Abstract
The mature HIV-1 protease (PR) bearing drug-resistance mutation L76V (PRL76V) is significantly less stable, with >7-fold higher dimer dissociation constant (Kd) of 71 ± 24 nM and twice the sensitivity to urea denaturation (UC50 = 0.85 M) relative to PR. Differential scanning calorimetry showed a decrease in Tm of 12 °C for PRL76V in the absence of inhibitors, and 5–7 °C in the presence of inhibitors darunavir (DRV), saquinavir (SQV) and lopinavir (LPV), relative to PR. Isothermal titration calorimetry gave a ligand dissociation constant of 0.8 nM for DRV, ~160-fold larger than that of PR, consistent with DRV resistance. Crystal structures of PRL76V complexed with DRV and SQV were determined at resolutions of 1.45–1.46 Å. Compared to the corresponding PR complexes, the mutated Val76 lacks hydrophobic interactions with Asp30, Lys45, Ile47, and Thr74, and exhibits closer interactions with Val32 and Val56. The bound DRV lacks one hydrogen bond with the main chain of Asp30 in PRL76V relative to PR, possibly accounting for resistance to DRV. SQV shows slightly improved polar interactions with PRL76V compared to PR. Although the L76V mutation significantly slows the N-terminal autoprocessing of the precursor TFR-PRL76V to give rise to the mature PRL76V, the co-selected M46I mutation counteracts by enhancing this rate but renders the TFR-PRM46I/L76V precursor less responsive to inhibition by 6 μM LPV while retaining inhibition by SQV and DRV. The correlation of lowered stability, higher Kd and impaired autoprocessing, with reduced internal hydrophobic contacts suggests a novel molecular mechanism for drug resistance.
Keywords: HIV-1 protease, crystal structure, structure-assisted design, drug resistance, differential scanning calorimetry, isothermal titration calorimetry, precursor autoprocessing, enzyme kinetics
Human immunodeficiency HIV-1 protease (PR) is a major drug target for treatment of AIDS. PR functions in the last step of the HIV-1 life cycle by catalyzing the cleavage of viral polyproteins produced in the host cell. (1). Hydrolysis of polyproteins into functional products is important for the maturation and production of infectious progeny virions. Protease inhibitors (PIs) act to prevent the maturation process. Currently, nine PIs are approved by the FDA as anti-viral drugs. However, the lack of a proofreading step of the viral reverse transcription leads to a high frequency of mutations, and under drug pressure, rapid selection of a combination of mutations confers drug resistance thus presenting a severe challenge in current anti-HIV treatment (2).
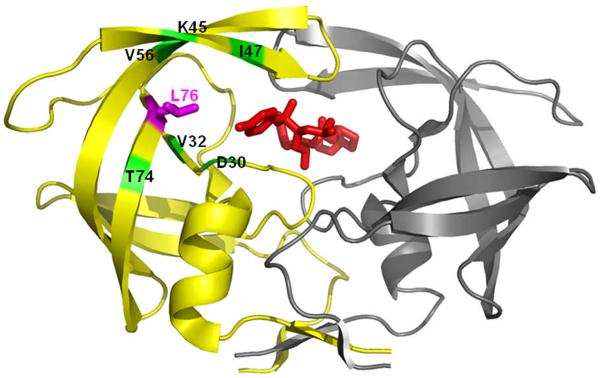
PR is a homodimer with 99 amino acids in each subunit (Figure 1) (3). The Asp25-Thr26-Gly27 triplets of both monomers form the catalytic site, where Asp25 and Asp25' (the prime indicates the second subunit of the dimer) serve as general acid-base catalysts of polyprotein cleavage in the proposed reaction mechanism (4). Two flexible interacting flaps are formed by residues 44–57 of both subunits. The two characteristic triplets and two flaps make important contributions to the active site cavity for the binding of substrates or inhibitors (5). The PR dimer is stabilized via noncovalent interactions of the residues at the dimer interface. Important intersubunit hydrogen bonds connect the catalytic triplets, flaps and beta sheet formed by the 4 terminal strands (6). The tertiary fold of each monomer is stabilized by hydrophobic interactions among the aliphatic residues in an internal hydrophobic core. Mutations occurring in over 30 of the 99 residues in each subunit are associated with drug resistance (7). Two major types of PI-resistance mutations are proposed to influence the PR activity (5). One type located near the active site can change the binding affinity and/or specificity of PR with the inhibitor by altering direct interactions (8). Other mutations do not change the binding cavity directly; however, they may affect PR stability and indirectly influence the binding of inhibitors via long-range structural perturbations (9).
Figure 1.
HIV-1 PR dimer structure (grey and yellow ribbons represent the two subunits) bound to darunavir (red sticks). The site of mutation is indicated by magenta sticks for Leu76 on one subunit. Positions of residues interacting with Leu76 are indicated and colored in green.
The L76V mutation has become more prevalent in datasets of HIV-1 mutants observed in patients (7). Presence of L76V as a single mutation was shown to hamper viral replication severely (10). L76V, generally accompanied by other mutations, is considered a major mutation providing 2-6-fold decreased susceptibility to darunavir (DRV), fosamprenavir, indinavir (IDV), and lopinavir (LPV) (11–12). It is also selected as a drug resistance mutation in patients receiving LPV therapy (10). This mutation, however, is not associated with resistance to atazanavir (ATV), nelfinavir (NFV), saquinavir (SQV) or tipranavir (TPV). Instead, mutants containing L76V exhibit increased susceptibility to these drugs (11, 13–14). Hence, this mutation shows opposing roles in drug resistance, acting to increase susceptibility to some drugs and decrease susceptibility to others (15).
We constructed PR with the single mutation of L76V in order to investigate its effect on the structure, stability and activity of the enzyme. Crystal structures of PRL76V with DRV and SQV were determined to investigate the molecular basis for the responses of this mutant to inhibitors. Leu76 is located in the hydrophobic core of PR close to the active site cavity, although its side chain has no van der Waals contacts with most substrates or inhibitors. Additionally, the effect of the mutation on the autocatalytic processing of the precursor, which is required for onset of the catalytic activity characteristic of the mature, dimeric enzyme, was assessed in vitro, and the effect of a second mutation, M46I, was examined in the precursor containing both mutations.
EXPERIMENTAL PROCEDURES
Protein expression and purification
The HIV-1 PR (Genbank HIVHXB2CG) clone optimized for structural and biochemical studies contains mutations Q7K, L33I and L63I to minimize autoproteolysis, and C67A and C95A to prevent cysteine-thiol oxidation (16). The L76V mutation was introduced in this PR template as well as the template encoding a PR precursor mimetic in which the full-length transframe region (TFR) is fused to the N-terminus of PR (TFR-PR) by use of the appropriate oligonucleotide primers using the QuikChange protocol (Stratagene, La Jolla, CA) and verified by DNA sequencing. A second construct, containing two mutations, M46I and L76V, in the PR domain of TFR-PR was also made using TFR-PRL76V as the template. Proteins were expressed using pET11a vector and E. coli BL21(DE3), purified and refolded using established protocols (17–19), and the identities of the products verified by ESI-MS.
Enzyme assays
The kinetic parameters were measured using a fluorescence assay as described (20) with the substrate Abz-Thr-Ile-Nle-pNO2Phe-Gln-Arg-NH2 (where pNO2Phe is para-nitrophenylalanine, Nle is norleucine and Abz is anthranilic acid) (Bachem Bioscience Inc., King of Prussia, PA), which is based on the p2/NC cleavage site of the viral polyprotein. 10 μl PR L76V at a final concentration of 34 nM from active site titration with SQV was mixed with 100 μl of reaction buffer [100 mM MES (4-morpholineethanesulfonic acid), pH 5.6, 400 mM NaCl, 1 mM ethylenediaminetetraacetic acid, and 5% glycerol] at 26 °C. The reaction was initiated by adding substrate from a stock solution of 186 μM to give a final concentration of 12–84 μM. PR activity was measured by monitoring the increase in fluorescence over 5 min using an excitation wavelength of 340 nm and emission wavelength of 420 nm (pNO2Phe as quencher-fluorescent acceptor; Abz as fluorescent donor) with a POLARstar OPTIMA 96-well microplate instrument (BMC Labtech). Values for kcat and Km were obtained by fitting the curves to the Michaelis-Menten equation using the program SigmaPlot (SPSS Inc., Chicago, IL).
For dimer dissociation and urea denaturation studies, enzyme activity was measured by following the initial rates of hydrolysis of the chromogenic peptide substrate IV that mimics the CA/p2 cleavage site (Lys-Ala-Arg-Val-Nle-pNO2Phe-Glu-Ala-Nle-NH2, California Peptide Research, Napa, CA) at 310 nm (Δε = 1797 M−1cm−1) in 50 mM sodium acetate buffer, pH 5.0, in 120 μL cuvettes at 28 °C. Substrate concentration (390 μM in the assay mixtures) was determined from the UV spectrum of substrate stock solutions (ε = 12,000 M−1 cm−1). The dimer dissociation constant, Kd, was determined by fitting the curve of activity (initial rate/protease concentration) vs. protease concentration (40–1200 nM as monomers) to an equation described previously (19). For urea denaturation studies, activity was measured at urea concentrations of 0–3.5 M (16, 21) and a enzyme concentration of 0.3–1.15 μM. The UC50 value is the urea concentration at which the PR activity is half of the maximum activity in the absence of urea.
Calorimetry
Inhibitor concentrations were determined kinetically by active site titration against the wild-type PR. Samples for differential scanning calorimetry (DSC) were prepared by the quench protocol from stock solutions in HCl as previously described (18) to give a final enzyme concentration of 14–15 μM (as dimer) in 50 mM sodium acetate buffer, pH 5.0. Final concentrations of inhibitors DRV, SQV and LPV were 28–29 μM (~2-fold molar excess relative to dimeric enzyme). DSC scans were run on a MicroCal VP-DSC microcalorimeter (GE Healthcare) at 90 °C/h from a starting temperature of 20 °C and terminated at 70–90 °C depending on the position of the transition. Data were processed using the instrument's Origin software as described (22). SDS-PAGE of the inhibitor-free enzyme on a 20% homogeneous PhastGel (GE Healthcare) before and after DSC showed no evidence for autoproteolysis during the course of the DSC experiment.
For isothermal titration calorimetry, PRL76V (1.9 mg/ml in 12 mM HCl) was folded by the quench protocol (18) to give final concentrations of 8–13.5 μM in 50 mM sodium acetate buffer, pH 5, and titrated with 16 injections of DRV in the same buffer (60–140 μM depending on the protein concentration) by use of a MicroCal iTC200 microcalorimeter (GE Healthcare) at 28 °C. Stock 160–200 μM inhibitor solutions were prepared in low ionic strength buffers at a final concentration of 0.5% (v/v) DMSO, diluted from a stock solution of inhibitor in 100% DMSO. All ITC experiments were performed at 0.5% or lower DMSO concentration. DMSO equivalent to that in the titrant was added to the protein solution in the cell to minimize thermal effects of changing DMSO concentration during the titration. As reduced autoproteolysis is intrinsic to the optimized PRs after folding at pH 5, concentrations of active PRL76V were verified from the known stoichiometry of inhibitor binding to the active site (N = 1) on titration. Data were processed using the instrument's Origin software. Three separate experiments at different concentrations of PRL76V and DRV were consistent with an upper limit of ~10 nM for the dissociation constant (KL). A displacement titration with 50 μM DRV in the presence of 16 μM RPB inhibitor (H-Arg-Val-Leu-(r)-Phe-Glu-Ala-Nle-NH2; Bachem Americas, Inc., Torrance, CA) added to the sample cell containing 4 μM PRL76V provided a more accurate value. Analogous displacement titrations under the same conditions with SQV or LPV did not give adequate thermal effect for reliable quantitation.
Autoprocessing of TFR-PRL76V Precursor
Isolation of PR precursors in significant quantities presents a substantial challenge because the intrinsic autoprocessing of the precursors to mature protease results in their depletion during expression. However, amounts of the full length precursors sufficient for small scale experiments can be recovered. Thus, remnants of the unprocessed precursor TFR-PRL76V and TFR-PRM46I/L76V were purified as described (16, 19). The precursor was folded by addition of 5.66 volumes of 5 mM sodium acetate buffer, pH 5.3, with or without added DRV, to TFR-PRL76V in 12 mM HCl, to give a final pH of 4.5. Samples (5 μl) were removed at times from 0–26 hours, mixed with 3 μl SDS-PAGE sample buffer and frozen immediately. Samples were subjected to electrophoresis on 20% homogeneous PhastGels (GE Healthcare), and visualized by staining with PhastGel Blue R.
Crystallographic Analysis
PRL76V at a concentration of 7 mg·ml−1 was mixed with the inhibitors DRV or SQV at 5-fold molar excess. Crystals were grown by the hanging-drop vapor-diffusion method at room temperature using 24 well VDX plates (Hampton Research, Aliso Viejo, CA). The crystallization drops had equal volumes of the protein and reservoir solutions. Crystals of PRL76V with DRV grew in solutions of 30 mM NaOAc buffer (pH 4.6–5.0), 1.4–1.8 M NaCl and 3% (v/v) DMSO. Crystals of PRL76V with SQV grew in solutions of 100 mM Tris-HCl buffer (pH 6.0–9.0) and 1.3 M NaCl with 3% (v/v) DMSO. The crystals were frozen in liquid nitrogen with the cryoprotectant of 20–30% (v/v) glycerol. X-ray diffraction data for the crystals were collected on the SER-CAT beamline of the Advanced Photon Source, Argonne National Laboratory in Chicago.
X-ray data were processed with HKL2000 (23). The structures were solved by molecular replacement using MolRep in the CPP4i suite of programs (24) and the starting model was the wild type PR complex with DRV (PDB code 2IEN) (25) in the same space group as the new structures. The structures were refined with SHELXL (26) and refitted using Coot 0.3.3 (27). Alternate conformations were modeled for PR residues, inhibitors and solvent molecules based on the observed electron density maps. The solvent was modeled with over 200 water molecules, ions and other solvent present in the crystallization solutions, as described (25). Anisotropic B, factors were applied for all the structures. Hydrogen atom positions were calculated in the last stage of refinement, using all data once all other parameters, including disorder, had been modeled. The mutant crystal structures were compared with the wild type PR by superimposing their Cα atoms as described (25). Structural figures were made using PyMol (28).
Protein Data Bank accession numbers
The structure coordinates and factors have been deposited in the RCSB Protein Data Bank with access number 3PWM for PRL76V-DRV and 3PWR for PRL76V-SQV.
RESULTS AND DISCUSSION
Properties and Stability of Mature PRL76V
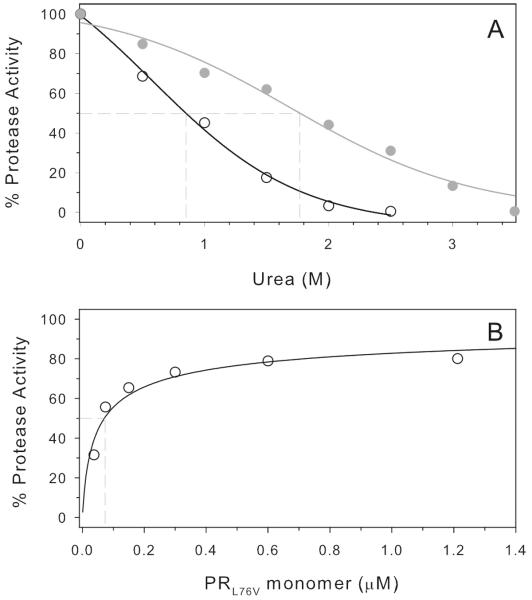
Purified mature PRL76V showed a Km of 37 ± 6 μM and kcat of 334 ± 23 min−1 at pH 5.6 and 200 mM NaCl. The catalytic efficiency, kcat/Km, of 9.0 μM −1 min−1 for hydrolysis of the fluorogenic substrate based on the p2/NC cleavage site is essentially the same (1.2-fold) as the value of 7.2 μM −1 min−1 determined for PR under the same conditions (20). A plot of the dependence of activity on urea concentration (Fig. 2A) shows a transition midpoint (UC50) of 0.85 M urea for PRL76V, which is approximately half the value of 1.78 M for PR (Table 1). Increased susceptibility to urea denaturation is consistent with the lower stability of the PRL76V dimer relative to the wild type enzyme as reflected by the Kd of 71 ± 24 nM for the PRL76V (Fig. 2B), which is at least 7-fold higher than that of PR (29).
Figure 2.
A. Urea denaturation of PRL76V and wild-type PR (UC50 = 0.85 M and 1.78 M, respectively, dashed intersect on X-axis). B. Kinetic determination of PRL76V dimer dissociation. A. dissociation constant, Kd = 71 ± 24 nM, indicated by the dashed vertical line at 50% activity, was determined as described (see Materials and Methods). For the wild type PR, Kd is < 10 nM [not shown, (29)].
Table 1.
Dimer dissociation, urea and thermal denaturation of PRL76V
Inhibitor Binding
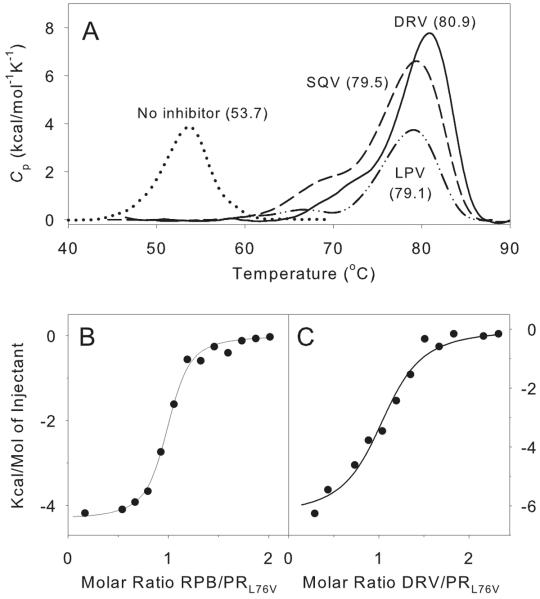
Differential scanning calorimetry (Fig. 3A) was used to assess the thermal denaturation of PRL76V in the absence and presence of inhibitors. The observed Tm values for PRL76V in Table 1 are compared with those previously reported for PR under the same conditions (30). In the absence of inhibitors PRL76V is markedly less stable to thermal denaturation than is PR, with a Tm that is 12 °C lower than that of PR. Although the mechanisms of heat- and urea-induced denaturations are not necessarily the same, the low Tm value for PRL76V is consistent with the observed effect of urea on its catalytic activity, as both heat- and urea- denaturations involve changes in the stability of the protein fold, which may precede and/or accompany dimer dissociation. Even when complexed with inhibitors, which should stabilize the proteases as dimers, the Tm value for PRL76V is about 7 °C lower than that for PR. The decreased thermal stability of PRL76V contrasts with results reported for several other PR mutants associated with drug resistance. For example, Tm values for ATV resistant mutant I50L/A71V (31) and multi-drug resistant mutant V82F/I84V (32) were both higher than the values observed for PR by 2.2 and 4 °C, respectively. A multi-drug resistant mutant bearing 11 mutations also exhibited a slightly higher Tm than the wild type enzyme (33).
Figure 3.
Thermochemical data for interactions of inhibitors with PRL76V. A. DSC thermograms of PRL76V in the presence and absence of inhibitors in 50 mM sodium acetate buffer, pH 5.0. The thermal transition temperatures (curve maxima) are in parentheses. Inhibitors were in a twofold molar excess relative to dimeric PRL76V. B. ITC trace of 5 μM PRL76V titrated with 80 μM RPB (KL = 61 nM) in 50 mM sodium acetate buffer, pH 5. C. Displacement of RPB (total concentration of 16 μM) bound to 4.2 μM PRL76V by titration with DRV (50 μM; KL for DRV = 0.79 nM).
An upper limit of ~10 nM for the dissociation constant (KL) of the PRL76V-DRV complex, with a range between 6.6 and 18 nM, was obtained by direct ITC titrations with the inhibitor. A substrate-analog inhibitor (RPB) with a reduced peptide bond exhibits a good negative thermal response (ΔH = −4.3 kcal/mol) on binding to PRL76V and a KL of 61 ± 13 nM (Fig. 3B). A displacement titration with DRV in the presence of RPB gave a value of 0.79 ± 0.29 nM (Fig. 3C), approximately 160–fold larger than KL for the PR-DRV complex (34–35). Lack of an adequate thermal response under the same conditions, either on direct titration or by displacement of RPB (10–15 μM), precluded KL measurements with SQV and LPV. Alternatively, a qualitative indication of the relative binding affinity of different inhibitors to the same protein can be obtained from the increase in thermal denaturation temperature (ΔTm) in the presence of inhibitor (22, 30, 31). The values of ΔTm for DRV, SQV and LPV binding to PRL76V are significantly larger (by 5–7 °C) than for PR, reflecting both the low thermal stability of the free PRL76V and significant stabilization of the protein-inhibitor complexes. ΔTm values for DRV, SQV and LPV binding to the PRL76V (Fig. 3A) differ by less than 2° C in the order DRV>SQV>LPV are consistent with large binding constants (small KL values), although the magnitude of their enthalpy values for thermal denaturation decrease significantly in the same order. The effects of these three inhibitors on autoprocessing of the TFR-PRL76V precursor (cf. Fig 4 and following section) suggest that KL for SQV and LPV will be larger than the 0.8 nM observed by ITC for DRV. By contrast, in vitro phenotype assays found the mutation L76V in PR derived from clinical samples to be associated with enhanced susceptibility to SQV and resistance to DRV and LPV (11, 14). However, in many, if not most, DRV resistant clinical isolates, a single resistance mutation does not occur alone but is associated with other mutations such as M46I and L90M (13, 36), which are selected along with L76V in clinical settings. It is possible that the relative responses to these inhibitors as well as the dimer stability may be altered by the presence of such additional mutations.
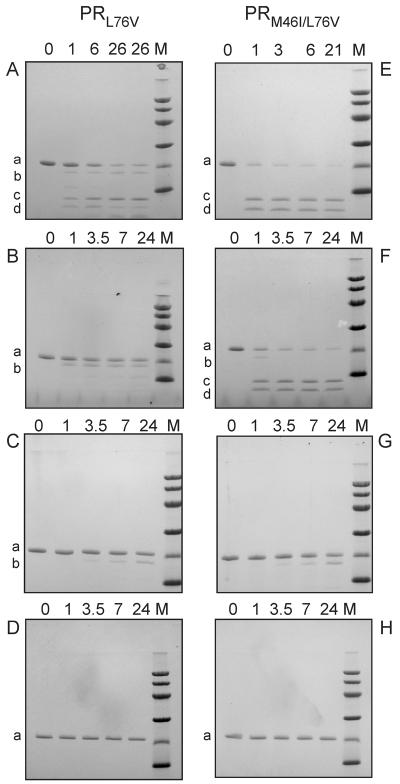
Figure 4.
Autocatalytic maturation of precursors (TFR-PRL76V and TFR-PR M46I/L76V) at pH 4.5 and its inhibition. Inhibitors were present at a 1:1 molar ratio (6 μM) to precursors (as dimers). (A and E) Controls without inhibitor. (B and F) In the presence of LPV. (C and G) In the presence of SQV. (D and H) In the presence of DRV. Numbers indicated on top of the gels denote autoprocessing reaction times in hours. Lane M corresponds to molecular weight markers of 97, 66, 45, 30, 20 and 14 kDa from the top. Letter designations for the bands are: a, full length precursor; b, TFR9–56-PR; c, mature, active PR; d, the fragment TFR9–56.
Autoprocessing of PRL76V and PRM46I/L76V Precursors
During viral replication, the autocatalytic processing (autoprocessing) of PR from its Gag-Pol polyprotein precursor is essential to generate the active mature PR and requisite structural and functional proteins of the infective virion (3). Cleavage between the transframe region (TFR), encoded in the Pol open reading frame, and the N terminus of PR domain (TFR/PR site) is crucial for the formation of stable PR dimers with full catalytic activity from the monomeric TFR-PR. Consequently, compromising this process will adversely affect the viability of the virus. Previous studies with the wild type precursor, TFR-PR, have identified the pathways for its maturation (3, 29). The time course of autoprocessing in the absence of inhibitors and in the presence of LPV, SQV and DRV is shown in Figure 4. At low pH an initial cleavage of TFR-PR (a) occurs between F8/L9 of the TFR to give an intermediate (b) with low catalytic activity similar to the full-length precursor. Subsequent cleavage at the TFR/PR site releases the fully active, mature PR (c) and the fragment TFR9–56 (d). Autoprocessing was assessed in vitro with a precursor comprising the 56-amino acid TFR fused to the N-terminus of the PR domain containing the L76V mutation (TFR-PRL76V). Reactions conducted at ~6 μM protein concentration permit monitoring the autoprocessing of the precursor in small volumes suitable for SDS-PAGE on PhastGels. In the absence of inhibitors, only ~50% of the autoprocessing reaction is complete at 6 h (Fig. 4A), in contrast to that of wild type TFR-PR, which is complete in < 1 h (3, 37). Thus, the slower rate of autoprocessing of TFR-PRL76V seems to correlate with the lower stability of the mature PRL76V, namely the higher Kd and increased susceptibility to denaturation by urea. Processing was only partially inhibited by LPV (Fig. 4B), but completely inhibited by DRV (Fig. 4D), using both inhibitors at a concentration of 6 μM. SQV is intermediate in its inhibition of TFR-PRL76V autoprocessing being slightly better than LPV and poorer than DRV. Internal cleavages within the TFR have been shown to be less responsive to inhibition than cleavages at the TFR-PR site (19). Thus in the presence of LPV and SQV some cleavage occurred at F8/L9 to give the intermediate TFR9–56-PRL76V, but very little or no subsequent cleavage was detected at the N-terminus of the protease domain to give mature PRL76V.
The single mutation L76V severely compromises viral replication in cell cultures, whereas coexistence of another mutation, M46I, was found to increase resistance to LPV while also partially restoring the ability of the virus to replicate (10). This presumably occurs by increasing the stability and/or catalytic efficiency of the protease or its precursor. Unfortunately, it was not possible to isolate the mature PRM46I/L76V because of its rapid autoproteolysis and very poor accumulation during its expression. However, limited quantities of the precursor TFR-PRM46I/L76V could be obtained for comparison with TFR-PRL76V under the same conditions. In the absence of inhibitors, TFR-PRM46I/L76V undergoes autoprocessing significantly faster (Fig. 4E) than TFR-PRL76V. The additional M46I mutation has no effect on inhibition by 6 μM SQV or DRV (Fig. 4G and 4H). Interestingly, however, TFR-PRM46I/L76V almost completely evades inhibition (Fig. 4F) under conditions where LPV inhibits processing of the single mutant TFR-PRL76V. These results are consistent with the observations of Nijhuis et al. (10), and underscore the potential importance of secondary mutations for precursor processing, both by improving the intrinsic autoprocessing activity of an otherwise compromised precursor, and by decreasing its susceptibility to inhibition.
PRL76V -Inhibitor Crystal Structures
Crystal structures of PRL76V in complexes with DRV and SQV were determined to identify any structural changes caused by the mutation. The crystallographic data collection and refinement statistics are displayed in Table 1. The crystal structures of PRL76V-DRV and PRL76V SQV were refined to R-factors of 0.14 at the resolutions of 1.45 and 1.46 Å. The crystal structures had one PR dimer with residues labeled 1–99 and 1'–99' in each crystallographic asymmetric unit in the space group of P21212. DRV and SQV were observed in two alternate conformations in the active site cavity of the dimer of PRL76V with relative occupancies of 0.63/0.37 and 0.73/0.27, respectively. Alternate conformations were modeled for 10 and 17 residues in the PRL76V-SQV and PRL76V-DRV structures, respectively. The flap residues 46, 50 and 51 exhibited alternative conformations in both subunits of the two structures. The DRV complex also showed alternative conformations of the side chains for Val82, Pro81', Val82' and I84' in the inhibitor binding site; however, these residues showed a single conformation in the SQV complex. The structures were refined with more than 200 water molecules. The solvent included one sodium ion, two chloride and 3 acetate molecules in the DRV complex, while the SQV complex showed 8 glycerol molecules.
Structural Changes at the Site of Mutation
The PRL76V complexes with DRV and SQV were compared with the corresponding PR-inhibitor complexes. The PR-DRV (2IEN) (25) structure was solved in the same space group P21212 at 1.30 Å resolution. The DRV complexes shared very similar backbone conformations with a low RMSD of 0.11 Å on Cα atoms. The PR-SQV (2NMW) (8) structure was refined at 1.16 Å resolution in the same space group, although the unit cell dimensions differed. Hence, the SQV complexes with PRL76V and PR superimposed with the larger RMSD value of 0.65 Å resulting from variations of up to 2.3 Å in residues 37–41 and 36'–45', as typically seen for PR complexes in different space groups.
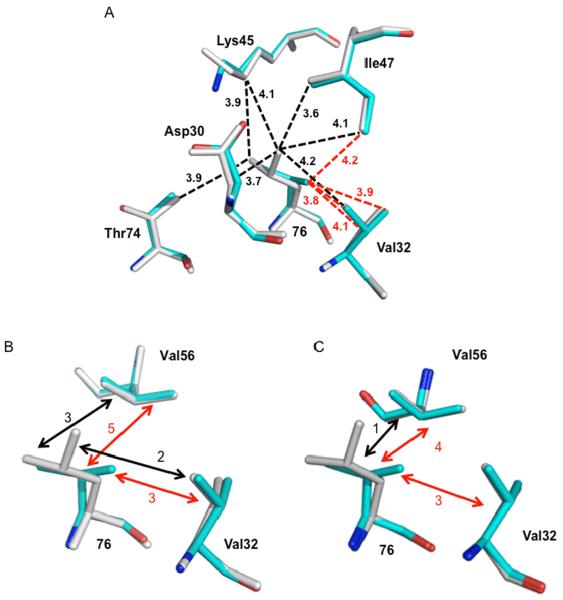
Leu76 lies in the inner hydrophobic cluster in each subunit of the PR dimer (38), and its side chain makes hydrophobic contacts with the side chains of Asp30, Val32, Lys45, Ile47, Val56, Gln58 and Thr74. The mutation of leucine 76 to valine gives a shorter side chain, which results in the loss of several van der Waals contacts. In the PRL76V-DRV complex, hydrophobic contacts with the side chains of Asp30, Lys45, and Thr74 and one contact with Ile47 are lacking in both subunits, as shown by an interatomic distance of more than 4.2 Å (Fig. 5A). Instead, the mutated Val76 forms more and closer hydrophobic interactions with the side chain of Val32, with interatomic distances of 3.8–4.0 Å rather than the longer 4.2 Å separation seen in the PR structure. Similar changes are observed in the PRL76V-SQV complex, with the exception of contacts with Val32 and Val56. The side chains of Val56 and residue 76 exhibit multiple hydrophobic contacts in the PRL76V complexes with both inhibitors and in the PR-DRV structure; however, the PR-SQV structure shows only a single van der Waals interaction between these residues (Fig. 5B and C). Similarly, the side chains of Leu76 and Val32 show no hydrophobic contacts in PR-SQV, while multiple contacts are seen in the PRL76V-SQV complex and both DRV complexes. Thus, PR-SQV shows fewer internal contacts around Leu76 compared to those of Val76 in the mutant complex, whereas similar contacts of residues 32, 56 and 76 are maintained in both DRV complexes.
Figure 5.
Hydrophobic interactions of residue 76 in the crystal structures of PRL76V (cyan), and wild type PR (grey). A. Interactions of residue 76 in subunit A of PRL76V-DRV and PR-DRV. Leu76 forms van der Waals contacts (3.6–4.2 Å) with the side chains of Asp30, Val32 Lys45, Ile47, and Thr74 (black dashed lines), while Val76 in PRL76V only has hydrophobic contacts with Val32 and Ile47 (red dashed lines). Interatomic distances are indicated in Å. Neighboring residue Gln58 is not shown since it forms similar interactions with residue 76 in PR and PRL76V. Val56 is omitted for clarity. B. Interactions of residue 76 with Val32 and Val56 in PRL76V-DRV and PRDRV. C. Interactions of residue 76 with Val32 and Val56 in PRL76V-SQV and PR-SQV. In B and C the number of hydrophobic contacts between the side chains is indicated by black (with Leu76) and red (with Val76) dashed arrows.
The decrease in internal van der Waals interactions is correlated with the lower stability of the mutant relative to the wild type enzyme described in the previous section. In contrast to other mutants exhibiting lower stability such as PRL24I, PRI50V and PRF53L (9, 39), however, no significant changes were seen at the dimer interface of the mutant structures. Therefore, the major molecular change associated with the significantly lower stability of PRL76V is the loss of internal hydrophobic contacts characteristic of Leu76. This loss of hydrophobic contacts at the mutated residue stands in strong contrast to the minimal changes reported for the majority of single mutants (9, 40). For example, our recent analysis of 6 mutants with single substitutions of residues in the hydrophobic clusters showed small structural adjustments that tended to conserve the hydrophobic interactions (40). Hence, the major loss of internal hydrophobic contacts for Val76 in PRL76V, coupled with reduced stability that is offset by secondary mutations, exemplifies a distinct mechanism for drug resistance.
Protease-Inhibitor Interactions
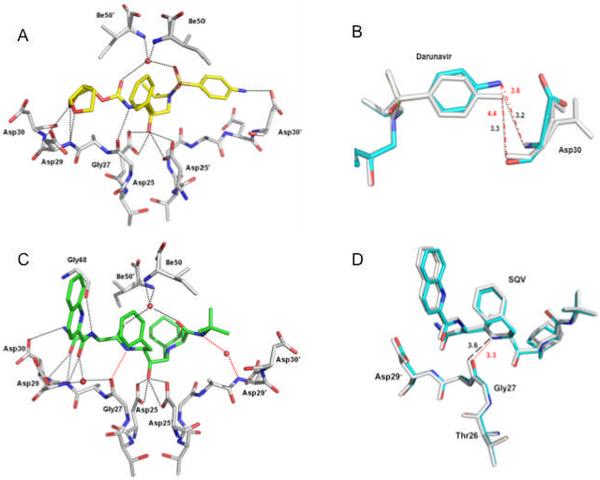
In the crystal structure of PRL76V-DRV, the inhibitor is observed in two orientations bound within the active site cavity by a set of hydrogen bonds, which are similar to those in the PR-DRV structure (25). Both orientations of DRV show similar hydrogen bond interactions with PRL76V. DRV forms 4 hydrogen bonds with the main chain atoms of Gly27', Asp29', and Asp30', and 5 hydrogen bonds with the side chain atoms of Asp25/Asp25' and Asp30 of PRL76V (Fig. 6A). The water-mediated hydrogen bonds between Ile50/50' and DRV are conserved in the majority of crystal structures of PR and its mutants with inhibitors and substrates. Compared with the wild type complex, DRV shows weaker interactions with the main chain of Asp30, which is associated with the shift of the aniline group of DRV away from the Asp30 of PRL76V (Fig. 6B). The interatomic distance representing a hydrogen bond between the aniline amino group of DRV and the amide nitrogen of Asp30 in PR-DRV increases from 3.2 to 3.6 Å in the A subunit of PRL76V-DRV, while the interatomic distance between the DRV amino group and the carbonyl oxygen of Asp30 increases from 3.3 to 4.4 Å indicating loss of a hydrogen bond in the PRL76V mutant. These changes may correlate with the loss of internal van der Waals contacts of Val76, especially those with Asp30, relative to those of the wild type Leu76.
Figure 6.
Protease-inhibitor hydrogen bond interactions. Hydrogen bonds are indicated by dotted lines. Water molecules are represented as red spheres. A. PRL76V hydrogen bond interactions with the major conformation of DRV (in yellow). B. Superposition of PRL76V-DRV (cyan bonds) and PR-DRV (grey bonds) showing PRL76V has fewer hydrogen bond interactions with the aniline group of DRV. The side chain of Asp30 has two alternate conformations in the PR-DRV structure. Interatomic distances are shown in Å with black dotted lines indicating the hydrogen bond interactions in the wild type complex, and red broken lines show the larger interatomic separation in the mutant. C. PRL76V hydrogen bond interactions with the major conformation of SQV (in green). The red dotted lines indicate the new hydrogen bond interactions formed by SQV in PRL76V relative to PR-SQV. D. Superposition of PRL76V-SQV (cyan bonds) and PR-SQV (grey bonds) showing the improved interaction of SQV with PRL76V arising from only slight structural changes. Red dotted lines indicate the new hydrogen bond interaction in the mutant, and black broken lines show the larger interatomic separation in the wild type SQV complex.
SQV is bound at the active site in two pseudosymmetric orientations in PRL76V-SQV. The major conformation of SQV forms 9 direct hydrogen bond interactions and four water-mediated hydrogen bonds with PRL76V (Fig. 6C). The mutant exhibits one improved direct and one more water-mediated interaction with SQV as compared with those in the wild type complex. Notably, a hydrogen bond interaction is formed between the carbonyl oxygen of Gly27 and the amide of SQV at 3.3 Å distance compared to the very weak 3.6 Å-long interaction observed in the wild type complex (Fig. 6D). The conserved water-mediated hydrogen bonds are seen between Ile50/50' and SQV, and another water-mediated interaction is conserved in reported high resolution crystal structures of PR and its mutants with SQV (8). Compared with the PR-SQV crystal structure, one more water-mediated hydrogen bond is observed between SQV and the amide of Asp29 in PRL76V. No water was visible at the equivalent position in the wild type structure; however, the observation of ordered solvent molecules partly depends on the crystallization conditions, quality and resolution of the diffraction data. Overall, SQV has slightly improved hydrogen bond interactions with PRL76V relative to the wild type PR suggesting that the mutant may retain high binding affinity for this inhibitor.
Implications for drug resistance
The structural changes in the PRL76V-inhibitor complexes suggest the molecular basis for the lowered stability of this mutant and slower autoprocessing of its precursor. The reduced hydrophobic interactions of Val76 can directly perturb the interactions of DRV with Asp30, although no direct effect on the SQV interactions was seen. Also, the mutation is likely to perturb the flaps through loss of contacts between residue 76 and flap residues Lys45 and Ile47. Movement of the flaps is required for binding of substrate and release of products (39). Inter-flap contacts also contribute to the dimer interface, so that destabilization of the flaps will likely contribute to the higher dimer dissociation constant of PRL76V and its increased susceptibility to thermal and urea-induced denaturations. Consequently, precursor processing, formation of the mature dimer and drug resistance depend on maintaining the correct flap conformations and their contacts with internal hydrophobic residues like Leu76.
The properties of this mutant are discussed in relation to the interactions of the nine clinical inhibitors in order to understand why the L76V mutation is associated with increased resistance to some drugs while retaining effective binding affinity for other clinical inhibitors. DRV forms no direct hydrogen bonds with flap residues suggesting that its binding may allow changes in flap conformation (Figure 6A). In contrast, SQV forms a hydrogen bond with the carbonyl oxygen of Gly48 in the flap, which suggests SQV binding may restrict such conformational changes (Figure 6C). This analysis of the PRL76V crystal structures and other PR complexes sheds light on the opposing clinical responses of this mutant to different drugs (10–11, 13). L76V is associated with resistance to the inhibitors LPV, DRV, APV, and IDV, which form hydrogen bond interactions with Gly27, Asp29 and/or Asp30 near the catalytic Asp25, as well as the conserved water-mediated interactions with Ile50/50' (25, 33, 40–41). On the other hand, inhibitors ATV, SQV and TPV, for which mutants containing L76V show increased susceptibility, all form direct or water-mediated hydrogen bond interactions with Gly48, while TPV is unique in forming direct, instead of water-mediated, hydrogen bonds with the amides of Ile50 and 50' (33). NFV is the only exception with no direct flap interactions (42), however, susceptibility to NFV is less strongly associated with the L76V mutation compared to other drugs (11–12).
The observed structural changes provide insight into the effects of combining mutation L76V with M46I, which has been reported to contribute strongly to competence of the virus to replicate and its clinical resistance to LPV (10). Analysis of the wild type PR crystal structures suggests that the P2 group of LPV is unusual in forming multiple close hydrophobic contacts with Ile47, whereas DRV and SQV show only 1 or 2 hydrophobic contacts with Ile47. Thus, altered contacts with flap residues 45 and 47 associated with mutation L76V may partially diminish the binding affinity of LPV and contribute to resistance to this drug while having less effect on the other drugs. Residues 46 and 76 have no direct contacts in the dimer structure, since residue 76 is part of the internal hydrophobic cluster, while the side chain of residue 46 points away from the protein surface of the flexible flap. Instead, Leu76 contacts the side chains of Lys45 and Ile47 on either side of Met46. In PRL76V, Lys45 and Ile47 show reduced interactions with Val76, suggesting that M46I may indirectly compensate for the destabilizing effects of L76V. Notably, although PRL76V shows a much slower rate of autoprocessing of its precursor relative to the wild type, in agreement with reported defective viral replication (10), the rate is increased upon introduction of M46I (Fig. 4D). Furthermore, the presence of both mutations significantly restores autoprocessing capability in the presence of LPV, although inhibition by DRV is retained. These results are consistent with previous data on viral replication in cell cultures (10) and suggest that investigating effects on precursor processing and its inhibition will provide a useful approach, which is complementary to studies of inhibitor interactions with the mature protease, for understanding the role and interactions of multiple mutations in drug resistance.
Table 2.
Crystallographic Data Collection and Refinement Statistics
| Protease | PRL76V | PRL76V |
|---|---|---|
|
| ||
| Inhibitor | DRV | SQV |
|
| ||
| Space group | P21212 | P21212 |
|
| ||
| Unit cell dimensions (Å) | ||
| a | 58.32 | 58.84 |
| b | 86.33 | 86.17 |
| C | 45.98 | 46.24 |
|
| ||
| Resolution range (Å) | 50–1.46 | 50–1.45 |
|
| ||
| Unique reflections | 37895 | 38308 |
|
| ||
| Rmerge (%) overall (final shell) | 5.2% (49.2%) | 6.4% (48.0%) |
|
| ||
| <I/sigma> overall (final shell) | 34.9 (2.1) | 27.3 (2.2) |
|
| ||
| Data range for refinement (Å) | 10–1.46 | 10–1.45 |
|
| ||
| Completeness (%) overall (final shell) | 91.9% (54.9%) | 90.1% (56.4%) |
|
| ||
| Rwork | 0.1404 | 0.1449 |
|
| ||
| Rfree | 0.1891 | 0.1971 |
|
| ||
| No. of solvent (total occupancies) | 278 (209.9) | 191 (151.7) |
|
| ||
| RMS Deviation from Ideality | ||
|
| ||
| bonds (Å) | 0.011 | 0.011 |
|
| ||
| angle distance (Å) | 0.032 | 0.029 |
|
| ||
| Average B-factors (Å2) | ||
| main-chain atoms | 14.1 | 18.9 |
| side-chain atoms | 19.6 | 23.9 |
| inhibitor | 10.9 | 15.4 |
| Solvent | 28.1 | 36.8 |
|
| ||
| Relative occupancy of inhibitor | 0.64/0.36 | 0.73/0.27 |
Acknowledgement
We thank Annie Aniana for technical assistance and the staff at SER-CAT beamline at the Advanced Photon Source, Argonne National Laboratory, for assistance during X-ray data collection. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. DRV, LPV and SQV were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH
This research was supported, in whole or in part, by National Institutes of Health grant GM062920, the Intramural Research Program of the NIDDK, National Institutes of Health (NIH) and Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- PR
pseudo wild type HIV-1 protease optimized for structural and biochemical studies by introduction of the mutations Q7K, L33I, L63I, C67A, and C95A
- PRL76V
PR with the L76V mutation
- DRV
darunavir
- SQV
saquinavir
- LPV
lopinavir
- Nle
norleucine
- DMSO
dimethylsulfoxide
- RMSD
root-mean-square deviation
- DSC
differential scanning calorimetry
- ITC
isothermal titration calorimetry
- RPB
H-Arg-Val-Leu-(r)-Phe-Glu-Ala-Nle-NH2 inhibitor with reduced peptide bond at position (r)
References
- 1.Turner BF, Summers MF. Structural biology of HIV. J. Mol. Biol. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 2.Cherry E, Wainberg MA. The Structure and Biology of HIV 1. In: Emini EA, editor. The human Inmmunodeficiency Virus: Biology, Immunology, and Therapy. Princiton University Press; 2002. pp. 1–43. [Google Scholar]
- 3.Louis JM, Weber IT, Tözsér J, Clore GM, Gronenborn AM. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 2000;49:111–146. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 4.Brik A, Wong CH. HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 2003;1:5–14. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 5.Weber IT, Agniswamy J. HIV-1 protease: structural perspectives on drug resistance. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber IT. Comparison of the crystal structures and intersubunit interactions of human immunodeficiency and Rous sarcoma virus proteases. J. Biol. Chem. 1990;265:10492–10496. [PubMed] [Google Scholar]
- 7.Johnson VA, Brun-Vézinet F, Clotet B, Gúnthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: December 2009. Top. HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 8.Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins: Struct., Funct., Bioinf. 2007;67:232–242. doi: 10.1002/prot.21304. [DOI] [PubMed] [Google Scholar]
- 9.Liu F, Boross PI, Wang YF, Tozser J, Louis JM, Harrison RW, Weber IT. Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S. J. Mol. Biol. 2005;354:789–800. doi: 10.1016/j.jmb.2005.09.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nijhuis M, Wensing AMJ, Bierman WFW, de Jong D, Kagan R, Fun A, Jaspers CAJJ, Schurink KAM, van Agtmael MA, Boucher CAB. Failure of treatment with first line lopinavir boosted with ritonavir can be explained by novel resistance pathways with protease mutation 76V. J. Infect. Dis. 2009;200:698–709. doi: 10.1086/605329. [DOI] [PubMed] [Google Scholar]
- 11.Young TP, Parkin NT, Stawiski E, Pilot-Matias T, Trinh R, Kempf DJ, Norton M. Prevalence, Mutation Patterns and Effects on Protease Inhibitor Susceptibility of the L76V Mutation in HIV-1 Protease. Antimicrob. Agents Chemother. 2010;54:4903–4906. doi: 10.1128/AAC.00906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller SM, Daeumer M, Kaiser R, Walter H, Colonno R, Korn K. Susceptibility to saquinavir and atazanavir in highly protease inhibitor (PI) resistant HIV-1 is caused by lopinavir-induced drug resistance mutation L76V. Antivir Ther. 2004;9:S44. [Google Scholar]
- 14.Vermeiren H, Van Craenenbroeck E, Alen P, Bacheler L, Picchio G, Lecocq P. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods. 2007;145:47–55. doi: 10.1016/j.jviromet.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Tartaglia A, Saracino A, Monno L, Tinelli C, Angarano G. Both a Protective and a Deleterious Role for the L76V Mutation. Antimicrob. Agents Chemother. 2009;53:1724–1725. doi: 10.1128/AAC.01693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 17.Wondrak EM, Louis JM. Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. Biochemistry. 1996;35:12957–12962. doi: 10.1021/bi960984y. [DOI] [PubMed] [Google Scholar]
- 18.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J. Biol. Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 19.Sayer JM, Agniswamy J, Weber IT, Louis JM. Autocatalytic maturation, physical/chemical properties and crystal structure of group N HIV 1 Protease: Relevance to drug resistance. Protein Sci. 2010;19:2055–2072. doi: 10.1002/pro.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Kovalevsky AY, Tie Y, Ghosh AK, Harrison RW, Weber IT. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J. Mol. Biol. 2008;381:102–115. doi: 10.1016/j.jmb.2008.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahalingam B, Louis JM, Reed CC, Adomat JM, Krouse J, Wang YF, Harrison RW, Weber IT. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. FEBS J. 1999;263:238–244. doi: 10.1046/j.1432-1327.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 22.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J. Biol. Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;267:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 25.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol. Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 26.Sheldrick GM, Schneider TR. SHELXL: high-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 29.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 protease: structure, dynamics, and inhibition. Adv. Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 30.Sayer JM, Louis JM. Interactions of different inhibitors with active site aspartyl residues of HIV 1 protease and possible relevance to pepsin. Proteins: Struct., Funct., Bioinf. 2009;75:556–568. doi: 10.1002/prot.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanchunas J, Jr., Langley DR, Tao L, Rose RE, Friborg J, Colonno RJ, Doyle ML. Molecular basis for increased susceptibility of isolates with atazanavir resistance-conferring substitution I50L to other protease inhibitors. Antimicrob. Agents Chemother. 2005;49:3825–3832. doi: 10.1128/AAC.49.9.3825-3832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd MJ, Luque I, Velázquez-Campoy A, Freire E. Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry. 2000;39:11876–11883. doi: 10.1021/bi001013s. [DOI] [PubMed] [Google Scholar]
- 33.Muzammil S, Armstrong AA, Kang LW, Jakalian A, Bonneau PR, Schmelmer V, Amzel LM, Freire E. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J. Virol. 2007;81:5144–5154. doi: 10.1128/JVI.02706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Béthune MP, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brower ET, Bacha UM, Kawasaki Y, Freire E. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem. Biol. Drug Des. 2008;71:298–305. doi: 10.1111/j.1747-0285.2008.00647.x. [DOI] [PubMed] [Google Scholar]
- 36.Mitsuya Y, Liu TF, Rhee SY, Fessel WJ. Prevalence of darunavir resistance and associated mutations: patterns of occurrence and association with past treatment. J. Infect. Dis. 2007;196:1177–1179. doi: 10.1086/521624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louis JM, Wondrak EM, Kimmel AR, Wingfield PT, Nashed NT. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 1999;274:23437–23442. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- 38.Ishima R, Louis JM, Torchia DA. Characterization of two hydrophobic methyl clusters in HIV-1 protease by NMR spin relaxation in solution. J. Mol. Biol. 2001;305:515–521. doi: 10.1006/jmbi.2000.4321. [DOI] [PubMed] [Google Scholar]
- 39.Liu F, Kovalevsky AY, Louis JM, Boross PI, Wang YF, Harrison RW, Weber IT. Mechanism of drug resistance revealed by the crystal structure of the unliganded HIV-1 protease with F53L mutation. J. Mol. Biol. 2006;358:1191–1199. doi: 10.1016/j.jmb.2006.02.076. [DOI] [PubMed] [Google Scholar]
- 40.Shen CH, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters. FEBS J. 2010;277:3699–3714. doi: 10.1111/j.1742-4658.2010.07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahalingam B, Wang YF, Boross PI, Tozser J, Louis JM, Harrison RW, Weber IT. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur. J. Biochem. 2004;271:1516–1524. doi: 10.1111/j.1432-1033.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- 42.Kaldor SW, Kalish VJ, Davies JF, Shetty BV, Fritz JE, Appelt K, Burgess JA, Campanale KM, Chirgadze NY, Clawson DK, Dressman BA, Hatch SD, Khalil DA, Kosa MB, Lubbehusen PP, Muesing MA, Patick AK, Reich SH, Su KS, Tatlock JH. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 1997;40:3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]