] [PubMed] [Google Scholar]
- van Zanten M, Snoek LB, Van Eck-Stouten E, et al. Ethylene-induced hyponastic growth in Arabidopsis thaliana is controlled by ERECTA. The Plant Journal. 2010;61:83–95. doi: 10.1111/j.1365-313X.2009.04035.x. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical analysis. Upper Saddle River, NJ: Prentice-Hall; 1996. [Google Scholar]
- Zhang L, Hu G, Cheng Y, Huang J. Heterotrimeric G protein α and β subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Developmental Biology. 2008;324:68–75. doi: 10.1016/j.ydbio.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Zhao L, Sack FD. Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. American Journal of Botany. 1999;86:929–939. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.