Summary
Argonaute (AGO) proteins are critical components of RNA silencing pathways that bind small RNAs and mediate gene silencing at their target sites. We found that Arabidopsis AGO2 is highly induced by the bacterial pathogen Pseudomonas syringae pv. tomato (Pst). Further genetic analysis demonstrated that AGO2 functions in antibacterial immunity. One abundant species of AGO2-bound small RNAs is miR393b*, which targets a Golgi-localized SNARE gene MEMB12. Pst infection down-regulates MEMB12 in a miR393b*-dependent manner. Loss-of-function of MEMB12 but not SYP61, another intracellular SNARE, leads to increased exocytosis of an antimicrobial pathogenesis-related protein PR1. Overexpression of miR393b* resembles memb12 mutant in resistance responses. Thus, AGO2 functions in antibacterial immunity by binding miR393b* to modulate exocytosis of antimicrobial PR proteins via MEMB12. Since miR393 also contributes to antibacterial responses, miR393*/miR393 represent an example of a miRNA*/miRNA pair that functions in immunity through two distinct AGOs - miR393* through AGO2 while miR393 through AGO1.
Introduction
Small RNA (sRNA)-mediated gene silencing has been recognized as an important regulatory mechanism in host immune responses of both plants and animals (Ding, 2010; Padmanabhan et al., 2009; Wessner et al., 2010). Plants have evolved multiple levels of immune responses, including basal defense triggered by virulent pathogens in susceptible hosts and resistance (R) gene-mediated resistance activated by avirulent pathogens in resistant hosts. Conserved microbial- or pathogen-associated molecular patterns (MAMPs or PAMPs) are recognized by host pattern recognition receptors and activate PAMP-triggered immunity (PTI), whereas pathogen-derived effector proteins, which can attenuate PTI, are recognized by host R proteins and trigger effector-triggered immunity (ETI) (Chisholm et al., 2006; Jones and Dangl, 2006). In Arabidopsis, microRNA393 (miR393) negatively regulates auxin signaling pathways and contributes to PTI (Navarro et al., 2006). Endogenous small interfering RNAs (siRNAs), nat-siRNAATGB2 and AtlsiRNA-1, are induced by the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 carrying an effector gene avrRpt2; these siRNAs play an important role in ETI by targeting negative regulators of the cognate resistance gene RPS2 signaling pathway (Katiyar-Agarwal et al., 2007; Katiyar-Agarwal et al., 2006).
sRNAs are loaded into Argonaute proteins (AGOs) and silence targets with complementary sequences. Many eukaryotes have evolved functionally diversified AGOs that regulate many cellular processes (Hutvagner and Simard, 2008; Mallory and Vaucheret, 2010). Specific AGOs from Drosophila, Arabidopsis, C. elegans, and Cryphonectria parasitica are essential for antiviral defenses by binding viral sRNAs and silencing viral genomes (Ding, 2010; Harvey et al., 2011). However, function of AGOs in antibacterial immunity is less clear. Arabidopsis encodes 10 AGO (Vaucheret, 2008). AGO1 is primarily associated with miRNAs and mainly regulates plant development and stress adaptations (Mallory and Vaucheret, 2010). AGO1 also contributes to PTI through several stress-related miRNAs (Li et al., 2010b). AGO7 mainly binds miR390 and triggers the generation of trans-acting siRNAs (Montgomery et al., 2008); it is required for the accumulation of AtlsiRNA-1 and contributes to ETI (Katiyar-Agarwal et al., 2007; Li et al., 2010b). AGO4, AGO6 and AGO9 predominantly bind heterochromatic siRNAs from different loci and function in RNA-directed DNA methylation (RdDM) (Havecker et al., 2010; Mallory and Vaucheret, 2010). AGO4 has been linked to antibacterial defense (Agorio and Vera, 2007). Whether these activities of AGO4 involve sRNAs or RdDM, however, is unknown because none of the other factors involved in the RdDM pathway has effects on antibacterial immunity.
To understand the differentiated function of Arabidopsis AGOs in plant immunity, we examined the expression of all the AGO genes in response to bacterial infection. We show that AGO2 is highly induced by Pst and that AGO2 mutation attenuates antibacterial immunity. Profiling of AGO2-bound sRNAs by high-throughput sequencing revealed that one of the most abundant sRNAs is miR393b*, which targets a gene encoding a Golgi-localized SDS-resistant soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) protein, MEMB12. Mutation in MEMB12 but not SYP61, another intracellular SNARE gene, leads to increased exocytosis of the antimicrobial pathogenesis-related protein PR1. miR393 is known to contribute to PTI. Here, we show that miR393* also contributes to immunity, mainly ETI, by modulating secretion of PR1. Thus, we provide an example of a functional pair of miRNA* and miRNA, each of which targets different regulators in host innate immunity through two distinct AGOs - miR393* through AGO2 while miR393 through AGO1.
Results
Arabidopsis AGO2 is highly induced by bacterial infection and plays an important role in innate immune responses
To further our understanding of the differentiated function of AGOs in antibacterial immunity, we first examined the expression of all AGO genes in response to the infection of Pst DC3000. Transcripts of AGO2, but no other AGOs, were induced by the virulent strain of Pst carrying an empty vector (EV). This induction was even more pronounced in response to an avirulent strain Pst (avrRpt2) at 6 and 14 hours post inoculation (hpi) (Fig. 1A and S1A). Western blot analysis confirmed that AGO2 protein level was also highly induced by both Pst (EV) and Pst (avrRpt2) (Fig. 1B).
Figure 1. AGO2 is highly induced by Pst and mutation in AGO2 attenuates plant resistance to both virulent and avirulent strains of Pst.
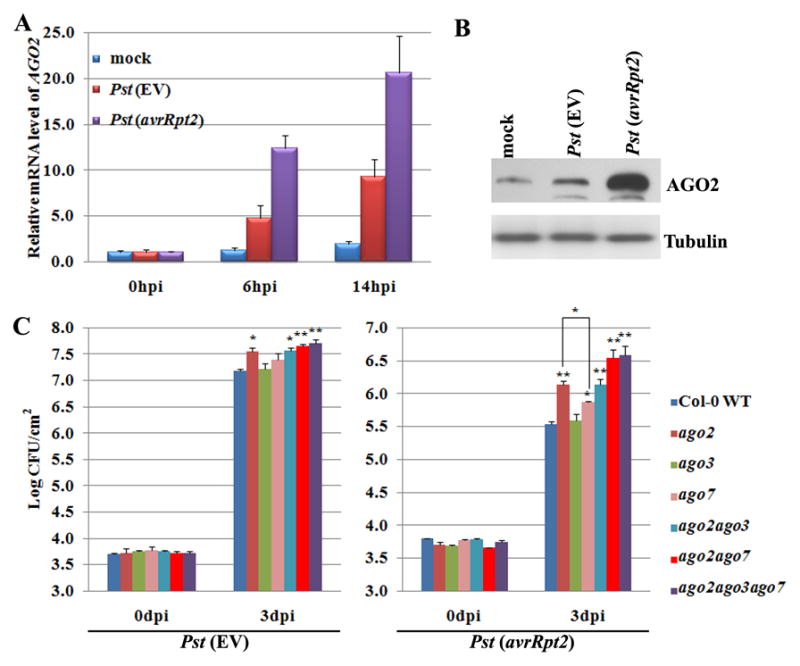
(A) AGO2 transcript was highly induced by Pst (EV) and Pst (avrRpt2). Sample leaves were collected at 6 and 14 hpi. Relative transcript levels were measured by real-time RT-PCR. Actin2 was used as an internal control. Standard deviations were calculated from three technical replicates. Similar results were obtained from three biological replicates. (B) Western blot analysis of AGO2 in response to Pst (EV) and Pst (avrRpt2) infection. The samples were collected at 14 hpi and AGO2 was detected by an peptide antibody (Harvey et al., 2011). Tubulin was used as a control. Similar results were observed from two replicates. (C) ago2 single, double, and triple mutants were more susceptible to Pst (EV) and Pst (avrRpt2) than WT plants. Four-week-old plants were infiltrated with Pst (EV) and Pst (avrRpt2) (5 × 105 cfu/ml). Error bars represent standard deviation of six leaf discs. Student’s t-test was performed to determine the significant differences between mutants and WT plants. Asterisks, “*” and “**”, indicate statically significant differences at a p-value of < 0.05 and <0.01, respectively. Similar results were obtained from three biological replicates. See figure S1 for expression levels of other Arabidopsis AGOs, characterization of ago2 mutants, and miR403 expression analysis.
To determine whether AGO2 regulates immune responses against bacterial infection, we carried out a bacterial growth assay on a loss-of-function mutant ago2-1 (Lobbes et al., 2006). ago2-1 displayed enhanced susceptibility to Pst (avrRpt2) and supported 5- to 6-fold more bacterial growth than wild type (WT) Col-0 at 3 days post inoculation (dpi) (Fig. 1C). It also consistently showed moderately enhanced susceptibility to Pst (EV) permitting 3- to 4-fold more bacterial growth than WT (Fig. 1C). Of the 10 Arabidopsis AGOs, AGO2 is closely related to AGO3 and AGO7. Consistent with previous reports (Katiyar-Agarwal et al., 2007; Li et al., 2010b), an ago7 mutant (zip-1 (Hunter et al., 2003)) was more susceptible to Pst (avrRpt2) but not to Pst (EV) (Fig. 1C). However, the effect of ago7 was weaker than that of ago2-1 (p<0.05). No alteration in bacteria growth was observed in ago3-1 (Fig. 1C) (Lobbes et al., 2006).
To test whether AGO2, AGO3, and AGO7 are partially redundant, double and triple mutants were generated. ago2ago7 was generated by crossing the relevant single mutants, ago2ago3 and ago2ago3ago7 were obtained by generating transgenic AGO3 RNAi lines in the background of ago2-1 and ago2ago7, respectively. Since AGO2 (At1g31280) and AGO3 (At1g31290) are closely linked on chromosome 1, it is impossible to obtain an ago2ago3 double mutant by crossing the single mutants. The transgenic AGO3 RNAi lines with the lowest AGO3 expression level were selected for functional analysis (Fig. S1B–C). ago2ago7 and ago2ago3ago7 behaved very similar, and showed more susceptibility to Pst (avrRpt2) than ago2 or ago7 single mutants, allowing 9- to 10-fold more bacterial growth than WT plants (Fig. 1C), confirming that both AGO2 and AGO7 contribute to ETI. Bacterial growth in ago2ago3 was similar to that in ago2, which confirmed that ago3 has little effect on the resistance (Fig. 1C). Pst (EV) grew to a similar level in the ago2 single, double, and triple mutants, suggesting that only AGO2 is involved in basal defense. The precise definition of basal defense is PTI plus weak ETI, minus effector-triggered susceptibility (Jones and Dangl, 2006). To determine whether AGO2 plays a role in PTI, we performed flg22-mediated protection assay (Zipfel et al., 2004). Inhibition of Pst (EV) growth by flg22 pretreatment was observed in both ago2 and WT plants (Fig. S1D). Flg22-treatment also caused growth inhibition of the ago2 mutant to a similar level as with WT (Fig. S1E). Furthermore, we still detected clear induction of AGO2 in response to Pst (EV) infection in bak1 mutant (Fig. S1F). BAK1 is a common signaling component in PTI. These data suggest that AGO2 may play a minor role, if any, in PTI.
The bacterial effector avrRpt2 is recognized by its cognate R protein RPS2 in Col-0 and triggers local cell death, which is referred to as the hypersensitive response (HR). Under the conditions we used, visible HR occurred at 15 hpi in WT plants, but not in ago2ago7 and ago2ago3ago7 (Fig. S1G). A delayed HR (at 17-18 hpi) to Pst (avrRpt2) was observed in ago2ago7 and ago2ago3ago7. Taken together, our results indicate that AGO2 plays an important role in plant innate immunity, especially in ETI against bacterial pathogens.
miR393b* is highly enriched in AGO2
We hypothesized that, as a critical component of the RNAi pathway, AGO2 regulates plant immunity through the action of its bound sRNAs. Therefore, we used Illumina deep sequencing to analyze the AGO2-associated sRNA population after Pst (avrRpt2) and mock treatments using a transgenic pAGO2:HA-AGO2 line (Montgomery et al., 2008). The dataset was deposited into NCBI (GSE26161). AGO1-associated sRNA libraries prepared under the same conditions were used as controls. Consistent with previous reports (Mi et al., 2008; Montgomery et al., 2008), AGO2-associated sRNAs were primarily 21 nt in length and had a bias for reads with the 5′-terminal A, whereas AGO1-bound sRNAs were mainly 21-nt miRNAs with 5′-terminal U. Strikingly, among the most abundant AGO2-bound sRNAs (Table 1 and S1), several miRNA*s, including miR165a*, miR393b*, miR396b*, and miR472*, had more than 1000 reads per million genome-matched sequences (mgs) (Table 1). miRNA*s are sRNA species that pair with corresponding miRNAs within the hairpin structure of the miRNA precursors, and are processed into miRNA:miRNA* duplexes by Dicer or Dicer-like proteins. They were often considered as non-functional byproducts because they are quickly degraded and are much less abundant than their corresponding miRNAs. However, we observed significant enrichment of several of these miRNA*s in the AGO2-immunoprecipitated (IP) fraction, suggesting that they may be biologically functional.
Table 1.
miRNA*s are highly enriched in the AGO2-IP fraction. Normalized miRNA* and miRNA (reads/mgs) from each sRNA library are listed. Total sRNAs without IP from our previous dataset (Chellappan et al., 2010; Zhang et al., 2010) are included as controls. AGO2-associated miRNA*s with more than 1000 reads/million are shown. See Table S1 for the full list of miRNA and miRNA* from the AGO-IP deep sequencing dataset.
| AGO2-IP
|
AGO1-IP
|
Total sRNA without IP
|
||||
|---|---|---|---|---|---|---|
| mock | Pst (avrRpt2) | mock | Pst (avrRpt2) | mock | Pst (avrRpt2) | |
| miR393b* | 2621.9 | 4546.6 | 0.3 | 24.4 | 36.6 | 89.6 |
| miR393 | 0.4 | 0.5 | 358.1 | 156.6 | 11.7 | 75.3 |
| miR165a* | 7943.4 | 15529.2 | 1.0 | 0.9 | 146.2 | 531.5 |
| miR165a | 0.3 | 6.3 | 30890.4 | 23823.2 | 300.3 | 177.2 |
| miR396b* | 1196.9 | 2864.6 | 188.2 | 214.6 | 76.4 | 10.2 |
| miR396b | 26.3 | 28.4 | 4121.9 | 6173.0 | 214.8 | 42.8 |
| miR472* | 508.7 | 1273.7 | 28.1 | 36.4 | 146.2 | 531.5 |
| miR472 | 0.9 | 2.0 | 246.9 | 61.5 | 300.3 | 177.2 |
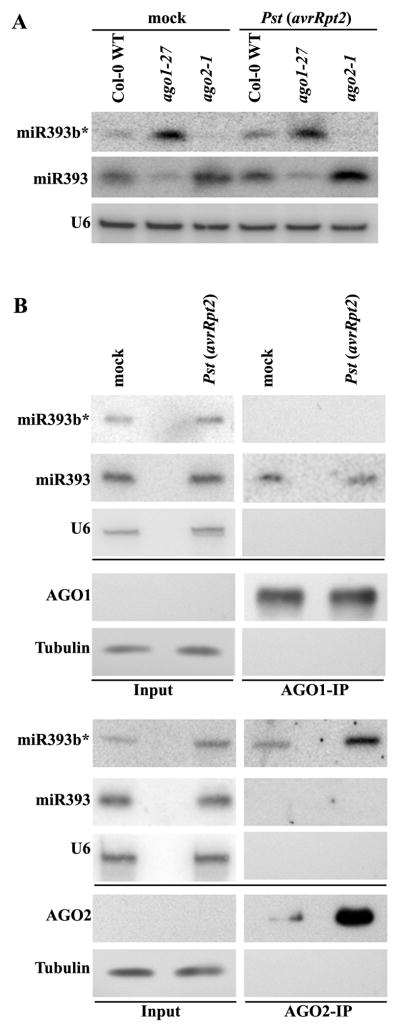
One of the most abundant miRNA*s that associated with AGO2 was miR393b*, which had more than 4500 reads/mgs in the AGO2-IP fraction after Pst (avrRpt2) treatment, but had only 24 reads/mgs in the AGO1-IP faction (Table 1). Interestingly, its corresponding miR393 was only present in the AGO1-IP but not AGO2-IP fraction (Table 1). miR393 has been shown to contribute to PTI by silencing auxin receptors (Navarro et al., 2006). Therefore, we were very interested in testing whether miR393b* has a regulatory role like its miRNA partner. Mature miR393a and miR393b have the same sequence, but miR393a* and miR393b* differ in one nucleotide and miR393b* is much more abundant than miR393a* (4546 versus 25 reads/mgs in the Pst (avrRpt2)-treated AGO2-IP libraries) (Table S1). To confirm that miR393b* and miR393 were incorporated into two different AGO proteins (AGO2 and AGO1, respectively), we performed sRNA Northern blot analysis using total RNA from ago mutants and sRNA fractions from the AGO-IPs. Expression of miR393b* was almost abolished in ago2-1, while its corresponding miR393 was reduced in ago1-27, a relatively weak allele (Fig. 2A) (Morel et al., 2002). miR393b* was detected only in AGO2- but not AGO1-IP fractions, whereas miR393 appeared only in AGO1- but not AGO2-IP fractions (Fig. 2B). These results confirmed that miR393b* and miR393 were bound to distinct AGOs. The level of miR393b* associated with AGO2 was significantly increased after Pst (avrRpt2) infection (Fig. 2B), most likely due to the induction of AGO2. These results strongly suggest that miR393b* is functional.
Figure 2.
miR393b* is associated with AGO2 whereas miR393 is associated with AGO1. Accumulation of miRNA393b* and miRNA393 in Col-0 WT, ago1-27, and ago2-1 mutants (A) or in AGO1-IP and AGO2-IP (B) was detected by miRNA393b* and miRNA393 probes. mock- or Pst (avrRpt2)-treated leaves (AGO2::3HA-AGO2) were collected at 12 hpi. 40 μg of total RNA was used for each genotype in (A). For RNA IP, cell lysate from 0.15g of tissue was incubated with AGO1 antibody or HA antibody for pulling down AGO1- and AGO2-fraction, respectively (See methods for details). Total RNA (without IP procedures) equivalents to 0.03g plant tissue was loaded as the input control. The IP efficiency was examined by detecting AGO1 and AGO proteins in the IP fractions (from 0.15g plant tissue) using AGO1 antibody and HA antibody, respectively. 30 μl original homogenized tissue (corresponding to 0.005 g) were loaded in the input panels. Tubulin was used as a loading control. Similar results were obtained in three biological replicates. See Table S1 for the rest of miRNA*s and miRNAs from the AGO pull-down sequencing dataset.
miR393b* targets MEMB12 encoding a SNARE protein
Recent studies in animals suggested that some miRNA*s may have regulatory roles (Ghildiyal et al., 2010; Guo and Lu, 2010; Okamura et al., 2008). However, in vivo functional analyses of these miRNA*s have not been reported. We proposed that miR393b* regulates plant immunity by targeting genes involved in plant defense responses. Therefore, we predicted potential targets of miR393b* (Table S2). Three of the putative targets, At5g50440, At4g19490, and At3g09530, have predicted roles in protein trafficking and secretion. At5g50440 encodes MEMB12, a Golgi-localized SNARE protein (Uemura et al., 2004), but its biological function has not been determined. At4g19490 encodes a Golgi/post-Golgi compartment localized protein VPS54, which is the homologue of a subunit of yeast Golgi-associated retrograde protein (GARP)/Vps Fifty Three (VFT) complex involved in retrograde transport from the vacuolar/late endosome compartment to the Golgi apparatus (Conibear and Stevens, 2000; Guermonprez et al., 2008). At3g09530 encodes EXO70H3, which belongs to the large EXO70 gene family and is a subunit of the exocyst complex predicted to be responsible for exocytosis (Li et al., 2010a). The expression of EXO70H3 in leaves is below the detection limit (Li et al., 2010a). Several plasma membrane (PM)-associated proteins involved in cell surface trafficking have been shown to be essential in plant immune responses, which indicates the importance of protein trafficking in plant defense (Bednarek et al., 2010; Collins et al., 2003; Kalde et al., 2007; Kwon et al., 2008). However, proteins responsible for intracellular vesicle transport and the early steps of protein secretion have not been identified in host immune responses. Two of the putative targets, MEMB12 and VPS54, are localized to the Golgi apparatus and have a predicted function in vesicle transport. Mutations in VPS54 and other GARP subunits display a transmission defect through the male gametophyte and could not yield homozygous mutants (Guermonprez et al., 2008), making it very difficult to test the role of VPS54 in plant defense. Therefore, in this study we focused on the characterization of MEMB12—a Golgi-localized SNARE protein—in plant immune responses.
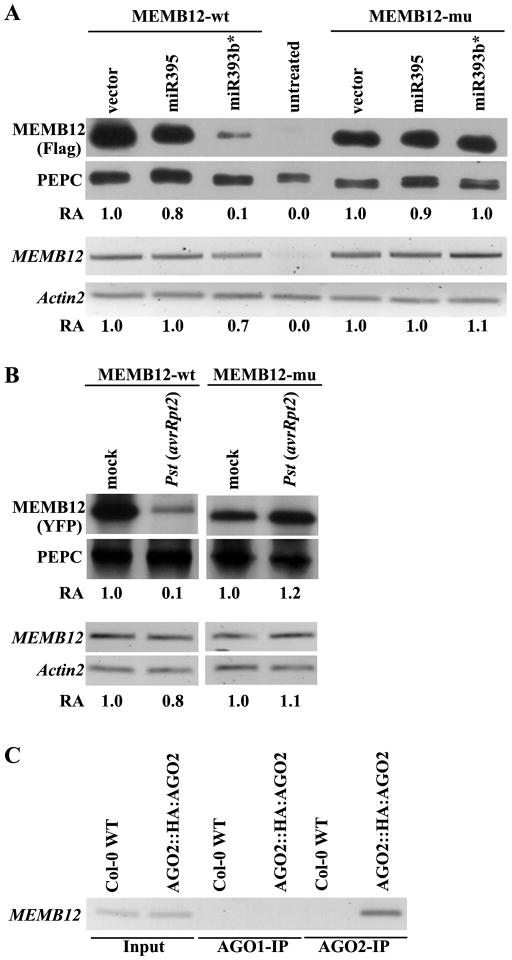
To test experimentally whether MEMB12 is a real target of miR393b*, we first performed Agrobacterium-mediated transient co-expression assays in Nicotiana benthamiana. A binary construct encoding the Flag-tagged MEMB12 with the wild-type miR393b* target site (Flag-MEMB12-wt) was co-expressed with miR393b* or miR395, a miRNA that cannot target MEMB12. The MEMB12 protein level was down-regulated by miR393b* but not miR395 (Fig. 3A). This down-regulation was abolished when the miR393b* target site in MEMB12 was mutated (Flag-MEMB12-mu, with no amino acid change) (Fig. 3A & S2). These results indicate that miR393b* is responsible for the down-regulation of MEMB12. The protein level of MEMB12 was significantly reduced while its transcript level was only reduced slightly, suggesting that miR393b* silences MEMB12 mainly by translational inhibition. We also generated transgenic Arabidopsis carrying YFP-MEMB12-wt or YFP-MEMB12-mu to test the regulatory role of miR393b* in vivo. MEMB12-wt protein was down-regulated after Pst (avrRpt2) treatment, whereas MEMB12-mu was not (Fig. 3B). The mRNA level of MEMB12-wt was not significantly reduced after infection of Pst (avrRpt2) (Fig. 3B), consistent with the notion that miR393b* mainly mediates translational inhibition rather than mRNA degradation. If MEMB12 is the target of miR393b*, MEMB12 transcripts should also be associated with AGO2, which is the AGO predominantly associated with miR393b*. Indeed, MEMB12 mRNA was enriched in the AGO2-IP fraction but was not detected in the AGO1-IP fraction (Fig. 3C). These results supported that MEMB12 is a real target of miR393b*.
Figure 3.
MEMB12 is a target of miR393b*. (A) miR393b* down-regulates MEMB12 in a transient co-expression assay in N. benthamiana. The Flag-tagged expression construct of MEMB12-wt or MEMB12-mu was co-expressed with miR393b* or miR395 in N.benthamiana. The protein levels (the upper panel) were detected by an anti-FLAG antibody. Phosphoenolpyruvate carboxylase (PEPC) was used as a loading control for measuring the relative abundance (RA) (shown below) using the ImageQuant software. The transcript levels of MEMB12 from the same treatment were measured by semi-quantitative RT-PCR (bottom panel). Actin2 was used for measuring the RA (shown below). Similar results were obtained in two biological replicates. (B) miR393b* down-regulates MEMB12 in stable transgenic Arabidopsis. Transgenic Arabidopsis carrying MEMB12-wt and MEMB12-mu (35S::YFP:MEMB12) were generated. The protein level of MEMB12 in the selected line after mock- or Pst (avrRpt2)-treatment was detected by an anti-GFP antibody. The transcript level of MEMB12 was measured as in A. The experiments were repeated three times and similar results were observed. (C) MEMB12 mRNA was co-immunoprecipitated with AGO2 but not AGO1. RNAs from input, AGO1-IP, and AGO2-IP fractions from both transgenic (AGO2::3HA-AGO2) and Col-0 WT plants were reverse transcribed and amplified using MEMB12 specific primers. The PCR bands were gel purified and confirmed by sequencing. Similar results were observed in two independent experiments. See Figure S2 for the alignment between miR393b* and MEMB12-wt and MEMB12-mu.
Loss-of-function of MEMB12 promotes secretion of PR1
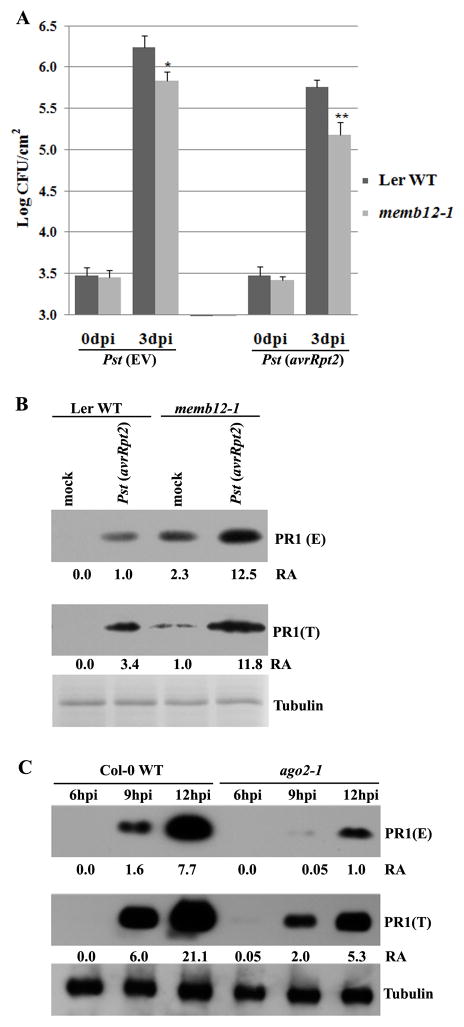
To test whether MEMB12 plays a role in antibacterial defense, we isolated the MEMB12 knockout mutant memb12-1 (GT22391), which is a transposon-tagged line with an insertion in the first exon (Fig. S3A-C) (Springer et al., 1995). memb12-1 had no obvious developmental defects, but displayed enhanced resistance to both avirulent and virulent strains of Pst. Growth of Pst (avrRpt2) in memb12-1 was reduced 5 to 6-fold compared with the wild-type control (Fig. 4A). memb12-1 also showed enhanced resistance to Pst (EV), although to a lesser degree (Fig. 4A).
Figure 4.
Mutation in MEMB12 leads to enhanced resistance to both Pst (EV) and Pst (avrRpt2) and promotes PR1 secretion. (A) Growth of Pst (EV) and Pst (avrRpt2) was measured in four-week-old memb12-1 and corresponding Ler WT as described in figure 1C. Error bars represent standard deviation of six leaf discs (*: p<0.05; **: p<0.01). Similar results were obtained in three biological replicates. (B) memb12-1 promotes PR1 secretion. The level of PR1 in total (T; whole leaf) and secreted proteins (E; intercellular wash fluid) from mock- and Pst (avrRpt2)-treated Ler WT and memb12-1 plants was detected by Western blot analysis. Equal volume of intercellular wash fluid was loaded for detecting secreted PR1. Tubulin was used as an equal loading control and RA measurement for detecting total PR1 protein. Similar results were observed from 4 biological repeats. (C) Secretion of PR1 was reduced and delayed in ago2-1 mutant plants. The total (T) and secreted (E) PR1 were compared between Col-0 WT and ago2 plants at 6, 9 and 12 hpi of Pst (avrRpt2). Similar results were obtained from 3 biological repeats (Fig. S3). See Fig S3 for the characterization of memb12-1 mutant.
To understand the mechanism of function of MEMB12 in innate immunity, we analyzed the localization of MEMB12 by examining the transgenic YFP-MEMB12-wt plants. We observed punctate structures in the cytosol (Fig. S3D), which is the characteristic feature of the plant Golgi apparatus (Chatre et al., 2005; Matheson et al., 2006). This observation is in accordance with previously reported MEMB12 localization to the Golgi (Geldner et al., 2009; Uemura et al., 2004). Secretion of antimicrobial proteins or small molecules is essential for effective defense against pathogens (Bednarek et al., 2010; Collins et al., 2003; Kalde et al., 2007; Kwon et al., 2008; Nomura et al., 2006). To examine whether MEMB12 suppresses defense responses by affecting protein secretion, we examined the level of one of the major secreted antimicrobial pathogenesis-related proteins—PR1—in the intercellular wash fluid of memb12-1 before and after bacterial infection as described (Wang et al., 2005a). After Pst (avrRpt2) infection, the level of intercellular secreted PR1 was more than 10-fold greater in memb12-1 than in WT (Fig. 4B), while total PR1 protein only increased by about 3-4 fold in memb12-1 compared with that in WT (Fig. 4B). These results indicate that the secretion of PR1 in memb12-1 is indeed increased. Secreted PR1 also accumulated in the untreated memb12-1 (Fig. 4B). Furthermore, the secretion of PR1 upon Pst (avrRpt2) infection was significantly reduced and delayed in the ago2-1 mutant (Fig. 4C and S3E-F), which is consistent with the result that ago2 was more susceptible to Pst (avrRpt2) (Fig. 1C).
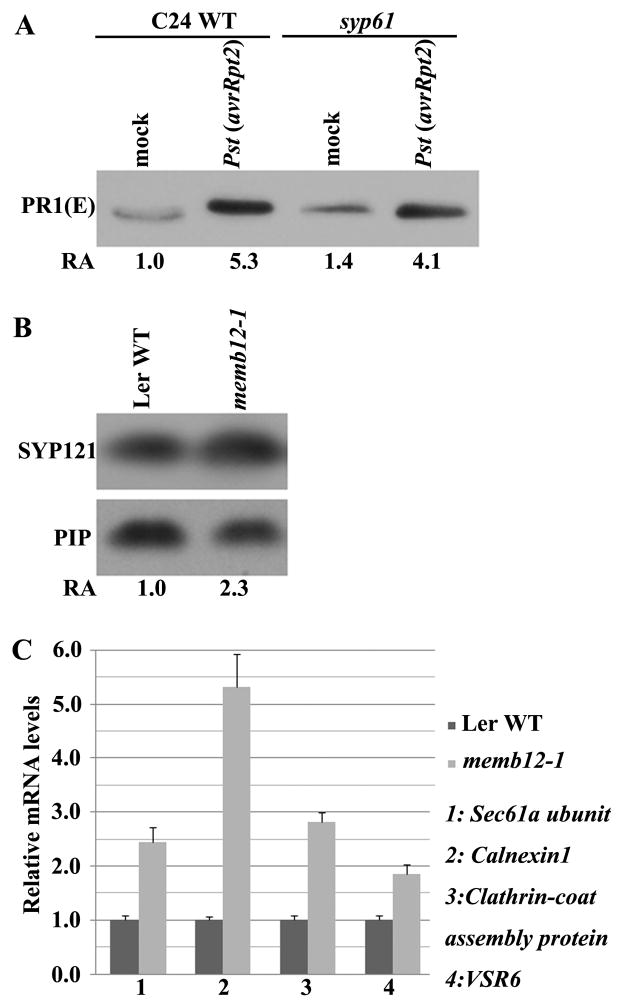
On the contrary, mutation in the trans-Golgi-network (TGN)/endosomal SNARE SYP61 gene did not alter the secretion level of PR1 (Fig. 5A) (Robert et al., 2008; Sanderfoot et al., 2001; Sanmartin et al., 2007; Zhu et al., 2002). These results suggest that intracellular SNARE MEMB12 but not SYP61 specifically controls the exocytosis of vesicles containing antimicrobial protein PR1.
Figure 5.
MEMB12 is likely to regulate secretion of PR proteins in a rather specific manner. (A) Mutation of another intracellular SNARE gene SYP61 does not interfere with PR1 secretion. Secreted proteins were isolated from syp61 and the corresponding C24 WT and detected as in Fig. 4B. Experiments were performed two times with similar results. (B) MEMB12 has weak effect on secretion of a PM-localized SNARE protein SYP121. PM proteins were fractionated from four-week-old mock- and Pst (avrRpt2)-treated tissue collected at 12 hpi. SYP121 was detected by SYP121 antibody (Collins et al., 2003). PIP-28RD (AT2G37180) was used as a loading control. Similar results were obtained from three biological repeats. (C) Genes involved in protein folding and secretory pathways were coordinately up-regulated in memb12-1. The transcript levels of some reported secretory-related genes (Wang et al., 2005) were detected by real-time RT-PCR. Expression levels in Ler WT plants were assigned as 1.0. Error bars indicate standard deviation from three technical repeats. Similar results were obtained in two biological replicates. See Table S3 for detailed primer and probe sequences.
To determine whether suppression of MEMB12 interferes with general protein secretion, we also examined the level of another secreted protein – SYP121, a PM-associated syntaxin required for resistance to powdery mildew fungus (Collins et al., 2003). The level of SYP121 was slightly increased in the PM factions in memb12-1 as compared with WT plants (Fig. 5B). Mutation in MEMB12 has a much smaller effect on SYP121 secretion than on PR1 secretion, suggesting that MEMB12 is responsible for trafficking of a specific group of proteins, such as PR1.
When massive accumulation and secretion of PR proteins occur during defense responses, a coordinated up-regulation of the whole protein secretory machinery is often accompanied to ensure efficient transport (Wang et al., 2005b). Indeed, we detected up-regulation in memb12-1 of several secretion pathway genes that encode translocon complex Sec61 alpha subunit, co-chaperone Calnexin 1, a Clathrin-coat assembly protein, and a Vacuolar sorting receptor VSR6; up-regulation occurred even without pathogen challenges (Fig. 5C), which suggests that suppression of MEMB12 leads to an up-regulation of the secretory pathway for transporting certain proteins. Taken together, our results suggest that, miR393b* contributes to AGO2-regulated innate immunity by suppressing MEMB12 and subsequently promoting effective secretion of antimicrobial PR proteins.
miR393b* overexpression plants resemble memb12 in disease resistance responses
To determine whether overexpression of miR393b* results in a phenotype similar to the loss-of-function mutant of its target MEMB12, we generated transgenic plants overexpressing the MIR393b gene controlled by the constitutive CaMV 35S promoter and selected the lines that expressed high level of miR393b* for bacterial growth assay and PR1 secretion analysis (Fig. S4A). These plants showed enhanced disease resistance to Pst (avrRpt2) (Fig. 6A) as well as increased PR1 protein accumulation (Fig 6B), which nicely resembled the phenotype of memb12-1 in resistance responses (Fig. 4). However, in these MIR393b overexpression plants, miR393 was also produced from the same MIR393b precursor (Fig S4A), which was evidenced by the curly leaf phenotype that reminiscent of the double and triple mutants of the miR393 targets - auxin receptors, tir1/afb2 and tir1/afb2/afb3 (Fig. 6C and Fig. 5I and 5J in (Dharmasiri et al., 2005)). Plants overexpressing MIR393a show enhanced resistance to virulent strain Pst DC3000 but has no obvious effect on Pst (avrRpt2)-triggered ETI (Navarro et al., 2006). Because MIR393a and MIR393b give rise to the same miR393, the positive regulatory effect of MIR393b on Pst (avrRpt2)-triggered ETI is due to the generation of miR393b*. However, it is impossible to tell whether the increased secretion of PR1 is due to the overexpression of miR393b* or miR393 in our MIR393b overexpression lines.
Figure 6.
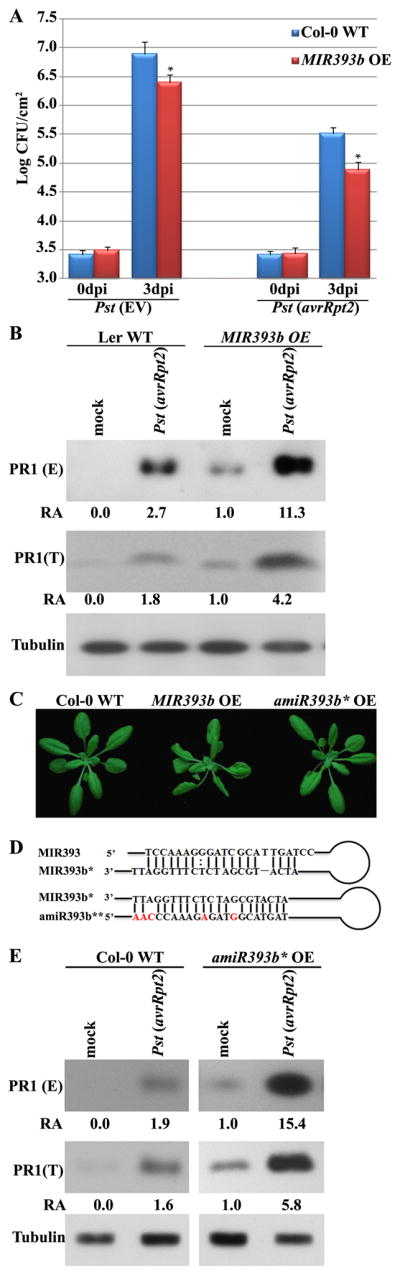
Overexpression of miR393b* resembles memb12-1 in disease resistance responses. (A) MIR393b overexpression plants (MIR393b OE) were more susceptible to both Pst (EV) and Pst (avrRpt2) than WT plants. Pathogen growth was measured as in figure 1C. Error bars represent standard deviation of six leaf discs (*: p<0.05). Similar results were obtained from three biological replicates. (B) Overexpression of MIR393b promotes PR1 secretion. The level of PR1 was measured as in Fig. 4B. Three biological repeats yielded similar results. (C) Developmental phenotypes of transgenic plants overexpressing MIR393b (middle) and amiR393b* (amiR393b* OE, right). Pictures were taken from three-week-old plants. (D) Sequences of miR393/393b* and miR393b*/393b** for generating transgenic MIR393b and amiR393b* plants. The nucleotides that are different between miR393 and miR393b** are in red. (E) amiR393b* transgenic plants show enhanced PR1 secretion. Experiments were repeated twice with similar results. See figure S4 for the characterization of MIR393b and amiR393b* transgenic plants.
To distinguish the effects of miR393b* from that of miR393 on PR1 secretion, we used Web microRNA Designer and made an artificial miR393b* (amiR393b*) construct that produced miR393b* and the reverse complementary strand of miR393b* (referred to as miR393b**), which contains 5 different nucleotides from miR393 sequences (Fig. 6D) (Schwab et al., 2006). The amiR393b* transgenic plants with high expression level of miR393b* were selected and no longer displayed the phenotype of altered auxin signaling (Fig. 6C and Fig. S4B). Thus, we succeeded in uncoupling the effect of miR393b* and miR393 in these plants. We still observed increased PR1 secretion in these transgenic lines expressing artificial miR393b* (Fig. 6E), which resembled the phenotype of memb12. These results support that MEMB12 is indeed one of the major targets of miR393b* and regulates PR1 secretion.
Discussion
miRNA* was once considered as a useless byproduct of miRNA biogenesis (Jones-Rhoades et al., 2006; Schwarz et al., 2003). Here, we identified a functional miRNA* that regulates plant immunity along with its cognate miRNA partner. Thus, miRNA genes have the potential to generate more than one functional sRNA under certain conditions. miRNA393* and miRNA393 represent an example of a pair of miRNA* and miRNA that function through different AGOs in vivo and regulate two different cellular pathways in innate immunity. miR393* is loaded into AGO2 and targets genes involved in protein trafficking and regulates plant immune responses, mainly ETI, by promoting exocytosis of PR proteins; whereas miR393 is loaded into AGO1 and contributes to PTI by targeting auxin receptors and suppressing the auxin signaling pathway.
Strand selection of miRNA/miRNA* duplex within AGO complexes is largely determined by asymmetrical thermodynamic stability of the duplex termini (Khvorova et al., 2003; Schwarz et al., 2003). The strand with less stability at its 5′-terminus is incorporated as the guide strand (miRNA), while the other strand (so-called passenger strand or miRNA*) is mostly degraded. However, here we show that different AGO proteins select different guide strand within the same miRNA/miRNA* duplex regardless of their thermodynamic stability. sRNA loading into AGOs in plants is known to conform to the “5′ first nucleotide recognition” model (Mi et al., 2008; Montgomery et al., 2008), in which different AGOs preferentially associate with sRNAs with distinct 5′ first nucleotides. For example, AGO1 prefers uridine (U), AGO2 and AGO4 prefer adenosine (A), and AGO5 prefers cytosine (C). The fact that miR393 and miR393* start with 5′-terminal U and A, respectively, fits this model well. However, the other miRNA*s that were highly enriched in AGO2-IP fraction do not have 5′-terminal A (Table 1), such as miR165a* and miR396b*, which have 5′-terminal G. On the contrary, miR390, which features a 5′-terminal A, binds predominantly to AGO7 but not AGO2 (Montgomery et al., 2008). Thus, there must be other factors that help determine sRNA loading. Although all Arabidopsis AGO proteins contain the three conserved functional PAZ, MID and PIWI domains, they have very different N-terminal regions. Even within the conserved domains, there are marked differences. For example, AGO2 and AGO3 are the only Arabidopsis AGOs that contain an Asp-Asp-Asp (DDD) motif instead of the conventional Asp-Asp-His (DDH) motif within the PIWI domain. This DDD motif is similar to that in bacterial RNaseH enzymes with cleavage activity (Nowotny et al., 2005; Vaucheret, 2008). It is likely that these sequence differences among AGO proteins may allow their interactions with different co-factors and direct distinct sRNA loading. The finding that the double-stranded RNA binding protein DRB1 assists AGO1-loading of some miRNAs supports the idea that additional components are required for determination of sRNA binding (Eamens et al., 2009).
Protein secretory systems appear to play an important role in plant defense (Bednarek et al., 2010; Collins et al., 2003; Kalde et al., 2007; Kwon et al., 2008; Nomura et al., 2006). The SNARE proteins that have been identified to be involved in immune responses are mostly associated with the PM (Bednarek et al., 2010; Collins et al., 2003; Kalde et al., 2007; Kwon et al., 2008). Here, we identified a previously uncharacterized intracellular SNARE, MEMB12, as a miR393b* target. Upon bacterial infection, suppression of MEMB12 by miR393b* promotes the secretion and accumulation of PR1 protein and contributes to resistance. MEMB12 is mainly localized to the Golgi and mediates protein trafficking between the Golgi and the ER (Uemura et al., 2004). Our results suggest that MEMB12 may be responsible for the retrograde trafficking from the Golgi to the ER for protein recycling and balance maintenance. MEMB12 and its close homologue MEMB11 are homologous to Bos1 in yeast and GS27 or membrin in mammals and regulates retrograde protein transport between Golgi and endoplasmic reticulum (ER) (Bubeck et al., 2008; Chatre et al., 2005), which is consistent with our results that down-regulation of MEMB12 promotes PR1 secretion likely through inhibiting retrograde transport and protein recycling. Mutation in another intracellular SNARE protein SYP61 had no effect on the secretion of PR1, indicating that not all the SNARE proteins affect secretion of PR proteins. Mutation in MEMB12 has weak effect on the secretion of the PM-associated syntaxin SYP121, suggesting that different SNARE proteins may be responsible for transporting different set of proteins. This specificity is also observed in other SNAREs. For example, VTI12 but not its close homolog VTI11 affects the transport of storage proteins (Sanmartin et al., 2007). These results suggest the specialization of MEMB12 in controlling the secretion of PR proteins for immune responses. In addition to targeting MEMB12, miR393b* was predicted to target two other proteins involved in trafficking, VPS54 and EXO70H3. Like MEMB12, VPS54 is also Golgi-localized and involved in the retrograde trafficking (Conibear and Stevens, 2000; Guermonprez et al., 2008). These results suggest that the immunity-specific secretory pathway is under the regulation and fine-tuning by sRNA-mediated RNAi in response to pathogen attacks.
Our work suggests that Arabidopsis employs AGO2 as an important RNAi effector in plant antibacterial immunity. AGO2 also contributes to antiviral defenses against two viruses carrying silencing suppressors that target AGO1 (Harvey et al., 2011). Repression of AGO1 by viral suppressors leads to the down-regulation of miR403 and subsequent induction of AGO2. It is unlikely that AGO2 induction by Pst is due to the same mechanism because strong induction of AGO2 by bacterial infection was still observed in ago1-27 mutant (Fig. S1H). Furthermore, we did not observe any suppression of miR403 by these bacterial strains (Fig. S1I). miR403 targets both AGO2 and AGO3 at their 3′ UTR, but only AGO2 was induced by Pst. We believe that induction of AGO2 by bacteria may mainly occur at the transcriptional level and is mechanistically different from the induction triggered by viruses. Several defense responsive cis-elements, including the W-box motif, elicitor-responsive element and gibberellin-responsive element, are present in the AGO2 promoter and future studies will elucidate their functions in AGO2 induction upon bacteria challenges.
AGO2 associates with a large array of sRNAs, among which miR393b* is one of the most abundant. It would be interesting to test whether other AGO2-associated sRNAs and their targets are also involved in plant immunity. We speculate that AGO2 may regulate and coordinate the expression of a group of genes involved in various pathways of plant immunity by binding to a group of functional sRNAs.
Experimental Procedures (see the supplemental procedures for details)
Bacterial infection
Bacterial growth assay and HR assay were performed as described (Katiyar-Agarwal et al., 2007; Katiyar-Agarwal et al., 2006). For RNA extraction and the HR assay, plants were inoculated with Pst strains at a concentration of 1×107 cfu/ml. For the bacterial growth assay, 5 × 105 cfu/ml was used. At least six leaf discs were collected for each experiment. Student’s t-test was employed to determine the significant difference between mutants and control plants.
Generation of transgenic plants
To generate the AGO3-RNAi construct, the 1-464 nt fragment of AGO3 coding sequence was amplified and cloned into pJawohl8-RNAi (EMBO database ID AF408413). The construct was transformed into ago2 and ago2ago7 to generate ago2ago3 and ago2ago3ago7 mutants, respectively. Transgenic 35S::YFP:MEMB12-wt or 35S::YFP:MEMB12-mu plants were generated by transformation of binary constructs pEG104 carrying the WT full-length MEMB12 CDS or the mutated version with 6 altered nucleotides in the miR393b* target site (Earley et al., 2006). Overexpression constructs of miR393b and amiR393b* were generated by cloning the miR393b precursor or artificial miRNA precursor using miR319 backbone (based on Web MicroRNA designer) into a Gateway destination vector pEG100 (Earley et al., 2006; Schwab et al., 2006). Primer sequences are listed in Table S3.
Cloning of AGO-associated sRNAs and sRNA library construction
Mock- or Pst (avrRpt2)-treated AGO2::3HA-AGO2 Arabidopsis plants were used for AGO1- and AGO2-IP at 12 hpi. Co-immunoprecipitation and sRNA purification were performed as described by Mi et al. (2008). To avoid saturation, we used excessive amount of protein A beads (Roche) and antibodies for the pull down, about 50 μl of protein A beads for the extracts from 0.15g plant tissue. For sRNA library construction, the miRCat® sRNA cloning linkers were used and manufacturer’s protocol was followed (miRCat® sRNA cloning kit; IDT). MEMB12 transcript was detected by nested RT-PCR from AGO2-associated RNAs.
Protein extraction and analysis
The intercellular wash fluid was collected from the same amount of tissue by centrifuging the leaves at 1500g for 5 min as described previously (Wang et al., 2005b). The proteins from the same volume of intercellular fluid for each sample were analyzed by Western blot analysis. Total proteins were examined by Western blot analysis using α-Tubulin as a loading control and RA measurement. For PM protein extraction, the PM fraction was isolated with a Dextran-PEG3500 two-phase system (Santoni, 2007). PM-associated PIP was used as a control.
Identification of target genes of miRNA*s
To identify miRNA* target genes, we followed the guideline for target prediction (Allen et al., 2005) with several modifications. Only one gap located in the sRNA was allowed, but with double penalty. Nucleotides at position 10 and 11 of the sRNA must be a perfect match with its target. Maximum of three continuous mismatches were allowed if the mismatch region contained at least two G:U pairs and the penalty score of the region was multiplied by 1.5. We used ≤ 5.5 as the cutoff score for selecting the miRNA* targets.
Supplementary Material
Acknowledgments
We thank Xinnian Dong, Jian-Kang Zhu, Thomas Eulgem, Isgouhi Kaloshian, Federica Brandizzi, Yifan Lii and Arne Weiberg for helpful comments; Dongdong Niu for experimental assistance. We thank Xinnian Dong, David Baulcombe, Paul Schulze-Lefert and Yijun Qi for providing antibodies; Jim Carrington for providing the pAGO2:HA-AGO2 line and mutant seeds; David Baulcombe, John Clarke, Herve Vaucheret for providing mutant seeds. This work was supported by an NIH R01 GM093008, an NSF Career Award MCB-0642843, and an AES-CE Research Allocation Award PPA-7517H to H. Jin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agorio A, Vera P. ARGONAUTE4 Is Required for Resistance to Pseudomonas syringae in Arabidopsis. Plant Cell. 2007;19:3778–3790. doi: 10.1105/tpc.107.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during transacting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bednarek P, Kwon C, Schulze-Lefert P. Not a peripheral issue: secretion in plant-microbe interactions. Curr Opin Plant Biol. 2010;13:378–387. doi: 10.1016/j.pbi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Bubeck J, Scheuring D, Hummel E, Langhans M, Viotti C, Foresti O, Denecke J, Banfield DK, Robinson DG. The syntaxins SYP31 and SYP81 control ER-Golgi trafficking in the plant secretory pathway. Traffic. 2008;9:1629–1652. doi: 10.1111/j.1600-0854.2008.00803.x. [DOI] [PubMed] [Google Scholar]
- Chatre L, Brandizzi F, Hocquellet A, Hawes C, Moreau P. Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 2005;139:1244–1254. doi: 10.1104/pp.105.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. Jul. 2010 Sep; doi: 10.1093/nar/gkq590. [Epub ahead of print]-online available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Huckelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. Rna. 2009;15:2219–2235. doi: 10.1261/rna.1646909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song KM, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Geldner N, Denervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant Journal. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. Rna. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez H, Smertenko A, Crosnier MT, Durandet M, Vrielynck N, Guerche P, Hussey PJ, Satiat-Jeunemaitre B, Bonhomme S. The POK/AtVPS52 protein localizes to several distinct post-Golgi compartments in sporophytic and gametophytic cells. J Exp Bot. 2008;59:3087–3098. doi: 10.1093/jxb/ern162. [DOI] [PubMed] [Google Scholar]
- Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstadt S, Carr JP, Baulcombe DC. An Antiviral Defense Role of AGO2 in Plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell. 2010 doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Current Biology. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kalde M, Nuhse TS, Findlay K, Peck SC. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci U S A. 2007;104:11850–11855. doi: 10.1073/pnas.0701083104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and rniRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–U810. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Li S, van Os GM, Ren S, Yu D, Ketelaar T, Emons AM, Liu CM. Expression and functional analyses of EXO70 genes in Arabidopsis implicate their roles in regulating cell type-specific exocytosis. Plant Physiol. 2010a doi: 10.1104/pp.110.164178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010b;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. Embo Reports. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A, Vaucheret H. Form, Function, and Regulation of ARGONAUTE Proteins. Plant Cell. 2010 doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson LA, Hanton SL, Brandizzi F. Traffic between the plant endoplasmic reticulum and Golgi apparatus: to the Golgi and beyond. Curr Opin Plant Biol. 2006;9:601–609. doi: 10.1016/j.pbi.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Mi SJ, Cai T, Hu YG, Chen Y, Hodges E, Ni FR, Wu L, Li S, Zhou H, Long CZ, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li DW, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA star species has substantial influence on microRNA and 3′ UTR evolution. Nature Structural & Molecular Biology. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan C, Zhang X, Jin H. Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Robert S, Chary SN, Drakakaki G, Li S, Yang Z, Raikhel NV, Hicks GR. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci U S A. 2008;105:8464–8469. doi: 10.1073/pnas.0711650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartin M, Ordonez A, Sohn EJ, Robert S, Sanchez-Serrano JJ, Surpin MA, Raikhel NV, Rojo E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:3645–3650. doi: 10.1073/pnas.0611147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V. Plant plasma membrane protein extraction and solubilization for proteomic analysis. Methods Mol Biol. 2007;355:93–109. doi: 10.1385/1-59745-227-0:93. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu ZS, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA. GENE TRAP TAGGING OF PROLIFERA, AN ESSENTIAL MCM2-3-5-LIKE GENE IN ARABIDOPSIS. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Plant ARGONAUTES. Trends in Plant Science. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005a;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong XN. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005b;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- Wessner B, Gryadunov-Masutti L, Tschan H, Bachl N, Roth E. Is there a role for microRNAs in exercise immunology? A synopsis of current literature and future developments. Exerc Immunol Rev. 2010;16:22–39. [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2010 doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JH, Gong ZZ, Zhang CQ, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA. OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell. 2002;14:3009–3028. doi: 10.1105/tpc.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.