Abstract
Biological cells harbor a variety of molecular machines that carry out mechanical work at the nanoscale. One of these nanomachines is the bacterial motor protein SecA which translocates secretory proteins through the protein-conducting membrane channel SecYEG. SecA converts chemically stored energy in the form of ATP into a mechanical force to drive polypeptide transport through SecYEG and across the cytoplasmic membrane. In order to accommodate a translocating polypeptide chain and to release transmembrane segments of membrane proteins into the lipid bilayer, SecYEG needs to open its central channel and the lateral gate. Recent crystal structures provide a detailed insight into the rearrangements required for channel opening. Here, we review our current understanding of the mode of operation of the SecA motor protein in concert with the dynamic SecYEG channel. We conclude with a new model for SecA-mediated protein translocation that unifies previous conflicting data.
Keywords: SecA, SecYEG, Protein translocation
Introduction
In bacteria, proteins are synthesized in the cytosol at the ribosomes. A major share of these proteins, i.e. about 30%, needs to be transported across or into the cytoplasmic membrane to function at the cell-surface. This process is essential for nutrition uptake, motility and energy conversion as well as pathogenesis. Secretory and membrane proteins are recognized by their N-terminal signal sequences or hydrophobic transmembrane segments (TMS) when emerging as nascent chains at the ribosome and are targeted via two separate routes to the evolutionary conserved protein-conducting channel SecYEG. This hetero–trimeric complex is embedded in the cytoplasmic membrane and allows both the insertion of membrane proteins into and passage of secretory proteins across the membrane barrier. Secretory proteins are mostly translocated posttranslationally after their synthesis is completed on the ribosome, but secretion can also occur co-translationally [1]. They appear from the ribosome as nascent polypeptide chains bearing a N-terminal signal sequence and are recognized by the molecular chaperone SecB which binds to the mature region of the preprotein [2]. SecB keeps the preproteins in a partially unfolded and therefore translocation competent state and targets them to the SecYEG bound motor protein SecA [3]. Subsequent binding of ATP to SecA causes the insertion of the signal sequence of the preprotein into the SecYEG channel and the release of SecB. Next, SecA catalyzes the step-wise translocation of the polypeptide chain through multiple cycles of ATP binding and hydrolysis [4–6]. Membrane proteins are targeted via a different route to SecYEG and are integrated into the cytoplasmic membrane in a co-translational manner. The hydrophobic core of TMS is recognized by the bacterial homologue of the signal recognition particle (SRP) when emerging from the ribosome. Subsequently, SRP targets the ribosome–nascent chain complex to SecYEG where translation at the ribosome provides the driving force for the insertion of the membrane protein (for review, see [7]). The Sec-translocase is a large machinery that involves not only SecA and SecYEG, but also another heterotrimeric complex SecDFyajC that stimulates protein translocation by an as yet unknown mechanism. Here, we will only focus on the core elements of the translocase and discuss our current insights in the mechanism of protein translocation.
The protein-conducting channel SecYEG
SecYEG structure
Protein secretion in bacteria and import of proteins into the endoplasmic reticulum is mediated by a protein-conducting channel that is conserved throughout the three kingdoms of life. This hetero–trimeric integral membrane protein complex is termed SecYEG in prokaryotes and the Sec61 complex in eukaryotes. It consists of one major subunit with ten TMS, SecY or Sec61α, and smaller subunits that are located at the exterior of the channel. Several crystal structures of archaeal and bacterial Sec-complexes provide detailed structural insight into organization and dynamics of SecYEG and its homologues. The TMS of SecY assemble into a clamshell-like fold with TMS 1–5 and 6–10 forming the two half-shells that are connected by a flexible hinge between TMS 5 and 6 (Fig. 1) which contains a short loop called HL-1 that was suggested by molecular dynamic simulations to be the flexible link that allows opening of the clamshell [8]. SecE appears to stabilize the channel by embracing the clamshell as a clamp. SecG is a non-essential subunit that associates peripherally with the channel [9]. The number of TMS of SecE and SecG varies among the organisms but the functional relevance of the additional TMS is unclear. SecE of Escherichia coli consists of three TMS while the homologue Sec61γ of the archaeon Methanocaldococcus jannaschii has only one TMS. However, an E. coli SecE mutant with the two extra TMS truncated is fully functional [10]. The tilt of the SecY TMS creates an hourglass-shaped pore with a funnel-like entrance of 20–25 Å that narrows down to a central constriction of around 4 Å. This pore ring consists of six hydrophobic isoleucine residues and restricts access to the periplasmic side in the closed conformation. Opening of this constriction is necessary to accommodate a translocating polypeptide chain and will result in the formation of a water-filled pore if no translocating polypeptide would occupy the channel. The trans-side of SecYEG is occluded by an α-helical segment of SecY, called the plug domain which folds back into the periplasmic cavity of the channel (Fig. 1). Point mutations that reduce the dependence on the signal-sequence recognition map to both pore ring and plug domain [11–13], but the full contribution of these structural elements to channel function and quality control is poorly understood.
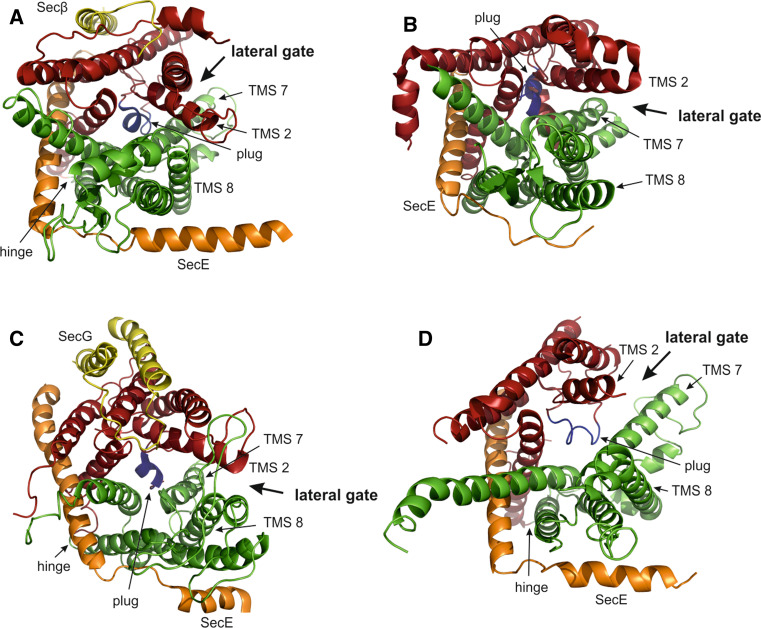
Fig. 1.
Crystal structures of SecYEG in top-view from the cytoplasm. SecY TMS 1–6 (red), TMS 7–10 (green), plug domain (blue), SecE (orange), SecG/β (yellow). a SecYEβ from M. jannaschii (PDB accession code: 1RH5). b SecYE from T. thermophilus co-crystallized with a Fab fragment (not shown) bound to the C5 loop of SecY (2ZJS). c SecYEG from T. maritima co-crystallized with SecA (not shown) (3DIN). d SecYE from P. furiosus. In the crystal, the C-terminal α-helix of a neighboring SecY molecule (not shown) inserts partially into the channel inducing opening of the lateral gate (3MP7)
SecYEG and its homologue Sec61αβγ were found to form higher oligomeric states in biochemical [14–16] and structural [17–20] studies. The functional relevance of the observed predominantly dimeric and tetrameric species remains unclear. Analysis of the first cryoEM structures of eukaryotic translocation channels led to the hypothesis that an aqueous pore is formed at the interface between oligomers [21–25]. This suggestion was further fueled by the hypothesis that large segments of the SecA motor protein inserts into the channel [4]. Based on the M. jannaschii SecYEβ X-ray structure, it was proposed that a single SecYEG heterotrimer forms the active translocation channel. Several biochemical and biophysical studies support this hypothesis. First, in a site-specific crosslinking study, a translocation substrate was located to the interior of a single SecY channel [26]. In a second study, introduction of different crosslinkers into the lateral gate prevented fusion of several SecYEG into a bigger pore and created single SecYEG channels with defined maximum pore size. Formation of disulfide bonds with short crosslinks abolished passage of polypeptide chains whereas crossing with longer crosslinking reagents permitted passage [27]. In analogy, cryoEM studies of the homologous Sec61p complex of mammals and yeast suggest that it is monomeric when associated with the ribosome and a translocating nascent chain [28].
SecYEG was also suggested to be dimeric, based on the observation that covalently linked SecYEG dimers are functional in protein translocation [26]. Osborne and co-workers proposed that SecA is anchored to one copy of SecYEG while translocating a polypeptide through the second protomer of a SecYEG dimer. A SecA–SecYEG co-crystal of components of the thermophilic bacteria Thermotoga maritima contradicts this view, as in this structure SecA is fully docked to monomeric SecYEG and only a few N-terminal residues are close to the presumed membrane interface (Fig. 2d) [29]. This structure indicates that SecA is anchored to the SecYEG channel that is promoting passage of the translocating polypeptide chain and that only a few SecA residues could interact with a neighboring SecYEG protomer in a putative SecYEG dimer. However, the corresponding residue on SecA that is crosslinked to SecY [26] is far away from the binding interface with SecYEG in the co-crystal suggesting that substantial conformation changes in SecA are needed to bring the two residues close enough to be crosslinked (Fig. 2c, d). As the membrane surface is absent in any crystal structure of SecYEG, Deville et al. [30] modeled a second SecYEG copy to the monomeric SecA–SecYEG co-crystal by tilting the two SecYEG protomers relative to each other to achieve an interaction with the SecA N-terminus.
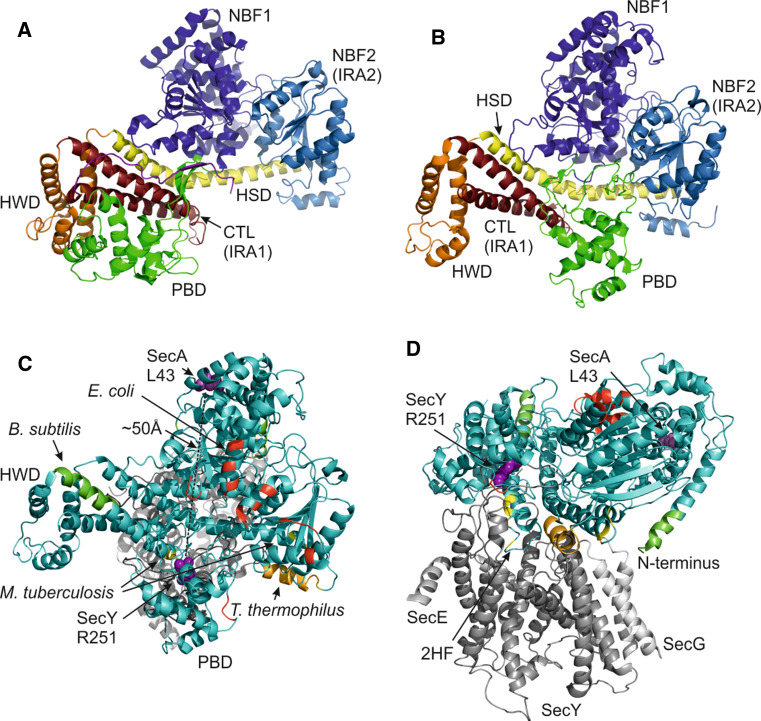
Fig. 2.
Conformational changes in SecA crystal structures and SecA dimerization interfaces. a SecA protomer from B. subtilis (1M6N). b SecA from T. maritima co-crystallized with SecYEG (not shown) (3DIN). c Top view (cytoplasmic side) of the SecA–SecYEG co-crystal as in (b) with residues implicated in dimerization in dimeric SecA structures of E. coli (red, 2FSF), B. subtilis (green, 1M6N), T. thermophilus (orange, 2IPC) and M. tuberculosis (yellow, 1NL3). Residues SecY251 and SecA43 that were previously crosslinked [26] are shown as purple spheres. SecA is displayed in cyan and SecYEG in gray. d Side view of (c)
SecYEG binding sites
The cytoplasmic side of SecYEG constitutes a combined binding interface for post- and co-translational protein translocation modes. The SecA ATPase was shown by in vivo photo-crosslinking to bind the cytoplasmic loops (C2 and C4–6) although to a different extent depending on the stage of the translocation cycle [31]. In two studies, site-specific disulfide formation between SecA and the C4/C5-loop of SecY was observed [32, 33], while a peptide scanning assay revealed a SecA binding site at the interface between TMS4 and the C3 loop of SecY that is close to SecG [32]. Direct contact of SecA and SecG was observed by crosslinking and protease protection [34], but this interaction seems to be non-essential as SecG merely stimulates protein translocation. The T. maritima SecA–SecYEG co-crystal provides insight into SecA binding in presence of ADP-BeFx, a structural analog of an intermediate step of ATP-hydrolysis [29]. Surprisingly, the majority of SecA residues identified by biochemical approaches to bind SecYEG are not in proximity to the respective SecY residues in the structure. One example is residue 255 on E. coli SecY that was crosslinked to residue 48 on B. subtilis SecA via disulfide bridge formation [26]. The corresponding residues in the T. maritima co-crystal (residues 251 on SecY and 43 on SecA) are about 50 Å apart while a disulfide bond is 2 Å in length (Fig. 2c, d) [26]. In this structure, SecA is crystalized in the closed conformation with the preprotein binding domain (PBD) moved towards the NBF2 (compare Fig. 2a, b), a conformational change that renders residue 255 of SecY hidden by the PBD (Fig. 2c). Thus, SecA must undergo a dramatic conformational change to allow for disulfide bridge formation. On the other hand, if SecA would be bound to SecYEG in the open conformation (Fig. 2a), the PBD would not hide residue 255 and would therefore render this position accessible from the cytoplasm. Thus, if needed, the binding of a second SecA protomer of a SecA dimer could occur, explaining the observed crosslink. Another example is Arg357 in the C5 loop of E. coli SecY that was found to be essential for the initiation of post-translational protein translocation [35] but not for SecA binding [32, 35, 36]. Although the corresponding residue of T. maritima SecY (Arg346) is only ~9 Å away from a SecA residue in the SecA–SecYEG co-crystal, no extensive contacts between SecA and SecY are made in this region of the structure. Taken together, these observations suggest that the occupation of SecYEG binding sites by SecA is dynamic during the protein translocation cycle. Ribosome binding occurs at similar sites on SecYEG as for SecA binding. Apparently, these sites play a general role in recruiting ligands, which are essential for channel functioning. The two SecY loops C4 and C5 [17, 37–40] as well as the cytoplasmic SecG loop and the N-terminal region of SecE [17] contact the ribosome. Point mutations in loops C4 and C5 including Arg357 substitutions eliminate ribosome binding [39]. In a recent study, the bacterial homologue of the signal recognition particle (SRP), FtsY, was found to bind to SecY loops C4 and C5 [40]. Hence, SecA, ribosome and SRP use overlapping binding sites on SecY. Occupation of these sites must be dynamic when all components cooperate during membrane protein insertion into the cytoplasmic membrane.
Opening of the channel
The interface between the tips of the two SecY clamshell-halves is shaped by TMS 2–3 and 7–8 and forms the so-called lateral gate (Fig. 1). Opening of this gate is indispensable to expand the channel to accommodate a translocating polypeptide chain during protein secretion [27] and to allow insertion of signal sequences and TMS of integral membrane proteins into the lipid bilayer [41]. Indeed, the signal-sequence of a precursor protein was found to crosslink to TMS 2 and TMS 7 of the yeast SecY homologue Sec61p during posttranslational protein import into the ER [42]. Insertion of a signal-sequence into the lateral gate would result in opening of the hydrophobic constriction as three of the six pore ring residues are located on TMS 2 and 7. However, to allow passage of an α-helical TMS, a gap in the lateral gate of 10–12 Å is required. Three X-ray structures provide different views of partially opened translocation channels. SecYE of Thermus thermophilus was co-crystallized with a Fab antibody fragment bound to the C5 loop of SecY that induced opening of the cytosolic part of the lateral gate (Fig. 1b) [33]. In the closed conformation represented by the M. jannaschii structure (Fig. 1a), TMS 2 is in close proximity to both TMS 7 and 8 while the TMS 8 of T. thermophilus SecY moved away from TMS 2 suggesting a ‘preopen’ state of the channel. Binding of SecA (ADP-BeFx) in the T. maritima SecYEG-SecA co-crystal induced separation of both cytosolic and periplasmic parts of the lateral gate although further opening would be required to allow insertion of a TMS in α-helical conformation (Fig. 1c) [29]. Apparently, the lateral gate opens by a rigid body movement of TMS 6–10 relative to TMS 1–5. This process may be initiated by insertion of a SecA segment, called the two-helix-finger (2HF), into SecYEG nearby SecY loops C4 and C5 (Fig. 2d). In a recent X-ray structure, two Pyrococcus furiosus SecYEβ crystallized as such that the C-terminal helix of one SecY partially inserts into the neighboring SecYEβ thereby functioning as a nascent chain mimic (Fig. 1d) [43]. As the result of this insertion, the lateral gate is opened throughout its entire length and the cytoplasmic entrance as well as the pore ring is expanded. While the plug domain still occludes the channel, the pore ring is widened to 13.6 Å where in the M. jannaschii structure a 4.6 Å diameter was determined (compare Fig. 1a, d) [11, 43].
In vivo [44] and in vitro [45] crosslinking experiments suggest extensive plug movement by around 20–27 Å to the C-terminal loop of SecE to allow unrestricted passage of polypeptide chains through the channel. However, immobilization of the plug inside the channel by chemical crosslinkers that allowed displacement of maximal 13 Å did not abolish protein translocation in vitro, indicating that movement of the plug to the periplasmic side of SecE is not necessary [46]. As for the physiological role of the plug domain, three hypotheses have been proposed. First, stabilization of the closed conformation has been suggested by instability of SecYEG upon plug deletion [47] and by molecular dynamics simulations [8]. Secondly, in the closed conformation, the plug domain prevents ion leakage through the SecYEG channel acting as a seal [48]. However, deletion of yeast Sec61p and E. coli plug domains resulted in thermolabile but viable cells with no significant growth or translocation defects [47, 49]. Third, deletion of the plug domain and many point mutations along the plug cause a reduced fidelity of signal sequence recognition (prlA phenotype) [47, 50, 51] and affect membrane protein topology [51]. Therefore, the plug may be involved in proofreading of preproteins and TMSs and mediate channel gating. Furthermore, the plug has been proposed to function as an adjustable flap to protect the emerging preprotein from periplasmic proteases or to assist in folding [52].
Many of the Prl mutations, identified by genetic studies to suppress translocation defects caused by defective signal sequences, reside inside the SecYEG channel [12, 13, 53]; for review, see [54]. Apparently, this prlA phenotype is caused by destabilization of the SecY-E interaction that facilitates channel opening [12, 55]. Inserted signal peptide, SecA and the ribosome are thought to stabilize the open state of the channel through multiple interactions with SecYEG [11]. Interestingly, PrlA mutants that were shown to bind SecA more tightly also exhibited an enhanced activity in protein translocation assays [56] and were less dependent on the proton motive force (PMF) [57]. Therefore, weaker SecY-E interactions may allow tighter SecA binding by facilitating channel opening leading to exposure of additional or stronger binding sites for SecA. As SecA binding to SecYEG results in a conformational change in SecA that stimulates its ATPase activity [58, 59], it appears that a tighter binding of SecA stimulates the initiation of protein translocation, thus allowing a more efficient translocation of wild-type preproteins as well as preproteins carrying a defective signal sequence. Therefore, the mechanism of proofreading of the translocation substrates may reside in the regulation of SecA’s ATPase activity by conformational states of SecYEG that responds to the physical properties of the signal peptide.
The translocation motor SecA
Cellular localization and binding partners
The ATP-dependent motor protein SecA is an essential element of the bacterial translocase engaged in transfer of polypeptides across the cytoplasmic membrane. As a soluble peripheral subunit, it associates with the membrane channel SecYEG and generates the driving force for the transport of secretory proteins and large periplasmic loops of membrane proteins [60]. Binding to signal sequences, unfolded preprotein substrates, anionic phospholipids, SecB and SecYEG allosterically stimulates the ATPase activity of SecA and couples the motor function to the translocation process [58, 61].
Localized to the cytosol, SecA is a targeting factor that recognizes signal sequences, and the SecB chaperone loaded with the preprotein and binds preproteins destined for secretion [62, 63]. On the other hand, SecA was shown to assist in folding of proteins lacking signal sequences [64]. Since preproteins are secreted essentially unfolded, such general chaperone function excludes cytoplasmic proteins from the secretion pathway and adds an additional quality control mechanism to the highly specific secretion process [64]. This suggested moonlight function may additionally be one reason for the relatively high cytoplasmic concentration of SecA (8 μM [65], corresponding to ~13,000 SecA copies per E. coli cell assuming an average volume of 2.75 μm3 per cell [66]), compared to the approximately 500 copies of SecYEG present in a bacterial cell [67].
Structure of SecA
The SecA protomer consists of functionally and structurally separate domains that act in concert to couple substrate recognition to cellular localization and motor action. The nucleotide-binding domain (NBD) consists of two essential nucleotide-binding folds, NBF1 and NBF2 (also termed intramolecular regulator of ATP hydrolysis 2, IRA2), that sandwich ATP for hydrolysis (Fig. 2). The two specificity domains are termed preprotein crosslinking domain (PPXD), or preprotein binding domain (PBD), and C-terminal domain (CTD) [68]. Preproteins bind to the PBD [69–71], the C-terminal linker (CTL), and to the α-helical scaffold domain (HSD) [72]. The CTD is involved in SecB [3, 73–75] and lipid binding [76] and consists of an α-helical wing domain (HWD), the CTL that is coordinated with a zinc-ion [75] and HSD that contacts all other domains of SecA (Fig. 2).
SecA is a member of the superfamily 2 DEAD (DExH/D) proteins because of its motor domain that has all nine conserved motifs of DNA/RNA helicases included in this family [62, 77–79]. Both NBF1 and NBF2 are essential for ATP binding and hydrolysis [80] and form RecA-like binding folds containing Walker A and B motifs [81]. NBF2 regulates the hydrolysis reaction at NBF1 [80, 82, 83] when a single ATP is bound between the motor sub-domains as common in DEAD-box helicases [84, 85]. The ATPase activity of cytosolic SecA is inhibited by the intramolecular regulator of ATP hydrolysis 1 (IRA1), a helix–loop–helix structure of the HSD that contacts both NBF2 and PBD. SecA mutants lacking IRA1 or bearing defects in IRA1/NBF2 interactions display increased ATPase activity independent of preprotein binding [86]. The mechanism of ATPase stimulation by preprotein binding utilizes a conserved electrostatic salt bridge called Gate1 that regulates the access to the nucleotide binding site [59]. Motions with the motor domain are induced by ATP binding and ADP release, respectively [83, 85, 87], and these conformational changes are thought to be transmitted to the two specificity domains, PBD and CTD, which directly contact the NBD [70, 83]. The preprotein binding domain inserts into NBF1 as a stem-like structure while the CTD contacts both NBF1 and NBF2 via an extended helix of the HSD (Fig. 2a, b) and inhibits the ATPase activity [86–89]. Motions of PBD [29, 59, 70, 90] and CTD [4, 29, 86, 88, 89] have been suggested to be transferred to the preprotein to push it through the SecYEG channel [4, 29, 77, 90]. PBD displacement relative to the HWD is a major conformational change observed in SecA crystal structures (compare Fig. 2a, b), but whether this motion is dependent on nucleotide or preprotein binding remains unclear [68]. Yet, it seems conceivable that SecA mediates the directed movement of polypeptide segments through the SecYEG channel by coupling motor action to specific substrate and SecYEG interaction.
The second protomer and the dimerization interface
Soluble SecA forms homo-dimers and is expected to be mostly dimeric in the cytosol [65, 91, 92]. Several high-resolution structures of SecA from various species have been solved: eight by X-ray crystallography and one by NMR [70]. Additionally, medium and low resolution structures obtained by cryo-EM [93–95], atomic force microscopy (AFM) [93], small angle X-ray scattering [96] and small angle neutron scattering [97] provide insight in SecA oligomerization and its conformational changes. Although the majority of structures show SecA as a homo-dimer, dimerization interface and orientation of the second protomer differ greatly among the structures [84, 93–100]. Furthermore, the diameter of observed SecA dimer particles in electron microscopy varies from 10 to 15 nm demonstrating significant conformational differences. In most of the structures, SecA is organized as an antiparallel dimer with its C-domains at the extreme ends. Exceptions are the parallel dimer structure from T. thermophilus [99] and one structure from B. subtilis that shows a single protomer in the asymmetric unit, although a crystallographic dimer can be recognized [85]. The variety of dimerization interfaces implies that either multiple dimeric conformations are possible or some of the observed dimers do not reflect the physiological state of cytosolic SecA. Since for some structures high salt concentrations were present during crystallization, monomerization and re-dimerization according to the crystal confinements could have occurred. This hypothesis is supported by the observation that dimerization occurs via electrostatic and hydrophobic interactions [91, 95, 98, 100, 101]. If cytosolic SecA would indeed form multiple dimeric states, they might either all support its role as targeting factor [68] or display SecA’s different functions as, for example, chaperone for cytosolic proteins. The hypothesis of diverse conformational states of SecA dimers is supported by a study suggesting the existence of multiple equilibriums of SecA dimers in solution [91]. It is tempting to speculate further that the observed dimerization interfaces are part of the catalytic cycle of dimeric SecA during protein translocation. Alternatively, the different dimeric conformations in solution may be irrelevant if only the SecYEG bound state is stabilized and critical for function [68]. Interestingly, in the T. maritima SecA–SecYEG co-crystal, residues that were implicated for dimerization in the E. coli and B. subtilis SecA crystal structures are exposed to the cytoplasmic surface and would allow binding of a second SecA protomer via this interface (Fig. 2c, d). The residues thought to be important for dimerization in the T. thermophilus and M. tuberculosis SecA crystals, however, are in close proximity to the SecY–SecA binding interface, and thus would not allow SecYEG binding of SecA dimers in a configuration as presented in these SecA crystals (Fig. 2c, d). In several studies, dimerization and orientation of the second protomer in solution was examined. Intermolecular crosslinking studies at the C-terminus confirm dimerization of SecA [102] and are in agreement with the antiparallel orientation of three of the observed structures [100, 103] as well as a FRET study [104].
Ligands modulate SecA dimerization
The oligomeric state of SecA in solution, bound to SecYEG and when engaged in protein translocation, has been investigated in a multitude of studies with various methods. Yet, contradicting results, their interpretations and physiological relevance caused a long-lasting controversy. Briefly, oligomerization of SecA in solution is dynamic and the dissociation constant K d that was previously estimated to be around 0.1 μM [91] has been determined by an equilibrium technique to 0.76 nM [105]. This monomer–dimer equilibrium of SecA in solution may be modulated by ligands such as signal peptides, anionic lipids or nucleotides, although contradicting results do not allow firm conclusions. However, the sensitive nature of the monomer–dimer equilibrium is apparent in the presence of factors such as detergents, salt or temperature [91, 106, 107]. The oligomeric state of SecYEG bound SecA has been investigated mainly in detergent or other artificial environments that may have compromised its native condition. However, an important contribution to this subject has been made by a model of SecA bound to SecYEG derived from a medium-resolution X-ray structure (Fig. 2c, d) [29]. Monomeric SecA from T. maritima is bound to a single SecYEG trimer of the same organism. The most prominent change is the rotation of the PBD of about 80° toward the NBF2 (Fig. 2b) compared to the B. subtilis SecA structure (Fig. 2a), which is considered to present the closed form of SecA [85]. Another structure of dimeric SecA from B. subtilis alone was suggested to be the open form and had the PBD rotated to a lesser extent [100]. In the SecA–SecYEG complex lacking a substrate, PBD movement from CTD to NBF2 results in a clamp-like structure that may enclose a preprotein. Moreover, the PBD rotation creates a pore in SecA that aligns with the SecYEG channel. Crystallization was achieved in presence of ADP-BeFx, a structural analog of an ATP hydrolysis intermediate step, and the model may present an intermediate state of protein translocation. However, the necessary use of detergent suggested to be responsible for the loss of the second SecYEG subunit may also have caused dissociation of the SecA dimer especially since detergents were shown to monomerize SecA [106, 107]. Therefore, interpretations derived from this model concerning the mechanism of SecA protein translocation activity need to be confirmed in a more physiological environment. In a recent study, the oligomeric state and binding of SecA to SecYEG is investigated in equilibrium under physiological buffer conditions and SecA is found to bind as a dimer [105]. During catalysis, SecA appears to be dimeric as demonstrated by in vitro protein translocation activity assays. Intermolecular crosslinked SecA dimers were shown to be fully active in protein translocation [102, 108, 109] and more active than non-crosslinked species under conditions that favor monomerization [105]. Furthermore, mutants of SecA with a monomer–dimer equilibrium shifted towards the monomeric form display low or no protein activity [103, 107, 110, 111], and hetero-dimers composed of active and non-active SecA monomers were completely inactive [92]. The translocation chaperone SecB interacts with dimeric SecA [73, 112] and with the C-termini of both protomers [73, 113]. Moreover, SecA reaches maximal coupling of ATP hydrolysis with preprotein translocation when two of its protomers bind SecB [112]. The two SecA protomers were shown to bind SecB separately by a disulfide crosslinking study [114]. The multitude of contradicting results, methods and interpretations causes an ongoing debate on the oligomeric state of SecA during protein translocation. Therefore, it is important to focus this discussion on the SecA that is actively engaged in translocation and bound to SecYEG.
Energy-fueling translocation
After targeting of a preprotein to SecYEG bound SecA, energy in the form of both ATP and PMF is consumed at different stages of the translocation reaction [5, 115, 116]. ATP hydrolysis occurs at the DEAD-motor domain of SecA with ADP-release being the rate-limiting step for the subsequent catalysis [83, 117]. The ATPase activity of cytosolic SecA [86] is stimulated by binding to SecYEG and preproteins [58, 59] and is inhibited by azide [118]. Thus, ATP is hydrolyzed throughout the reaction and is essential for the initiation of protein translocation [5]. Although protein translocation through the Sec-system requires polypeptide chains that are largely unfolded, tightly folded protein domains can be translocated by SecA accompanied by elevated ATP consumption. Therefore, a chaperone-like and ATP-dependent unfolding activity has been proposed for SecA [64, 119].
The PMF is not essential for protein translocation but has stimulating effects on different stages of the reaction cycle. During initiation of preprotein translocation, the insertion and possibly also orientation of the signal-sequence in the translocation channel is affected by the PMF [120]. Protein translocation can be driven by the PMF alone when the inserted polypeptide chain is not attached to SecA [5, 6, 115, 121]. Furthermore, SecA de-insertion from the membrane (i.e. a SecA conformational change) is accelerated in presence of the PMF either by stimulating ADP release from SecA [122, 123] or by promoting conformational changes in SecY [14, 57, 115]. In the absence of PMF and SecA, translocating preproteins can slide backwards towards the cytoplasmic side of the channel [5, 120]. However, the PMF does not promote preprotein movement in an electrophoretic manner as stretches of both positively and negatively charged amino acids inhibit protein translocation [124]. Clusters of particularly positive amino acids appear to inhibit SecA’s ATPase activity by an as yet unknown mechanism. Overall, it seems as if the PMF supports the unidirectional movement of preproteins at stages where SecA does not contact the translocating polypeptide chain, and promotes the catalytic cycle by stimulating conformational changes in the translocon. In this respect, protein translocation by PrlA mutants shows a remarkably reduced PMF-dependence, and this has led to the suggestion that the PMF modulates the channel opening [57].
On the mechanism of SecA-mediated protein translocation
Brownian ratchet, power stroke, peristalsis and subunit recruitment models
Despite the multitude of structural and biochemical details revealed over the last two decades, the exact mechanism of SecA-mediated protein translocation is unknown. The suggested mechanisms can be summarized in four models: (1) Brownian ratchet; (2) power stroke; (3) peristalsis; and (4) subunit recruitment model. Besides this general mode of SecA function, the oligomeric states of SecA and SecYEG in these models are a matter of controversy. After reviewing the four conflicting previous models, we propose a new model combining core elements of all four models into one unifying hypothesis that is supported by recent data.
In analogy to protein import into the endoplasmatic reticulum, a Brownian ratchet mechanism was proposed for protein secretion [125]. Here, SecA would only mediate channel opening thereby enabling the preprotein to diffuse into the SecYEG pore. Spontaneous backsliding of the preprotein is prevented by ATP-dependent trapping by SecA, thus allowing diffusion only in one direction similar to the action suggested for BiP (for review, see [126]). This hypothesis was recently revived by a study that suggests that preprotein translocation occurs in a single rate-limiting step which is dependent on preprotein length [127]. This idea is opposed to another study in which the translocation rate is independent of the length of the polypeptide chain [128], but also by observations that translocation occurs in steps [4–6]. Obviously, the Brownian ratchet model cannot explain the observed step-size of protein translocation.
Based on early observations that preproteins are translocated in steps of defined size, SecA was suggested to act as a power stroke motor [5, 6, 128, 129]. According to this model, ATP hydrolysis and the resulting conformational changes are directly coupled to mechanical pushing of the polypeptide chain through the SecYEG channel. To accomplish mechanical pushing, multiple contacts between preprotein and SecA are comprehensible and support this hypothesis [69–71]. Furthermore, a structural domain of SecA, the so-called two-helix finger, was shown to interact with the preprotein during protein translocation [130]. In the SecA–SecYEG co-crystal of T. maritima this two-helix finger is partially inserted into the cytoplasmic opening of the channel (Fig. 2d) [29]. Furthermore, a tyrosine residue in this domain that can be substituted only by another bulky hydrophobic residue is important for protein translocation. It has been suggested that the tyrosine residue interacts with the translocating protein through side-chain interactions. However, the observation that long stretches of glycine residues can be translocated rather suggests main chain interactions as a key step [124]. Moreover, since the possible conformational change of the two-helix finger and the resulting lever arm are relatively small, a single movement of the two-helix finger alone could not drive the translocation of the observed 2- to 2.5-kDa segments [5]. Surprisingly, residues suggested to mediate dimerization in the M. tuberculosis SecA crystal reside in the two-helix finger (Fig. 2d).
A more defined power stroke model is the inchworm model previously proposed for monomeric DEAD helicases that move along nucleic acids by means of their two nucleotide binding sites (NBS). One of the NBS (henceforth called NBS1) is bound tightly to the DNA while the other NBS (NBS2) is bound weakly. Upon ATP binding and hydrolysis, the weakly bound NBS2 dissociates from the DNA and is moved to a position ahead on the DNA strand by a power stroke motion. At its new position, NBS2 forms a tight interaction with the DNA and the previously tightly bound NBS1 loosens its interaction. A new cycle of ATP binding and hydrolysis would move the NBS1 ahead on the DNA [81, 131, 132]. Since the DEAD motor of SecA shares a high homology with monomeric helicases, a similar model was proposed for SecA-mediated protein translocation [133]. In analogy to the helicases, the translocation machinery would transport the preprotein by means of two substrate binding sites of which one would anchor the polypeptide chain by a tight interaction and the other would dissociate from the substrate to grab a consecutive segment. Monomeric SecA, however, has only one preprotein binding site and thus could not move the preprotein alone. Therefore, the second binding site was proposed to reside on SecYEG [26, 107] although experimental evidence is missing. Importantly, in the absence of SecA, translocating polypeptides can move freely within the channel [5]; thus, SecYEG does not seem to form a stable anchor for the preprotein. Moreover, for the observed translocation of preprotein segments of approximately 85 Å length [5, 6], a very large conformational change of monomeric SecA would be necessary to provide a lever arm of suitable length. Dimeric SecA, on the other hand, has two peptide binding domains and movement of one protomer relative to the other may allow SecA to move along the polypeptide chain similarly to DEAD helicases with their two substrate binding sites. One SecA protomer could trap the polypeptide chain in the channel while the other protomer moves back to grab a consecutive segment of the preprotein.
A combination of power stroke and Brownian ratchet model is the peristalsis model. It suggests binding of dimeric SecA to a SecYEG dimer such that a large vestibule is created between channel and motor protein [60, 126]. A central opening as observed in the crystal structures of dimeric SecA of B. subtilis [85], M. tuberculosis [98] and T. thermophius [99] was proposed to trap the polypeptide chain of a preprotein after ATP binding to SecA. Subsequently, an ATP-driven power stroke motion would reduce the volume of the vestibule and thereby force the channel to open, enabling the trapped preprotein segments to diffuse into the SecYEG channel. Thus, the actual translocation reaction is driven by Brownian motion while the power stroke merely promotes channel opening. Backsliding of the preprotein is prevented by closure of the central opening in the SecA dimer. This peristalsis model assumes the symmetric docking of the two SecA protomers onto two copies of the SecYEG channel. The orientation of SecA and SecYEG in the co-crystal, however, is opposed to the idea of a symmetric SecA2–SecYEG2 complex. Thus, new structural data are required to support this hypothesis.
A different model based on numerous observations on the sensitive nature of the SecA monomer–dimer equilibrium suggests a subunit recruitment mechanism [106]. This model is similar to the ‘active rolling’ model proposed for ATP-dependent helicases that are homologous to SecA. In this ‘rolling’ model, a monomeric helicase is bound to the nucleic acid polymer and recruits another protomer to bind the upstream segment of the nucleic acid strand. Thus, movement along the polymer is mediated by oligomerization of the helicases that only appears in presence of nucleic acids [134–136]. The observation that phospholipids induce monomerization of SecA while the presence of signal-peptides causes SecA to dimerize has lead to the hypothesis that polypeptide transport is mediated by recruitment of a second SecA protomer onto a SecYEG bound SecA monomer [106]. However, the presence of synthetic signal peptides caused monomerization of SecA in two other studies [107, 137], but it should be emphasized that these were observations in the absence of SecYEG involving non-physiological amounts of synthetic signal peptide. Rather, in the presence of SecYEG and a translocating preprotein, SecA does not seem to monomerize [105]. Yet, the sensitive nature of the SecA monomer–dimer equilibrium that is maintained in the SecYEG bound state [105] may support a subunit recruitment mechanism.
The reciprocating piston model
A unifying model would have to include a protein translocation cycle comprising two discrete steps, one ATP-dependent and one that only requires SecA binding to the translocon [5, 6]. ATP binding to SecA is thought to fuel a power stroke motion whereas the translocation event upon SecA (re-)binding likely underlies a different mechanism. Another element of our model are conformational changes in SecA that have been interpreted as insertion–deinsertion cycle as large parts of SecA become accessible from the periplasm for small probes [138] and are protected against proteases [4, 139]. Considering the inner dimensions of SecYEG and the bulkiness of SecA, it is unlikely that the latter penetrates entirely and deep into the channel or even reaches the periplasm. The conformational changes in SecA may trigger a partial channel opening, thus allowing small molecules to diffuse through SecYEG. The previously interpreted inserted state could result from a densely packed conformation that is resistant against the applied proteases. However, the two-helix finger of SecA inserts partially into the entry funnel of SecYEG in the SecA–SecYEG co-crystal [29] which may represent an intermediate state of the conformational cycle that SecA undergoes during protein translocation. As previously mentioned, several studies have come to the conclusion that SecA functions as a dimer [92, 102, 108, 110] while other studies suggest that monomeric SecA plays a role in protein translocation [29, 103, 107]. SecA was found to bind SecYEG in both monomeric [15, 29, 107] and dimeric form [15, 16, 102, 110], and the SecA monomer–dimer equilibrium is sensitive to a wide range of ligands and agents such as salt and detergents (see previous section). Together with the observation that SecA dissociates from the preprotein upon ATP hydrolysis [5], these data suggest that protein translocation includes cycling of SecA via a monomeric intermediate.
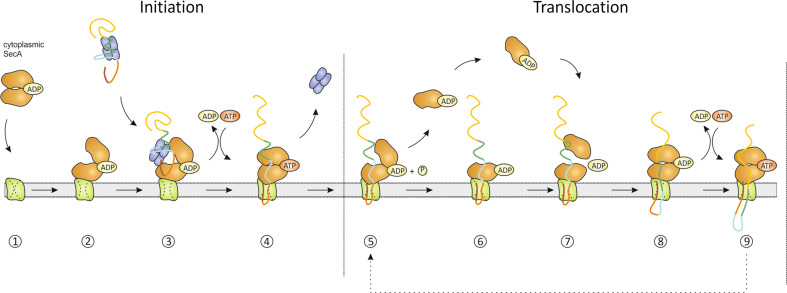
In an attempt to unify previous conflicting studies while integrating recent developments, we propose a reciprocating piston model for protein translocation (Fig. 3). First, a cytosolic SecA dimer binds with high affinity to the SecYEG channel (step 1) [102, 110] leading to a conformational change and an elevated ATPase activity of SecA (step 2) [58, 59]. Based on recent data [105], we propose a model of dimeric SecA binding to SecYEG where one SecA is anchored to SecYEG while the other protomer interacts with the bound SecA alone. Interestingly, the preprotein binding domain (PBD) of SecYEG-bound SecA undergoes a dramatic conformational change resulting in a clamp-like structure (compare Fig. 2a, b) [29]. Docking of a second SecA protomer onto the SecA–SecYEG structure brings both PBDs into close proximity enabling the transfer of polypeptide segments from one SecA protomer to the other [105]. In the cytosol, the molecular chaperone SecB maintains the translocation competent state of newly synthesized preproteins by binding to their mature region thereby keeping them partially unfolded (for SecB review, see [140]). Homo-tetrameric SecB targets the preprotein to SecYEG bound SecA [2] by interacting with both C-termini of the SecA dimer (step 3) [73, 112, 113]. The SecA protomers bind SecB separately (step 3) [114]. Upon binding of ATP to SecA, signal sequence and adjacent mature region of the preprotein are inserted into the SecYEG channel in a hairpin-like structure (step 4) [141–143]. During this step, dependent on ATP binding to SecA, SecB is released from the complex [73] which completes the initiation of protein translocation. ATP hydrolysis causes dissociation of SecA from the preprotein [5] and possibly also monomerization of the SecA dimer whereas one SecA protomer remains bound to SecYEG preventing backsliding of the partially translocated polypeptide chain (step 5). Additionally, ATP hydrolysis leads to the conformational change in the SecYEG bound SecA that has been interpreted as de-insertion (step 6) [4], a step that is accelerated by the PMF [14, 57, 115, 122, 123]. The dissociated SecA monomer can exchange with the soluble SecA pool or re-bind to the trapped preprotein (step 7) and the SecYEG bound SecA protomer would cause the observed ATP-independent translocation of a 2- to 2.5-kDa preprotein segment (step 8) [5, 6]. This step may resemble elements of the previously proposed peristalsis model [60] and subunit recruitment mechanism [106]. Binding of an ‘upstream’ preprotein segment by the soluble SecA protomer and subsequent dimerization could compress the polypeptide chain in the complex thereby forcing it into the channel. Here, Brownian motion would drive movement of the polypeptide chain while directionality is given by the bound, previously soluble SecA protomer. Next, binding of ATP to SecA results in a power stroke motion accompanied by insertion of SecA into SecYEG [4, 29, 139] and translocation of another 2- to 2.5-kDa preprotein segment (step 9). The steps 5–9 are repeated until the preprotein is fully translocated across the membrane. In total, 4–5 kDa of the polypeptide chain are transported in each catalytic cycle, which corresponds to around 25–30 amino acids [5, 6]. To elucidate the exact mechanism of SecA function, in particular the ATP-independent translocation step, new biophysical techniques are required that enable the analysis of protein translocation at the single molecule level with time resolution.
Fig. 3.
The reciprocating piston model of SecA-mediated protein translocation. The polypeptide chain of the preprotein is colored differently to illustrate distinct segments that are translocated in steps of defined size
Acknowledgments
This work was funded by NanoNed, a national nanotechnology program coordinated by the Dutch Ministry of Economical Affairs and the Zernike Institute for Advanced Materials, and by the Chemical Sciences division of The Netherlands Organization for Scientific Research (NWO-CW). We would like to thank A. Kedrov for valuable comments on this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Josefsson LG, Randall LL. Processing in vivo of precursor maltose-binding protein in Escherichia coli occurs post-translationally as well as co-translationally. J Biol Chem. 1981;256(5):2504–2507. [PubMed] [Google Scholar]
- 2.Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63(2):269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 3.Fekkes P, de Wit JG, van der Wolk JP, Kimsey HH, Kumamoto CA, Driessen AJ. Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29(5):1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 4.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78(5):835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 5.Schiebel E, Driessen AJ, Hartl FU, Wickner W. Delta mu H + and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64(5):927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 6.van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16(24):7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr Opin Struct Biol. 2005;15(2):213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Gumbart J, Schulten K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 2007;46(39):11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 9.Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418(6898):662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 10.Murphy CK, Beckwith J. Residues essential for the function of SecE, a membrane component of the Escherichia coli secretion apparatus, are located in a conserved cytoplasmic region. Proc Natl Acad Sci USA. 1994;91(7):2557–2561. doi: 10.1073/pnas.91.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427(6969):36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 12.Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187(18):6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flower AM. The SecY translocation complex: convergence of genetics and structure. Trends Microbiol. 2007;15(5):203–210. doi: 10.1016/j.tim.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Bessonneau P, Besson V, Collinson I, Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21(5):995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22(17):4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tziatzios C, Schubert D, Lotz M, Gundogan D, Betz H, Schagger H, Haase W, Duong F, Collinson I. The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J Mol Biol. 2004;340(3):513–524. doi: 10.1016/j.jmb.2004.04.076. [DOI] [PubMed] [Google Scholar]
- 17.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, 3rd, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438(7066):318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer TH, Menetret JF, Breitling R, Miller KR, Akey CW, Rapoport TA. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol. 1999;285(4):1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- 19.Manting EH, van Der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19(5):852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheuring J, Braun N, Nothdurft L, Stumpf M, Veenendaal AK, Kol S, van der Does C, Driessen AJ, Weinkauf S. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J Mol Biol. 2005;354(2):258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 21.Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87(4):721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 22.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278(5346):2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 23.Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107(3):361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 24.Menetret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6(5):1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 25.Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89(4):535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 26.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129(1):97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 27.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284(23):15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326(5958):1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455(7215):936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deville K, Gold VA, Robson A, Whitehouse S, Sessions RB, Baldwin SA, Radford SE, Collinson I. The oligomeric state and arrangement of the active bacterial translocon. J Biol Chem. 2011;286(6):4659–4669. doi: 10.1074/jbc.M110.175638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA. 2006;103(44):16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Sluis EO, Nouwen N, Koch J, de Keyzer J, van der Does C, Tampe R, Driessen AJ. Identification of two interaction sites in SecY that are important for the functional interaction with SecA. J Mol Biol. 2006;361(5):839–849. doi: 10.1016/j.jmb.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455(7215):988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagamori S, Nishiyama K, Tokuda H. Membrane topology inversion of SecG detected by labeling with a membrane-impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J Biochem. 2002;132(4):629–634. doi: 10.1093/oxfordjournals.jbchem.a003266. [DOI] [PubMed] [Google Scholar]
- 35.Mori H, Ito K. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc Natl Acad Sci USA. 2001;98(9):5128–5133. doi: 10.1073/pnas.081617398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Keyzer J, Regeling A, Driessen AJ. Arginine 357 of SecY is needed for SecA-dependent initiation of preprotein translocation. FEBS Lett. 2007;581(9):1859–1864. doi: 10.1016/j.febslet.2007.03.081. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol. 2005;168(1):67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raden D, Song W, Gilmore R. Role of the cytoplasmic segments of Sec61alpha in the ribosome-binding and translocation-promoting activities of the Sec61 complex. J Cell Biol. 2000;150(1):53–64. doi: 10.1083/jcb.150.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menetret JF, Schaletzky J, Clemons WM, Jr, Osborne AR, Skanland SS, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, Rapoport TA, Akey CW. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol Cell. 2007;28(6):1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B, Sourjik V, Koch HG (2011) The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic (in press) [DOI] [PubMed]
- 41.Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102(2):233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 42.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94(6):795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 43.Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci USA. 2010;107(40):17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris CR, Silhavy TJ. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol. 1999;181(11):3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24(19):3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lycklama ANJA, Bulacu M, Marrink SJ, Driessen AJ. Immobilization of the plug domain inside the SecY channel allows unrestricted protein translocation. J Biol Chem. 2010;285(31):23747–23754. doi: 10.1074/jbc.M110.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maillard AP, Lalani S, Silva F, Belin D, Duong F. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J Biol Chem. 2007;282(2):1281–1287. doi: 10.1074/jbc.M610060200. [DOI] [PubMed] [Google Scholar]
- 48.Saparov SM, Erlandson K, Cannon K, Schaletzky J, Schulman S, Rapoport TA, Pohl P. Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell. 2007;26(4):501–509. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 49.Junne T, Schwede T, Goder V, Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol Biol Cell. 2006;17(9):4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26(4):511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem. 2007;282(45):33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- 52.Bol R, de Wit JG, Driessen AJ. The active protein-conducting channel of Escherichia coli contains an apolar patch. J Biol Chem. 2007;282(41):29785–29793. doi: 10.1074/jbc.M702140200. [DOI] [PubMed] [Google Scholar]
- 53.Bieker-Brady K, Silhavy TJ. Suppressor analysis suggests a multistep, cyclic mechanism for protein secretion in Escherichia coli . EMBO J. 1992;11(9):3165–3174. doi: 10.1002/j.1460-2075.1992.tb05393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danese PN, Silhavy TJ. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli . Annu Rev Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 55.Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 1999;18(12):3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Wolk JP, Fekkes P, Boorsma A, Huie JL, Silhavy TJ, Driessen AJ. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 1998;17(13):3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci USA. 1996;93(12):5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60(2):271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 59.Karamanou S, Gouridis G, Papanikou E, Sianidis G, Gelis I, Keramisanou D, Vrontou E, Kalodimos CG, Economou A. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26(12):2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 61.Miller A, Wang L, Kendall DA. SecB modulates the nucleotide-bound state of SecA and stimulates ATPase activity. Biochemistry. 2002;41(16):5325–5332. doi: 10.1021/bi025639p. [DOI] [PubMed] [Google Scholar]
- 62.Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004;1694(1–3):67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 63.Gouridis G, Karamanou S, Gelis I, Kalodimos CG, Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462(7271):363–367. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eser M, Ehrmann M. SecA-dependent quality control of intracellular protein localization. Proc Natl Acad Sci USA. 2003;100(23):13231–13234. doi: 10.1073/pnas.2234410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akita M, Shinkai A, Matsuyama S, Mizushima S. SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer. Biochem Biophys Res Commun. 1991;174(1):211–216. doi: 10.1016/0006-291x(91)90507-4. [DOI] [PubMed] [Google Scholar]
- 66.Moran U, Phillips R, Milo R. SnapShot: key numbers in biology. Cell. 2010;141(7):1262–1262. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 67.Matsuyama S, Kimura E, Mizushima S. Complementation of two overlapping fragments of SecA, a protein translocation ATPase of Escherichia coli, allows ATP binding to its amino-terminal region. J Biol Chem. 1990;265(15):8760–8765. [PubMed] [Google Scholar]
- 68.Sardis MF, Economou A. SecA: a tale of two protomers. Mol Microbiol. 2010;76(5):1070–1081. doi: 10.1111/j.1365-2958.2010.07176.x. [DOI] [PubMed] [Google Scholar]
- 69.Musial-Siwek M, Rusch SL, Kendall DA. Selective photoaffinity labeling identifies the signal peptide binding domain on SecA. J Mol Biol. 2007;365(3):637–648. doi: 10.1016/j.jmb.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gelis I, Bonvin AM, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131(4):756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Papanikou E, Karamanou S, Baud C, Frank M, Sianidis G, Keramisanou D, Kalodimos CG, Kuhn A, Economou A. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280(52):43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- 72.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382(1):74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fekkes P, van der Does C, Driessen AJ. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16(20):6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou J, Xu Z. Structural determinants of SecB recognition by SecA in bacterial protein translocation. Nat Struct Biol. 2003;10(11):942–947. doi: 10.1038/nsb980. [DOI] [PubMed] [Google Scholar]
- 75.Fekkes P, de Wit JG, Boorsma A, Friesen RH, Driessen AJ. Zinc stabilizes the SecB binding site of SecA. Biochemistry. 1999;38(16):5111–5116. doi: 10.1021/bi982818r. [DOI] [PubMed] [Google Scholar]
- 76.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270(14):7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 77.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5(11):839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 78.Koonin EV, Gorbalenya AE. Autogenous translation regulation by Escherichia coli ATPase SecA may be mediated by an intrinsic RNA helicase activity of this protein. FEBS Lett. 1992;298(1):6–8. doi: 10.1016/0014-5793(92)80009-6. [DOI] [PubMed] [Google Scholar]
- 79.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 80.Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10(3):483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 81.Ye J, Osborne AR, Groll M, Rapoport TA. RecA-like motor ATPases–lessons from structures. Biochim Biophys Acta. 2004;1659(1):1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 82.Sato K, Mori H, Yoshida M, Mizushima S. Characterization of a potential catalytic residue, Asp-133, in the high affinity ATP-binding site of Escherichia coli SecA, translocation ATPase. J Biol Chem. 1996;271(29):17439–17444. doi: 10.1074/jbc.271.29.17439. [DOI] [PubMed] [Google Scholar]
- 83.Sianidis G, Karamanou S, Vrontou E, Boulias K, Repanas K, Kyrpides N, Politou AS, Economou A. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20(5):961–970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol. 2007;366(5):1545–1557. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 85.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297(5589):2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 86.Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34(5):1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 87.Keramisanou D, Biris N, Gelis I, Sianidis G, Karamanou S, Economou A, Kalodimos CG. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat Struct Mol Biol. 2006;13(7):594–602. doi: 10.1038/nsmb1108. [DOI] [PubMed] [Google Scholar]
- 88.Vrontou E, Karamanou S, Baud C, Sianidis G, Economou A. Global co-ordination of protein translocation by the SecA IRA1 switch. J Biol Chem. 2004;279(21):22490–22497. doi: 10.1074/jbc.M401008200. [DOI] [PubMed] [Google Scholar]
- 89.Mori H, Ito K. The long alpha-helix of SecA is important for the ATPase coupling of translocation. J Biol Chem. 2006;281(47):36249–36256. doi: 10.1074/jbc.M606906200. [DOI] [PubMed] [Google Scholar]
- 90.Economou A. Structural biology: clamour for a kiss. Nature. 2008;455(7215):879–880. doi: 10.1038/455879a. [DOI] [PubMed] [Google Scholar]
- 91.Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11(4):875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Driessen AJ. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32(48):13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 93.Wang HW, Chen Y, Yang H, Chen X, Duan MX, Tai PC, Sui SF. Ring-like pore structures of SecA: implication for bacterial protein-conducting channels. Proc Natl Acad Sci USA. 2003;100(7):4221–4226. doi: 10.1073/pnas.0737415100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Y, Tai PC, Sui SF. The active ring-like structure of SecA revealed by electron crystallography: conformational change upon interaction with SecB. J Struct Biol. 2007;159(1):149–153. doi: 10.1016/j.jsb.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen Y, Pan X, Tang Y, Quan S, Tai PC, Sui SF. Full-length Escherichia coli SecA dimerizes in a closed conformation in solution as determined by cryo-electron microscopy. J Biol Chem. 2008;283(43):28783–28787. doi: 10.1074/jbc.C800160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shilton B, Svergun DI, Volkov VV, Koch MH, Cusack S, Economou A. Escherichia coli SecA shape and dimensions. FEBS Lett. 1998;436(2):277–282. doi: 10.1016/s0014-5793(98)01141-7. [DOI] [PubMed] [Google Scholar]
- 97.Bu Z, Wang L, Kendall DA. Nucleotide binding induces changes in the oligomeric state and conformation of Sec A in a lipid environment: a small-angle neutron-scattering study. J Mol Biol. 2003;332(1):23–30. doi: 10.1016/s0022-2836(03)00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR, Jr, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100(5):2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vassylyev DG, Mori H, Vassylyeva MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol. 2006;364(3):248–258. doi: 10.1016/j.jmb.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 100.Zimmer J, Li W, Rapoport TA. A novel dimer interface and conformational changes revealed by an X-ray structure of B subtilis SecA. J Mol Biol. 2006;364(3):259–265. doi: 10.1016/j.jmb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 101.Wowor AJ, Yu D, Kendall DA, Cole JL (2011) Energetics of SecA Dimerization. J Mol Biol (in press) [DOI] [PMC free article] [PubMed]
- 102.de Keyzer J, van der Sluis EO, Spelbrink RE, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJ. Covalently dimerized SecA is functional in protein translocation. J Biol Chem. 2005;280(42):35255–35260. doi: 10.1074/jbc.M506157200. [DOI] [PubMed] [Google Scholar]
- 103.Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280(10):9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 104.Ding H, Mukerji I, Oliver D. Nucleotide and phospholipid-dependent control of PPXD and C-domain association for SecA ATPase. Biochemistry. 2003;42(46):13468–13475. doi: 10.1021/bi035099b. [DOI] [PubMed] [Google Scholar]
- 105.Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Driessen AJ. Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure. 2011;19(3):430–439. doi: 10.1016/j.str.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 106.Benach J, Chou YT, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J Biol Chem. 2003;278(6):3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]
- 107.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21(17):4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jilaveanu LB, Oliver D. SecA dimer cross-linked at its subunit interface is functional for protein translocation. J Bacteriol. 2006;188(1):335–338. doi: 10.1128/JB.188.1.335-338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H, Na B, Yang H, Tai PC. Additional in vitro and in vivo evidence for SecA functioning as dimers in the membrane: dissociation into monomers is not essential for protein translocation in Escherichia coli . J Bacteriol. 2008;190(4):1413–1418. doi: 10.1128/JB.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jilaveanu LB, Zito CR, Oliver D. Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci USA. 2005;102(21):7511–7516. doi: 10.1073/pnas.0502774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJ. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol. 2005;348(2):479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 112.Mao C, Hardy SJ, Randall LL. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J Bacteriol. 2009;191(3):978–984. doi: 10.1128/JB.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Randall LL, Crane JM, Liu G, Hardy SJ. Sites of interaction between SecA and the chaperone SecB, two proteins involved in export. Protein Sci. 2004;13(4):1124–1133. doi: 10.1110/ps.03410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suo Y, Hardy SJ, Randall LL. Orientation of SecA and SecB in complex, derived from disulfide cross-linking. J Bacteriol. 2011;193(1):190–196. doi: 10.1128/JB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tani K, Tokuda H, Mizushima S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli effect of membrane energization. J Biol Chem. 1990;265(28):17341–17347. [PubMed] [Google Scholar]
- 116.Driessen AJ. Precursor protein translocation by the Escherichia coli translocase is directed by the protonmotive force. EMBO J. 1992;11(3):847–853. doi: 10.1002/j.1460-2075.1992.tb05122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fak JJ, Itkin A, Ciobanu DD, Lin EC, Song XJ, Chou YT, Gierasch LM, Hunt JF. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43(23):7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 118.Chun SY, Randall LL. In vivo studies of the role of SecA during protein export in Escherichia coli . J Bacteriol. 1994;176(14):4197–4203. doi: 10.1128/jb.176.14.4197-4203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nouwen N, Berrelkamp G, Driessen AJ. Bacterial sec-translocase unfolds and translocates a class of folded protein domains. J Mol Biol. 2007;372(2):422–433. doi: 10.1016/j.jmb.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 120.van Dalen A, Killian A, de Kruijff B. Delta psi stimulates membrane translocation of the C-terminal part of a signal sequence. J Biol Chem. 1999;274(28):19913–19918. doi: 10.1074/jbc.274.28.19913. [DOI] [PubMed] [Google Scholar]
- 121.Driessen AJ, Wickner W. Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc Natl Acad Sci USA. 1991;88(6):2471–2475. doi: 10.1073/pnas.88.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nishiyama K, Fukuda A, Morita K, Tokuda H. Membrane deinsertion of SecA underlying proton motive force-dependent stimulation of protein translocation. EMBO J. 1999;18(4):1049–1058. doi: 10.1093/emboj/18.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shiozuka K, Tani K, Mizushima S, Tokuda H. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli . J Biol Chem. 1990;265(31):18843–18847. [PubMed] [Google Scholar]
- 124.Nouwen N, Berrelkamp G, Driessen AJ. Charged amino acids in a preprotein inhibit SecA-dependent protein translocation. J Mol Biol. 2009;386(4):1000–1010. doi: 10.1016/j.jmb.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 125.Simon SM, Peskin CS, Oster GF. What drives the translocation of proteins? Proc Natl Acad Sci USA. 1992;89(9):3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tomkiewicz D, Nouwen N, Driessen AJ. Pushing, pulling and trapping–modes of motor protein supported protein translocation. FEBS Lett. 2007;581(15):2820–2828. doi: 10.1016/j.febslet.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 127.Liang FC, Bageshwar UK, Musser SM. Bacterial Sec protein transport is rate-limited by precursor length: a single turnover study. Mol Biol Cell. 2009;20(19):4256–4266. doi: 10.1091/mbc.E09-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tomkiewicz D, Nouwen N, van Leeuwen R, Tans S, Driessen AJ. SecA supports a constant rate of preprotein translocation. J Biol Chem. 2006;281(23):15709–15713. doi: 10.1074/jbc.M600205200. [DOI] [PubMed] [Google Scholar]
- 129.Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli . J Biol Chem. 1995;270(52):30862–30868. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 130.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455(7215):984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281(27):18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 132.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97(1):75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 133.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA. 2004;101(30):10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90(4):635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 135.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 136.Wong I, Lohman TM. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256(5055):350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 137.Musial-Siwek M, Rusch SL, Kendall DA. Probing the affinity of SecA for signal peptide in different environments. Biochemistry. 2005;44(42):13987–13996. doi: 10.1021/bi050882k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jilaveanu LB, Oliver DB. In vivo membrane topology of Escherichia coli SecA ATPase reveals extensive periplasmic exposure of multiple functionally important domains clustering on one face of SecA. J Biol Chem. 2007;282(7):4661–4668. doi: 10.1074/jbc.M610828200. [DOI] [PubMed] [Google Scholar]
- 139.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83(7):1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 140.Bechtluft P, Nouwen N, Tans SJ, Driessen AJ. SecB–a chaperone dedicated to protein translocation. Mol Biosyst. 2010;6(4):620–627. doi: 10.1039/b915435c. [DOI] [PubMed] [Google Scholar]
- 141.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 142.Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169(2):219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joly JC, Wickner W. The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J. 1993;12(1):255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]