Abstract
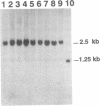
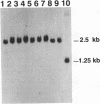
We established a mouse Ltk- cell line that contains within its genome a herpes simplex virus thymidine kinase gene (tk) that had been disrupted by the insertion of the recognition sequence for yeast endonuclease I-SceI. The artificially introduced 18 bp I-SceI recognition sequence was likely a unique sequence in the genome of the mouse cell line. To assess whether an induced double-strand break (DSB) in the genomic tk gene would be repaired preferentially by gene targeting or non-homologous recombination, we electroporated the mouse cell line with endonuclease I-SceI alone, one of two different gene targeting constructs alone, or with I-SceI in conjunction with each of the two targeting constructs. Each targeting construct was, in principle, capable of correcting the defective genomic tk sequence via homologous recombination. tk+ colonies were recovered following electroporation of cells with I-SceI in the presence or absence of a targeting construct. Through the detection of small deletions at the I-SceI recognition sequence in the mouse genome, we present evidence that a specific DSB can be introduced into the genome of a living mammalian cell by yeast endonuclease I-SceI. We further report that a DSB in the genome of a mouse Ltk- cell is repaired preferentially by non-homologous end-joining rather than by targeted homologous recombination with an exogenous donor sequence. The potential utility of this system is discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett C. B., Lewis A. L., Baldwin K. K., Resnick M. A. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R. J., Waldman A. S., Liskay R. M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- Bryant P. E., Johnston P. J. Restriction-endonuclease-induced DNA double-strand breaks and chromosomal aberrations in mammalian cells. Mutat Res. 1993 May;299(3-4):289–296. doi: 10.1016/0165-1218(93)90105-m. [DOI] [PubMed] [Google Scholar]
- Colleaux L., D'Auriol L., Galibert F., Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Thomas K. R., Capecchi M. R. Location of crossovers during gene targeting with insertion and replacement vectors. Mol Cell Biol. 1993 Apr;13(4):2134–2140. doi: 10.1128/mcb.13.4.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire M. K., Epstein L. H., Young C. S., Munz P. L., Fishel R. Nonhomologous recombination in human cells. Mol Cell Biol. 1994 Jan;14(1):156–169. doi: 10.1128/mcb.14.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead C., Dujon B. Consequences of unique double-stranded breaks in yeast chromosomes: death or homozygosis. Mol Gen Genet. 1993 Aug;240(2):170–178. doi: 10.1007/BF00277054. [DOI] [PubMed] [Google Scholar]
- Fairman M. P., Johnson A. P., Thacker J. Multiple components are involved in the efficient joining of double stranded DNA breaks in human cell extracts. Nucleic Acids Res. 1992 Aug 25;20(16):4145–4152. doi: 10.1093/nar/20.16.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R., Derbyshire M. K., Moore S. P., Young C. S. Biochemical studies of homologous and nonhomologous recombination in human cells. Biochimie. 1991 Feb-Mar;73(2-3):257–267. doi: 10.1016/0300-9084(91)90211-i. [DOI] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974 Sep;24(3):281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Harris R. S., Longerich S., Rosenberg S. M. Recombination in adaptive mutation. Science. 1994 Apr 8;264(5156):258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992 Nov 13;71(4):543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- Kramer K. M., Brock J. A., Bloom K., Moore J. K., Haber J. E. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994 Feb;14(2):1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L., Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harb Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci U S A. 1980 Jan;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezard C., Nicolas A. Homologous, homeologous, and illegitimate repair of double-strand breaks during transformation of a wild-type strain and a rad52 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 1994 Feb;14(2):1278–1292. doi: 10.1128/mcb.14.2.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás A. L., Young C. S. Characterization of DNA end joining in a mammalian cell nuclear extract: junction formation is accompanied by nucleotide loss, which is limited and uniform but not site specific. Mol Cell Biol. 1994 Jan;14(1):170–180. doi: 10.1128/mcb.14.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North P., Ganesh A., Thacker J. The rejoining of double-strand breaks in DNA by human cell extracts. Nucleic Acids Res. 1990 Nov 11;18(21):6205–6210. doi: 10.1093/nar/18.21.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington S. L., Wilson J. H. Gene targeting in Chinese hamster ovary cells is conservative. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9498–9502. doi: 10.1073/pnas.88.21.9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Thode S., Hancke J., Vielmetter W. Mechanisms of overlap formation in nonhomologous DNA end joining. Mol Cell Biol. 1994 Feb;14(2):888–895. doi: 10.1128/mcb.14.2.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. W., Morgan W. F. Illegitimate recombination induced by DNA double-strand breaks in a mammalian chromosome. Mol Cell Biol. 1994 Sep;14(9):5794–5803. doi: 10.1128/mcb.14.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thacker J., Chalk J., Ganesh A., North P. A mechanism for deletion formation in DNA by human cell extracts: the involvement of short sequence repeats. Nucleic Acids Res. 1992 Dec 11;20(23):6183–6188. doi: 10.1093/nar/20.23.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Valancius V., Smithies O. Double-strand gap repair in a mammalian gene targeting reaction. Mol Cell Biol. 1991 Sep;11(9):4389–4397. doi: 10.1128/mcb.11.9.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Sharp J. A., Summers W. C. Nucleotide sequence of the thymidine kinase gene of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Liskay R. M. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S. The search for homology does not limit the rate of extrachromosomal homologous recombination in mammalian cells. Genetics. 1994 Feb;136(2):597–605. doi: 10.1093/genetics/136.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Waldman B. C. Stimulation of intrachromosomal homologous recombination in mammalian cells by an inhibitor of poly(ADP-ribosylation). Nucleic Acids Res. 1991 Nov 11;19(21):5943–5947. doi: 10.1093/nar/19.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman B. C., Waldman A. S. Illegitimate and homologous recombination in mammalian cells: differential sensitivity to an inhibitor of poly(ADP-ribosylation). Nucleic Acids Res. 1990 Oct 25;18(20):5981–5988. doi: 10.1093/nar/18.20.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiffenbach B., Haber J. E. Homothallic mating type switching generates lethal chromosome breaks in rad52 strains of Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):522–534. doi: 10.1128/mcb.1.6.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Leung W. Y., Bosco G., Dieu D., Haber J. E. The frequency of gene targeting in yeast depends on the number of target copies. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):177–181. doi: 10.1073/pnas.91.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winegar R. A., Phillips J. W., Youngblom J. H., Morgan W. F. Cell electroporation is a highly efficient method for introducing restriction endonucleases into cells. Mutat Res. 1989 Jan-Feb;225(1-2):49–53. doi: 10.1016/0165-7992(89)90032-8. [DOI] [PubMed] [Google Scholar]
- Zheng H., Wilson J. H. Gene targeting in normal and amplified cell lines. Nature. 1990 Mar 8;344(6262):170–173. doi: 10.1038/344170a0. [DOI] [PubMed] [Google Scholar]