Abstract
The relationship between efflux system overexpression and cross-resistance to cefoxitin, quinolones, and chloramphenicol has recently been reported in Klebsiella pneumoniae. In 3 previously published clinical isolates and 17 in vitro mutants selected with cefoxitin or fluoroquinolones, mutations in the potential regulator genes of the AcrAB efflux pump (acrR, ramR, ramA, marR, marA, soxR, soxS, and rob) were searched, and their impacts on efflux-related antibiotic cross-resistance were assessed. All mutants but 1, and 2 clinical isolates, overexpressed acrB. No mutation was detected in the regulator genes studied among the clinical isolates and 8 of the mutants. For the 9 remaining mutants, a mutation was found in the ramR gene in 8 of them and in the soxR gene in the last one, resulting in overexpression of ramA and soxS, respectively. Transformation of the ramR mutants and the soxR mutant with the wild-type ramR and soxR genes, respectively, abolished overexpression of acrB and ramA in the ramR mutants and of soxS in the soxR mutant, as well as antibiotic cross-resistance. Resistance due to efflux system overexpression was demonstrated for 4 new antibiotics: cefuroxime, cefotaxime, ceftazidime, and ertapenem. This study shows that the ramR and soxR genes control the expression of efflux systems in K. pneumoniae and suggests the existence of efflux pumps other than AcrAB and of other loci involved in the regulation of AcrAB expression.
INTRODUCTION
Klebsiella pneumoniae is an important pathogen, both in the community and in the hospital setting, and is responsible for a variety of infections, including urinary tract infections, pneumonia, liver abscesses and bacteremia (18, 34). Acquired resistance to β-lactams in this bacterial species can be due to expression of different enzymes, such as extended-spectrum β-lactamases (ESBLs) (39), plasmid-mediated AmpC β-lactamases (30), and, more recently, carbapenemases (45). Moreover, two other nonenzymatic mechanisms participate in resistance to β-lactams in K. pneumoniae. One of them is porin alteration, which has been involved in resistance to cefoxitin and, more recently, to ertapenem in mutants producing the ESBL CTX-M-15 (20). The second is efflux system overexpression, which has recently been shown to induce low-level cross-resistance to different antibiotic families, including quinolones, chloramphenicol, and β-lactams (28, 29). This efflux-related antibiotic cross-resistance has been observed in almost 5% of the non-plasmid-mediated β-lactamase-producing K. pneumoniae clinical isolates in our hospital since 2000 (29).
The efflux system most extensively studied to date is the AcrAB pump, which belongs to the resistance-nodulation-division (RND) family and is associated with the outer membrane protein TolC. AcrAB-TolC is present in various bacterial species belonging to the family Enterobacteriaceae, such as Salmonella enterica, Enterobacter aerogenes, Enterobacter cloacae, Escherichia coli, Morganella morganii, Proteus mirabilis, and K. pneumoniae (21, 26, 31, 33, 35, 36, 41). Substrates for this efflux system include quinolones, chloramphenicol, macrolides, tetracycline, tigecycline, trimethoprim, and β-lactams (3, 23, 32, 36, 37, 38). Besides its implication in antibiotic resistance, the participation of the AcrAB efflux system in bacterial virulence has been demonstrated in S. enterica, E. coli, and, recently, K. pneumoniae (3, 6, 15, 28).
More insight into the regulatory pathways that control expression of the AcrAB-TolC system in some Enterobacteriaceae species has been gained over the last few years. Expression of the acrAB operon is regulated by its local repressor, AcrR (32). At a global level, it is also influenced by several transcriptional activators belonging to the AraC/XylS family, especially MarA, RamA, SoxS, and Rob (2, 12). Transcription of the marRAB locus is activated by the binding of MarA to the marO operator region, whereas it is repressed by the binding of MarR (9, 40). More recently, the local repressor of RamA, called RamR, whose gene is found directly upstream of ramA in the opposite orientation, has been identified in S. enterica (1). SoxS is the effector of the soxRS global superoxide response regulon, and its expression can be activated by its regulator, SoxR, present in its oxidized form (43). It has also been shown that mutations in the regulator genes acrR, marR, ramR, and soxR can result in overexpression of the acrAB operon and contribute to a multidrug-resistant phenotype (1, 19, 24, 27, 38, 42). However, these systems have been studied mainly in E. coli and S. enterica, and little is known about their roles in K. pneumoniae, and the effects of mutations in the regulator genes on resistance to β-lactams are still unclear.
Therefore, the present study had a triple objective: (i) assessing the effect of antibiotic pressure by cefoxitin, on one hand, and by fluoroquinolones, on the other hand, on in vitro selection of resistant mutants overexpressing an efflux system from fully susceptible K. pneumoniae strains; (ii) searching for the presence of mutations in the regulator genes acrR, ramR, ramA, marR, marA, soxR, soxS, and rob in clinical isolates and in in vitro-selected mutants displaying an efflux-related antibiotic cross-resistance pattern; and (iii) determining the impacts of these mutations on the susceptibilities of the different strains to a large panel of antibiotics.
MATERIALS AND METHODS
Bacterial strains.
The collection studied comprised (i) 3 previously published K. pneumoniae clinical isolates shown to have cross-resistance to cefoxitin, quinolones, and chloramphenicol (KPBj E+), with increased expression of the acrB gene for 2 of them (KPBj1 E+ and KPBj3 E+) (3, 29); (ii) their spontaneous revertants (KPBj Rev) susceptible to the three antibiotic families (3); and (iii) mutants selected from these 3 revertant strains with cefoxitin, ciprofloxacin, or levofloxacin. For all experiments, K. pneumoniae strain ATCC 138821 was used as a control.
In vitro selection of mutants.
Mutants were selected by using the procedure previously described by Miller et al. (22). Briefly, 5 μl of an overnight culture was transferred into 9 ml of fresh Mueller-Hinton (MH) broth containing ciprofloxacin (Sigma Aldrich, Saint-Quentin-Fallavier, France), levofloxacin (Sigma Aldrich), or cefoxitin (Panpharma, Fougères, France) at 0.25× MIC for the strain tested and incubated for 18 h at 37°C. Such growth conditions were repeated until mutants were obtained from overnight culture aliquots plated after each broth culture cycle on MH agar containing 4× the original MIC.
Antibiotic susceptibility.
The susceptibilities of the in vitro-selected mutants to the well-known markers of efflux system overexpression (i.e., cefoxitin, nalidixic acid, and chloramphenicol) were determined by using the agar disk diffusion method. Only the mutants displaying cross-resistance to these 3 antibiotics were further studied. Susceptibility to a large panel of antibiotics, including β-lactams (cefoxitin, amoxicillin, and amoxicillin associated with clavulanic acid, ticarcillin, cefazolin, cefuroxime, cefotaxime, ceftazidime, ertapenem, imipenem, meropenem, and doripenem), quinolones (nalidixic acid, ciprofloxacin, and levofloxacin), chloramphenicol, tetracycline, and tigecycline, was determined by the agar dilution method. MIC determination, replicated 3 times, was performed and interpreted following the recommendations of the French Antibiogram Committee [http://www.sfm.asso.fr/nouv/general.php?pa=2].
Real-time RT-PCR for analysis of gene expression.
The transcription (mRNA) levels of the acrB, ramA, and soxS genes were determined according to the method previously described by Doumith et al. (10). Briefly, total cellular RNA was extracted using the RNeasy Mini kit (Qiagen, Courtaboeuf, France) and treated with RNase-free DNase (Qiagen) for 30 min at 37°C, after which a second step of purification was performed. Reverse transcription (RT)-PCR was carried out in a LightCycler using the one-step LightCycler RNA Master SYBR green I kit (Roche Applied Science, Meylan, France) and the primers listed in Table 1. The specificity of the generated products was tested by melting-point analysis. Amplifications were performed in duplicate from two different RNA preparations. The cycle threshold (CT) values of the target genes were compared with the CT values of the housekeeping rpoB gene, chosen as an endogenous reference for normalizing the transcription levels of the target genes. Strain ATCC 138821 was used as a control, and the normalized relative expression of the acrB, ramA, and soxS genes was determined for each strain according to the following formula: 2−ΔΔCT, where ΔΔCT = (CT − CTrpoB)studied strain − (CT − CTrpoB)control strain.
Table 1.
Primers used in this study
| Primer use and target region | Primer | Oligonucleotide sequence | Tm (°C)a | Reference |
|---|---|---|---|---|
| Sequencing | ||||
| gyrA | gyrA6 | 5′-CGACCTTGCGAGAGAAAT-3′ | 56 | 7 |
| gyrA631 | 5′-GTTCCATCAGCCCTTCAA-3′ | |||
| parC | parC F | 5′-CTGAACGCCAGCGCGAAATT-3′ | 58 | 7 |
| parC R | 5′-TGCGGTGGAATATCGGTCGC-3′ | |||
| acrR | acrR F | 5′-GCTAAGCTGCCTGAGAGCAT-3′ | 58 | This study |
| acrR R | 5′-ATGCAAATGCCGGAGAATAC-3′ | |||
| ramR | ramR F | 5′-CACGGTTCATATCCTGACCA-3′ | 60 | This study |
| ramR R | 5′-CCRTCGACCTTAAACACGTC-3′ | |||
| ramA | ramA F | 5′-TGGGATGAACCGTATCAACG-3′ | 58 | This study |
| ramA R | 5′-ATCTTACTGCTGGCCCTGCT-3′ | |||
| marA-marR | loc mar F | 5′-CATAGCTGAGGCTGGAGRCC-3′ | 56 | This study |
| loc mar R | 5′-TCGGCCAATTCATAATGTTG-3′ | |||
| soxS-soxR | loc sox F | 5′-CGGAACCTCCATCAACAGATT-3′ | 60 | This study |
| loc sox R | 5′-GCAGGTAAGCTGGCTCTACAA-3′ | |||
| rob | rob F | 5′-TCACGCACTTAGCAGAAAAGG-3′ | 60 | This study |
| rob R | 5′-ACTATCAGCAAAGCCCGTGG-3′ | |||
| RT-PCR | ||||
| acrB | acrB F | 5′-CGATAACCTGATGTACATGTCC-3′ | 60 | 10 |
| acrB R | 5′-CCGACAACCATCAGGAAGCT-3′ | |||
| ramA | ramA F | 5′-ATCGTCGAGTGGATTGATGA-3′ | 60 | 5 |
| ramA R | 5′-AGATGCCATTTCGAATACCC-3′ | |||
| soxS | soxS F | 5′-GCATCACGGTACGGAACAT-3′ | 60 | 5 |
| soxS R | 5′-AGTCGCCAGAAAGTCAGGAT-3′ | |||
| rpoB | rpoB F | 5′-AAGGCGAATCCAGCTTGTTCAGC-3′ | 60 | 10 |
| rpoB R | 5′-TGACGTTGCATGTTCGCACCCATCA-3′ | |||
| Cloning | ||||
| ramR | ramR F | 5′-CACGGTTCATATCCTGACCA-3′ | 60 | This study |
| ramR R | 5′-CCRTCGACCTTAAACACGTC-3′ | |||
| soxR | soxR F | 5′-AACCAGCGAGATAATGCGAA-3′ | 53 | This study |
| soxR R | 5′-ATAAAGCGGCCTCTCTCAAT-3′ |
Analysis of gene sequences.
The presence of mutations in the gyrA and parC genes, which encode targets for quinolones, and in the acrR, ramR, ramA, marR, marA, soxR, soxS, and rob genes was assessed by PCR and sequencing. The sequences of the primers used, as well as the annealing temperature (Tm), for each gene are shown in Table 1 (7). PCR was performed with a 0.2 μM concentration of each primer, a 500 μM concentration of the deoxynucleoside triphosphates (dNTPs), 1× PCR buffer with MgCl2, and 2.5 U of Taq DNA polymerase per 50-μl reaction mixture. After a 5-min denaturation at 95°C, amplification was performed for 35 cycles of 1 min at 95°C, 1 min at Tm, and 1 min at 72°C, with a final extension of 10 min at 72°C. After purification of the PCR products on Bio-Gel P-100 polyacrylamide gel (Bio-Rad, Marne-la-Coquette, France) columns, the presequencing amplification reaction was performed with a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Courtaboeuf, France) according to the manufacturer's protocol. The PCR products obtained were then purified on Sephadex G-50 (Amersham Biosciences, Orsay, France) columns and sequenced on the 3130xl Genetic Analyzer (Applied Biosystems). The presence of mutations in the amplified regions was assessed by multiple-sequence alignments using CLUSTALW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Nucleic acid sequences were then translated into corresponding protein sequences using the EMBOSS Transeq application (http://www.ebi.ac.uk/Tools/emboss/transeq/), in order to further analyze the consequences of the mutations observed.
Complementation with the wild-type ramR or soxR gene.
The 895-bp ramR fragment, generated by PCR using primers RamR F and RamR R (Table 1) from the genomic DNA of strain ATCC 138821, was cloned into the pSC-A-amp/kan plasmid vector using a StrataClone PCR Cloning Kit (Agilent Technologies, Massy, France). The same procedure was carried out for the 617-bp soxR fragment, using primers SoxR F and SoxR R (Table 1). The empty plasmid pSC-A-amp/kan-E1, used as a negative control, was generated from pSC-A-amp/kan-ramR after digestion with EcoRI enzyme (Ozyme, Saint-Quentin-en-Yvelines, France) and ligation with the LigaFast Rapid DNA Ligation System (Promega, Charbonnières-les-Bains, France). All constructs were verified by sequencing them. The cloned wild-type ramR and soxR genes were transferred into electrocompetent ramR and soxR mutant strains, respectively, by transformation with the recombinant plasmids pSC-A-amp/kan-ramR and pSC-A-amp/kan-soxR.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank, and their corresponding accession numbers are presented in Table 2.
Table 2.
GenBank accession numbers
| Strain | Accession number |
|||||
|---|---|---|---|---|---|---|
| acrR | ramR | ramA | marA-marR | soxS-soxR | rob | |
| KPBj1 Rev | GU985172 | HM036711 | GU985178 | HM010916 | HM036717 | HQ992823 |
| KPBj1 E+ | GU985173 | HM036712 | GU985179 | HM010917 | HM036718 | HQ992824 |
| KPBj3 Rev | GU985174 | HM036713 | GU985180 | HM010918 | HM036719 | HQ992825 |
| KPBj3 E+ | GU985175 | HM036714 | GU985181 | HM010919 | HM036720 | HQ992826 |
| KPBj5 Rev | GU985176 | HM036715 | GU985182 | HM010920 | HM036721 | HQ992827 |
| KPBj5 E+ | GU985177 | HM036716 | GU985183 | HM010921 | HM036722 | HQ992828 |
RESULTS
Antibiotic resistance pattern of the mutants.
Resistant mutants were obtained after one or two passages in low antibiotic concentrations from the susceptible KPBj Rev strains with a frequency of approximately 10−6 when ciprofloxacin or levofloxacin was used as a selector and 2 × 10−6 when cefoxitin was used. Four of the 9 mutants selected with cefoxitin presented a low level of resistance only to that molecule and remained susceptible to nalidixic acid and chloramphenicol. The remaining 5 mutants, similarly to the mutants selected with ciprofloxacin (n = 6) or levofloxacin (n = 6), displayed cross-resistance to the 3 antibiotics, suggesting the selection of mutants overexpressing an efflux system. These 17 mutants were retained for further analysis, after having been checked for the absence of mutations in both the gyrA and parC genes.
Transcription of the acrB gene.
The relative transcription levels of acrB for the 3 KPBj Rev strains and their respective mutants are shown in Table 3. There was a notable increase in acrB transcription for all the mutants selected by both cefoxitin and fluoroquinolones from strains KPBj1 Rev and KPBj3 Rev compared with the parental strains: 2.9 to 6.1 times and 1.8 to 4.2 times, respectively. This increase (2.8 to 10 times) was observed for all of the mutants originating from strain KPBj5 Rev except one, mutant KPBj5 M1 Cip, for which the level of transcription of acrB was equivalent to that of the parental strain.
Table 3.
Relative transcription levels of the acrB, ramA, and soxS genes in the in vitro-selected mutants, their parental strains, and their wild-type ramR or soxR transformants (for ramR and soxR mutants, respectively) in comparison with strain ATCC 138821
| Straina | Gene transcription level (fold change)b |
||
|---|---|---|---|
| acrB | ramA | soxS | |
| ATCC 138821 | 1 | 1 | 1 |
| KPBj1 Rev | 1.15 (±0.15) | 1.05 (±0.44) | ND |
| ramR mutants | |||
| KPBj1 M2 Cip | 7.05 (±1.58) | 31.62 (±16.67) | ND |
| KPBj1 M2 Cip T0 | 5.49 (±3.87) | 7.68 (±1.02) | ND |
| KPBj1 M2 Cip TramR | 1.93 (±0.03) | 0.19 (±0.07) | ND |
| KPBj1 M1 Cip | 3.38 (±0.92) | 43.91 (±30.32) | ND |
| KPBj1 M2 Lev | 4.34 (±2.20) | 38.10 (±9.24) | ND |
| KPBj1 M2 Lev TramR | 1.27 (±0.25) | 0.12 (±0.01) | ND |
| KPBj1 M3 Fox | 5.81 (±2.41) | 87.53 (±42.78) | ND |
| KPBj1 M3 Fox TramR | 1.37 (±0.08) | 0.14 (±0.04) | ND |
| Mutant with unknown mutation | |||
| KPBj1 M1 Lev | 4.01 (±0.40) | ND | ND |
| KPBj3 Rev | 2.20 (±0.57) | 2.31 (±2.92) | ND |
| ramR mutants | |||
| KPBj3 M2 Fox | 9.18 (±0.85) | 37.50 (±18.97) | ND |
| KPBj3 M2 Fox TramR | 1.52 (±0.36) | 0.09 (±0.03) | ND |
| KPBj3 M3 Fox | 6.48 (±1.32) | 13.64 (±8.37) | ND |
| KPBj3 M3 Fox TramR | 1.40 (±0.27) | 0.06 (±0.001) | ND |
| Mutants with unknown mutations | |||
| KPBj3 M1 Cip | 4.04 (±0.40) | ND | ND |
| KPBj3 M2 Cip | 4.94 (±0.07) | ND | ND |
| KPBj3 M1 Lev | 4.54 (±0.26) | ND | ND |
| KPBj3 M2 Lev | 5.50 (±0.06) | ND | ND |
| KPBj5 Rev | 0.97 (±0.05) | 2.59 (±0.34) | 2.78 (±3.17) |
| ramR mutants | |||
| KPBj5 M2 Lev | 5.09 (±2.18) | 84.21 (±72.24) | ND |
| KPBj5 M2 Lev TramR | 1.45 (±0.34) | 0.14 (±0.12) | ND |
| KPBj5 M3 Fox | 3.33 (±0.82) | 80.29 (±60.10) | ND |
| KPBj5 M3 Fox TramR | 1.39 (±0.01) | 0.10 (±0.06) | ND |
| soxR mutant | |||
| KPBj5 M1 Cip | 1.37 (±0.07) | 3.38 (±0.97) | 7.21 (±4.73) |
| KPBj5 M1 Cip TsoxR | 1.66 (±0.11) | 3.99 (±2.70) | 2.34 (±0.33) |
| Mutants with unknown mutations | |||
| KPBj5 M2 Cip | 2.72 (±0.11) | ND | ND |
| KPBj5 M1 Lev | 3.28 (±0.83) | ND | ND |
| KPBj5 M1 Fox | 9.70 (±3.92) | ND | ND |
T0, strain complemented with plasmid pSC-A-amp/kan-E1; TramR, strain complemented with plasmid pSC-A-amp/kan-ramR bearing the wild-type ramR gene; TsoxR, strain complemented with plasmid pSC-A-amp/kan-soxR bearing the wild-type soxR gene.
ND, not determined. For gene expression levels, the results of two different experiments are indicated as mean (±standard deviation).
Sequences of the regulator genes.
We analyzed the sequences of the regulator genes acrR, ramR, ramA, marR, marA, soxR, soxS, and rob of the 3 clinical isolates (KPBj E+), the 3 revertant strains (KPBj Rev), and the 17 mutants. First, we compared the sequences of the genes of each KPBj Rev strain with those of the 3 K. pneumoniae strains available in the GenBank database, i.e., K. pneumoniae subsp. pneumoniae MGH 78578 (accession number NC_009648) (25), K. pneumoniae 342 (accession number NC_011283) (11), and K. pneumoniae NTUH-K2044 (accession number NC_012731) (44). Whatever the gene, in strains KPBj1 Rev and KPBj5 Rev, the sequences were either strictly identical to those of strains MGH 78578 and NTUH-K2044 or displayed only one or two silent polymorphisms. For strain KPBj3 Rev, all the genes studied had sequences considerably different from those of strains MGH 78578 and NTUH-K2044 but very similar to those of strain 342, except for the ramR gene. Indeed, in strain 342, this gene showed a 96-bp deletion, resulting in the deletion of 32 amino acids approximately in the middle of the RamR protein. Such a deletion was not observed in the ramR gene of strain KPBj3 Rev, although the other polymorphisms characteristic of strain 342, compared with strain MGH 78578, were present.
Second, we compared the sequences of the regulator genes of the 3 clinical isolates and of the 17 mutants, which all showed cross-resistance to cefoxitin, nalidixic acid, and chloramphenicol, with those of the corresponding revertant strain. No mutation was detected in the 8 regulator genes studied from the 3 clinical isolates and 8 of the 17 mutants. However, point mutations within the ramR gene were detected in 8 of the 9 remaining mutants. In 7 of the ramR mutants, there were single-nucleotide substitutions, occurring at 6 different positions, whereas there was a deletion of 1 nucleotide in the last one. These genetic changes were distributed all along the ramR gene and led either to an amino acid exchange or to occurrence of a premature stop codon (directly or resulting from a frameshift mutation) and synthesis of a truncated RamR protein (Fig. 1). Finally, in the 9th mutant, KPBj5 M1 Cip, we found a point mutation in the soxR gene (C375G) that resulted in an amino acid substitution (N125K) in the SoxR protein (Fig. 2).
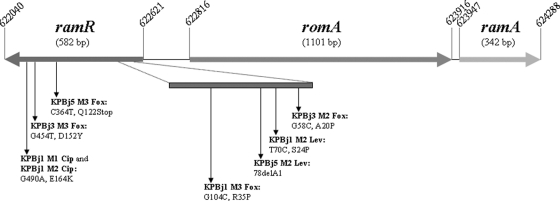
Fig. 1.
Schematic diagram of the genomic region comprising the ramR and ramA genes in K. pneumoniae strain MGH 78578 (CP000647; accession number NC_009648). The mutations identified in the 7 ramR mutants and the deduced protein changes are clarified.
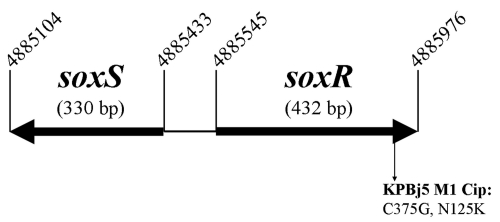
Fig. 2.
Schematic diagram of the genomic region comprising the soxR and soxS genes in K. pneumoniae strain MGH 78578 (CP000647; accession number NC_009648). The mutation identified in the soxR mutant and the deduced protein are clarified.
Functional impacts of the mutations observed in the ramR and soxR genes.
The expression levels of the genes ramA and soxS, encoding the transcriptional activators RamA and SoxS, were measured for the mutants in which genetic changes were detected in ramR and soxR, respectively. In all the ramR mutants, we observed a dramatic increase in the ramA transcription level, up to 80 times that of the parental strain (Table 3). The soxR mutant, KPBj5 M1 Cip, showed an increase in the soxS transcription level compared to the parental strain, KPBj5 Rev, although this increase was lower than for the ramA gene (Table 3).
In order to demonstrate that the mutations found in the ramR and soxR regulator genes caused the observed efflux system overexpression, phenotypically characterized by antibiotic cross-resistance, we complemented the ramR and soxR mutants with the cloned wild-type ramR and soxR genes, respectively. For all the ramR mutants, introduction of the wild-type ramR gene strikingly lowered the transcription level of ramA and also suppressed the overexpression of the acrB gene (Table 3). For the soxR mutant, complementation with the wild-type soxR gene normalized the transcription level of soxS. However, it had no effect on acrB expression, consistent with the fact that KPBj5 M1 Cip was the only mutant that did not overexpress the acrB gene (Table 3).
Impact of efflux system overexpression on antibiotic susceptibility.
The MICs of members of different antibiotic families, including quinolones, chloramphenicol, cyclines, and β-lactams, among which were different penicillins, cephalosporins, and carbapenems, for the 3 clinical isolates, their 3 revertants, and the different types of mutants (with identified and with unknown mutations) that all displayed an overexpressed efflux-related resistance phenotype were measured (Table 4). When mutants selected with a given antibiotic from the same parental strain had the same sequence for their regulator genes (KPBj1 M1 Cip and KPBj1 M2 Cip, KPBj3 M1 Cip and KPBj3 M2 Cip, and KPBj3 M1 Lev and KPBj3 M2 Lev), MICs were determined for only one of them. As had previously been shown (3), the spontaneously reverted KPBj Rev strains obtained from the KPBj E+ clinical isolates were fully susceptible to all the antibiotics tested except amoxicillin and ticarcillin, consistent with the natural resistance profile of the K. pneumoniae species. For the 14 mutants tested, the MICs of quinolones were 4 to 32 times higher than for the parental strains. All of the mutants showed a significant increase in chloramphenicol (8 to 64 times), tetracycline (4 to 16 times), and tigecycline (4 to 16 times) MICs compared to the susceptible parental strains. β-Lactam MICs were also increased for the mutants, except for amoxicillin, alone or associated with clavulanic acid, imipenem, meropenem, and doripenem. This increase was always significant, whatever the mutant, for cefoxitin and cefuroxime (MICs 4 to 16 times higher than for the parental strains) and inconsistently significant for ticarcillin, cefazolin, cefotaxime, and ceftazidime (2 to 8 times) and for ertapenem (2 to 4 times). It is noteworthy that the MICs of all the antibiotics tested were similar for all the strains sharing an overexpressed efflux-related resistance phenotype and the in vitro-selected mutants, as well as the 3 clinical isolates.
Table 4.
Antibiotic susceptibilities of strain ATCC 138821, the 3 clinical isolates, their 3 revertants, the in vitro-selected mutants overexpressing an efflux system, and the ramR and soxR mutants complemented with the wild-type ramR and soxR genes, respectively
| Strain | MIC (μg/ml)a |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL (≤8, >16) | CIP (≤0.5, >1) | LEV (≤1, >2) | CMP (≤8, >8) | TET (≤4, >8) | TGC (≤1, >2) | FOX (≤8, >32) | AMX (≤4, >8) | AMC (≤4/2, >8/2) | TIC (≤8, >16) | CF (≤8, >32) | CXM (≤8, >8) | CTX (≤1, >2) | CAZ (≤1, >8) | ETP (≤0.5, >1) | IMP (≤2, >8) | MEM (≤2, >8) | DOR (≤1, >4) | |
| ATCC 138821 | 2 | 0.03 | 0.03 | 2 | 1 | 0.25 | 2 | 128 | 2 | 512 | 2 | 4 | 0.06 | 0.125 | 0.007 | 0.25 | 0.015 | 0.125 |
| KPBj1 E+b | 32 | 0.5 | 1 | 128 | 4 | 2 | 8 | 128 | 2 | 512 | 4 | 8 | 0.125 | 0.25 | 0.007 | 0.06 | 0.015 | 0.015 |
| KPBj1 Rev | 2 | 0.015 | 0.03 | 2 | 1 | 0.5 | 2 | 64 | 2 | 128 | 2 | 2 | 0.06 | 0.125 | 0.007 | 0.125 | 0.03 | 0.03 |
| KPBj1 M2 Cipc | 32 | 0.25 | 0.5 | 32 | 8 | 4 | 32 | 128 | 4 | 512 | 4 | 16 | 0.25 | 0.5 | 0.015 | 0.125 | 0.015 | 0.03 |
| KPBj1 M2 Cipc T0e,h | 32 | 0.25 | 0.5 | 16 | 8 | 2 | 32 | >1,024 | >128 | >1,024 | 16 | 16 | 0.25 | 0.5 | 0.03 | 0.125 | 0.015 | 0.03 |
| KPBj1 M2 Cip TramRf | 2 | 0.0075 | 0.03 | 1 | 0.5 | 0.25 | 2 | >1,024 | >128 | >1,024 | 4 | 1 | 0.03 | 0.06 | 0.007 | 0.125 | 0.06 | 0.06 |
| KPBj1 M2 Levc | 16 | 0.125 | 0.5 | 32 | 8 | 4 | 32 | 128 | 4 | 512 | 8 | 16 | 0.5 | 1 | 0.03 | 0.06 | 0.015 | 0.015 |
| KPBj1 M2 Lev TramR | 4 | 0.03 | 0.03 | 2 | 1 | 0.25 | 4 | >1,024 | >128 | >1,024 | 8 | 2 | 0.06 | 0.125 | 0.015 | 0.125 | 0.06 | 0.06 |
| KPBj1 M3 Foxc | 16 | 0.125 | 0.5 | 32 | 16 | 4 | 32 | 128 | 4 | 512 | 8 | 16 | 0.5 | 1 | 0.03 | 0.06 | 0.015 | 0.015 |
| KPBj1 M3 Fox TramR | 2 | 0.015 | 0.03 | 1 | 0.5 | 0.25 | 2 | >1,024 | >128 | >1,024 | 8 | 2 | 0.06 | 0.06 | 0.015 | 0.125 | 0.03 | 0.06 |
| KPBj1 M1 Levb | 16 | 0.25 | 0.5 | 32 | 16 | 4 | 32 | 128 | 4 | 512 | 8 | 32 | 0.5 | 1 | 0.03 | 0.06 | 0.015 | 0.015 |
| KPBj3 E+b | 32 | 0.25 | 0.5 | 128 | 4 | 2 | 4 | 64 | 2 | 256 | 4 | 8 | 0.06 | 0.25 | 0.007 | 0.125 | 0.015 | 0.015 |
| KPBj3 Rev | 2 | 0.015 | 0.03 | 4 | 1 | 0.5 | 2 | 32 | 2 | 64 | 1 | 2 | 0.06 | 0.125 | 0.007 | 0.125 | 0.015 | 0.03 |
| KPBj3 M2 Foxc | 16 | 0.125 | 0.25 | 32 | 16 | 4 | 16 | 64 | 4 | 256 | 2 | 16 | 0.25 | 1 | 0.03 | 0.06 | 0.015 | 0.015 |
| KPBj3 M2 Fox TramR | 2 | 0.0075 | 0.03 | 2 | 1 | 0.25 | 2 | >1,024 | >128 | >1,024 | 4 | 2 | 0.06 | 0.125 | 0.015 | 0.125 | 0.03 | 0.03 |
| KPBj3 M3 Foxc | 16 | 0.125 | 0.25 | 64 | 16 | 4 | 16 | 64 | 4 | 256 | 2 | 16 | 0.25 | 1 | 0.03 | 0.06 | 0.015 | 0.015 |
| KPBj3 M3 Fox TramR | 2 | 0.0075 | 0.03 | 2 | 1 | 0.25 | 2 | >1,024 | >128 | >1,024 | 4 | 2 | 0.06 | 0.125 | 0.015 | 0.125 | 0.03 | 0.03 |
| KPBj3 M1 Cipb | 32 | 0.25 | 0.5 | 64 | 4 | 2 | 8 | 64 | 2 | 256 | 2 | 8 | 0.06 | 0.5 | 0.015 | 0.06 | 0.015 | 0.015 |
| KPBj3 M1 Levb | 32 | 0.25 | 0.5 | 128 | 8 | 2 | 8 | 64 | 2 | 256 | 2 | 8 | 0.06 | 0.25 | 0.007 | 0.125 | 0.015 | 0.03 |
| KPBj5 E+ | 8 | 0.06 | 0.125 | 8 | 4 | 2 | 8 | 256 | 2 | 1,024 | 4 | 8 | 0.125 | 0.25 | 0.015 | 0.125 | 0.03 | 0.06 |
| KPBj5 Rev | 2 | 0.015 | 0.06 | 2 | 1 | 0.5 | 2 | 128 | 2 | 256 | 2 | 2 | 0.06 | 0.125 | 0.007 | 0.125 | 0.03 | 0.06 |
| KPBj5 M2 Levc | 16 | 0.125 | 0.5 | 32 | 16 | 4 | 32 | 256 | 4 | 1,024 | 8 | 16 | 0.5 | 1 | 0.03 | 0.06 | 0.015 | 0.03 |
| KPBj5 M2 Lev TramR | 2 | 0.03 | 0.03 | 2 | 1 | 0.25 | 2 | >1,024 | >128 | >1,024 | 8 | 2 | 0.06 | 0.06 | 0.015 | 0.125 | 0.06 | 0.06 |
| KPBj5 M3 Foxc | 16 | 0.125 | 0.5 | 64 | 16 | 4 | 32 | 256 | 4 | 1,024 | 4 | 16 | 0.5 | 1 | 0.03 | 0.06 | 0.015 | 0.03 |
| KPBj5 M3 Fox TramR | 2 | 0.03 | 0.03 | 2 | 1 | 0.5 | 2 | >1,024 | >128 | >1,024 | 8 | 2 | 0.06 | 0.06 | 0.015 | 0.125 | 0.06 | 0.06 |
| KPBj5 M1 Cipd | 16 | 0.125 | 0.25 | 16 | 8 | 2 | 16 | 256 | 2 | 512 | 4 | 8 | 0.125 | 0.5 | 0.015 | 0.06 | 0.015 | 0.03 |
| KPBj5 M1 Cip TsoxRg | 2 | 0.03 | 0.03 | 2 | 1 | 0.25 | 2 | >1,024 | >128 | >1,024 | 8 | 2 | 0.06 | 0.06 | 0.015 | 0.125 | 0.03 | 0.03 |
| KPBj5 M2 Cipb | 64 | 0.5 | 1 | 128 | 8 | 2 | 16 | 256 | 2 | 1,024 | 4 | 8 | 0.125 | 0.5 | 0.015 | 0.06 | 0.015 | 0.03 |
| KPBj5 M1 Levb | 64 | 0.5 | 1 | 128 | 8 | 2 | 16 | 256 | 2 | 1,024 | 4 | 8 | 0.125 | 0.25 | 0.015 | 0.125 | 0.015 | 0.03 |
| KPBj5 M1 Foxb | 64 | 0.125 | 0.25 | 128 | 16 | 4 | 32 | 128 | 2 | 512 | 4 | 16 | 0.25 | 0.5 | 0.03 | 0.03 | 0.015 | 0.007 |
NAL, nalidixic acid; CIP, ciprofloxacin; LEV, levofloxacin; CMP, chloramphenicol; TET, tetracycline; TGC, tigecycline; FOX, cefoxitin; AMX, amoxicillin; AMC, amoxicillin associated with clavulanic acid; TIC, ticarcillin; CF, cefazolin; CXM, cefuroxime; CTX, cefotaxime; CAZ, ceftazidime; ETP, ertapenem; IMP, imipenem; MEM, meropenem; DOR, doripenem. The breakpoints according to the French Antibiogram Committee are given in parentheses.
Overexpression of acrB.
Overexpression of gees acrB and ramA.
Overexpression of soxR and soxS.
T0, strain complemented with plasmid pSC-A-amp/kan-E1.
TramR, strain complemented with plasmid pSC-A-amp/kan-ramR bearing the wild-type ramR gene.
TsoxR, strain complemented with plasmid pSC-A-amp/kan-soxR bearing the wild-type soxR gene.
For the strains bearing plasmid pSC-A-amp/kan with or without a cloned gene, the MICs of AMX, AMC, TIC, and CEF are greatly increased because the plasmid harbors a penicillinase-encoding gene.
Table 4 also shows that the transformation of the ramR and soxR mutants with the wild-type ramR and soxR genes, respectively, restored the strains' susceptibility to all the antibiotic families that are substrates of the efflux systems. The contribution of the wild-type regulator genes to susceptibility to amoxicillin, alone or associated with clavulanic acid, ticarcillin, and cefazolin, could not be evaluated because the pSC-A-amp/kan plasmid used for transformation harbored a bla gene responsible for high-level resistance to penicillins and first-generation cephalosporins.
DISCUSSION
It has previously been shown in K. pneumoniae clinical isolates that efflux system overexpression results in cross-resistance to different families of antibiotics, including quinolones, chloramphenicol, tetracycline, tigecycline, and β-lactams (13, 21, 28, 29, 37). In this study, we showed the possibility of obtaining in vitro mutants displaying this cross-resistance as a result of antibiotic pressure exerted either with cefoxitin (the most impaired molecule within β-lactams) or with a fluoroquinolone (i.e., ciprofloxacin or levofloxacin). To obtain such mutants, we followed the procedure described by Miller et al. for the molecules with low endogenous resistance potential (22). However, it is noteworthy that, when cefoxitin was used, mutants with another kind of antibiotic resistance pattern could be selected: a low level of resistance to cefoxitin and full susceptibility to nalidixic acid and chloramphenicol. The mechanism explaining this single resistance to cefoxitin was not investigated in this study, but according to recently published data, porin alteration seems to be the most plausible hypothesis (3).
We had previously shown in K. pneumoniae clinical isolates that cefoxitin, quinolone, and chloramphenicol cross-resistance was not always associated with an increase in the transcription level of the genes encoding AcrAB, the most frequently investigated efflux pump in Enterobacteriaceae (3). Interestingly, such a feature was also found in one of our 17 mutants, KPBj5 M1 Cip. The analysis of the sequences of the genes regulating efflux pump expression revealed that the soxR gene was mutated in this strain, leading to soxS overexpression. We demonstrated the involvement of this genetic modification in the expression of a multidrug-resistant efflux phenotype through the complementation of the mutant with the wild-type soxR gene. The soxR mutation observed led to the replacement of an asparagine residue by a lysine residue at position 125, which is located close to the C-terminal cluster of 4 cysteine residues (Cys110, Cys113, Cys115, and Cys121) that has been demonstrated to be essential for the binding of the [2Fe-2S] centers and for the activity of SoxR in E. coli (4). The fact that the SoxR protein is a transcriptional activator of soxS and not a repressor may explain why a single soxR mutant (versus 8 ramR mutants) was obtained among the 17 mutants studied. Indeed, a mutation leading to constitutive activation of a regulator protein is probably more difficult to achieve that an inactivating one. A point mutation in this region of the soxR gene has already been described in a multidrug-resistant clinical isolate of S. enterica, but whether this mutation influenced the expression of AcrAB or another efflux pump was not explored (19). To our knowledge, this is the first time that the involvement of the soxR gene in the regulatory pathways controlling the expression of an efflux pump in K. pneumoniae has been shown. However, a mutated soxR gene was not observed in the K. pneumoniae clinical isolate in which the transcription level of the acrB gene was not increased (3). Overall, these results strongly suggest the existence of an efflux pump(s) other than AcrAB in K. pneumoniae.
As previously reported in 5 clinical isolates of K. pneumoniae with reduced susceptibility to tigecycline (14), we found point mutations in the ramR genes of 8 of our 17 mutants, and these mutations were shown to be responsible for overexpression of both the ramA and acrB genes. However, their locations in the gene sequence were different from those described by Hentschke et al. (14). Still, by complementation experiments, we demonstrated that RamR, the local repressor of ramA, is really involved in the regulation of AcrAB expression in K. pneumoniae, as has been demonstrated in S. enterica (1). Whether RamA controls AcrAB expression directly or indirectly through interplay with other global regulators, as previously suggested by some authors (5, 8, 27), was not studied here.
In vitro ramR mutants were obtained at a high frequency, irrespective of the selector used, i.e., cefoxitin or fluoroquinolones. Such a result might suggest that ramR mutants could easily be obtained in vivo. Nevertheless, we did not observe ramR mutants among the 3 clinical isolates that we analyzed, as we did not observe soxR mutants. The most surprising observation was the absence of genetic modification in the 8 regulator genes studied among the 3 clinical isolates. Alignment of the sequences of their regulator genes with those available in the GenBank database revealed that 2 of our clinical isolates were very similar to the 2 published K. pneumoniae strains of clinical origin: MGH 78578, isolated from a patient with pneumonia (25), and NTUH-K2044, isolated from a patient with liver abscess and meningitis (44). On the other hand, our third clinical isolate strongly resembled K. pneumoniae 342, a nitrogen-fixing endophyte strain isolated from the interior of maize plants (11), except for the 96-bp deletion in the ramR gene. Also, we did not observe any mutation in 8 of the 17 mutants studied, although all showed a typical overexpressed efflux-related antibiotic resistance pattern, like the 3 clinical isolates, and all had an increased acrB transcription level, like 2 clinical isolates. This finding highlights the complexity of the regulatory pathways that are involved in the control of AcrAB expression in K. pneumoniae. We hypothesize that mutations in other regulator regions, first in the binding sites of the transcriptional activators and/or repressors, could explain the overexpression of AcrAB and possibly of other efflux pumps not yet identified.
Finally, this study allowed us to specify the β-lactam antibiotics whose activities are impaired by efflux system overexpression. Considering our results, it seems clear that, besides cefoxitin, first-generation (cefazolin), second-generation (cefuroxime), and even third-generation (cefotaxime and ceftazidime) cephalosporins are substrates of efflux pumps. Our findings seem to contradict a study by Källman et al. (17) in which the authors concluded that the nonsusceptibility to cefuroxime observed in multidrug-resistant K. pneumoniae isolates was not related to efflux. They based this conclusion on the absence of cefuroxime activity restoration when the efflux pump inhibitor phenylalanine arginine β-naphthylamide (PAβN) was used. However, this argument appears inadequate, since our team showed recently that the use of both cloxacillin and PAβN was required to inhibit the efflux of β-lactams (29). Regarding carbapenems, MICs of imipenem, meropenem, and doripenem were not significantly increased in both clinical isolates and the different types of mutants, whereas ertapenem MICs were significantly increased in some mutants, notably in the ramR mutants.
In conclusion, beyond suggesting the complex regulation of efflux system expression in K. pneumoniae, this study demonstrated that a large panel of antibiotics, including those widely used for a long time for treating human infectious diseases (fluoroquinolones and various β-lactams) and those more recently commercialized (tigecycline and ertapenem), are substrates of efflux systems in this species. Although we showed that mutants overexpressing efflux systems display a low level of resistance to the majority of the antibiotics studied, it is reasonable to think that this is a threatening mechanism of resistance. Indeed, its association with other mechanisms of resistance, notably to β-lactams (β-lactamase production and/or porin alteration), could provide advantages for bacterial survival, as suggested by our previous study on K. pneumoniae virulence in the Caenorhabditis elegans model and by the cefoxitin resistance that was present in the first ESBL-producing K. pneumoniae strain responsible for outbreaks in France (3, 16).
Footnotes
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Abouzeed Y. M., Baucheron S., Cloeckaert A. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52:2428–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alekshun M. N., Levy S. B. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bialek S., et al. 2010. Membrane efflux and influx modulate both multidrug resistance and virulence of Klebsiella pneumoniae in a Caenorhabditis elegans model. Antimicrob. Agents Chemother. 54:4373–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradley T. M., Hidalgo E., Leautaud V., Ding H., Demple B. 1997. Cysteine-to-alanine replacements in the Escherichia coli SoxR protein and the role of the [2Fe-2S] centers in transcriptional activation. Nucleic Acids Res. 25:1469–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bratu S., Landman D., George A., Salvani J., Quale J. 2009. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J. Antimicrob. Chemother. 64:278–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buckley A. M., et al. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8:847–856 [DOI] [PubMed] [Google Scholar]
- 7. Chen J. Y., et al. 2001. Molecular epidemiology and mutations at gyrA and parC genes of ciprofloxacin-resistant Escherichia coli isolates from a Taiwan medical center. Microb. Drug Resist. 7:47–53 [DOI] [PubMed] [Google Scholar]
- 8. Chollet R., Chevalier J., Bollet C., Pages J. M., Davin-Regli A. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48:2518–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohen S. P., Hachler H., Levy S. B. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doumith M., Ellington M. J., Livermore D. M., Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659–667 [DOI] [PubMed] [Google Scholar]
- 11. Fouts D. E., et al. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. George A. M., Hall R. M., Stokes H. W. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909–1920 [DOI] [PubMed] [Google Scholar]
- 13. Hasdemir U. O., Chevalier J., Nordmann P., Pages J. M. 2004. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J. Clin. Microbiol. 42:2701–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hentschke M., Wolters M., Sobottka I., Rohde H., Aepfelbacher M. 2010. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob. Agents Chemother. 54:2720–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imuta N., et al. 2008. The Escherichia coli efflux pump TolC promotes aggregation of enteroaggregative E. coli 042. Infect. Immun. 76:1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jarlier V., Nicolas M. H., Fournier G., Philippon A. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer ß-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867–878 [DOI] [PubMed] [Google Scholar]
- 17. Källman O., et al. 2008. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J. Antimicrob. Chemother. 62:986–990 [DOI] [PubMed] [Google Scholar]
- 18. Keynan Y., Rubinstein E. 2007. The changing face of Klebsiella pneumoniae infections in the community. Int. J. Antimicrob. Agents 30:385–389 [DOI] [PubMed] [Google Scholar]
- 19. Koutsolioutsou A., Martins E. A., White D. G., Levy S. B., Demple B. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob. Agents Chemother. 45:38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez-Martinez L. 2008. Extended-spectrum beta-lactamases and the permeability barrier. Clin. Microbiol. Infect. 14(Suppl. 1):82–89 [DOI] [PubMed] [Google Scholar]
- 21. Mazzariol A., Zuliani J., Cornaglia G., Rossolini G. M., Fontana R. 2002. AcrAB efflux system: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrob. Agents Chemother. 46:3984–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller K., O'Neill A. J., Chopra I. 2002. Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J. Antimicrob. Chemother. 49:925–934 [DOI] [PubMed] [Google Scholar]
- 23. Nikaido H., Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oethinger M., Podglajen I., Kern W. V., Levy S. B. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogawa W., et al. 2005. Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol. Pharm. Bull. 28:1505–1508 [DOI] [PubMed] [Google Scholar]
- 26. Okusu H., Ma D., Nikaido H. 1996. AcrAB efflux pump plays a major rôle in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Regan E., et al. 2009. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar enteritidis: involvement of RamA and other global regulators. Antimicrob. Agents Chemother. 53:1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Padilla E., et al. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 54:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pages J. M., et al. 2009. Efflux pump, the masked side of β-lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One 4:e4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papanicolaou G. A., Medeiros A. A., Jacoby G. A. 1990. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and a-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 34:2200–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pérez A., et al. 2007. Cloning, nucleotide sequencing, and analysis of the AcrAB-TolC efflux pump of Enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolate. Antimicrob. Agents Chemother. 51:3247–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piddock L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piddock L. J., White D. G., Gensberg K., Pumbwe L., Griggs D. J. 2000. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:3118–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Podschun R., Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pradel E., Pages J. M. 2002. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 46:2640–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruzin A., Keeney D., Bradford P. A. 2005. AcrAB efflux pump plays a role in decreased susceptibility to tigecycline in Morganella morganii. Antimicrob. Agents Chemother. 49:791–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruzin A., Visalli M. A., Keeney D., Bradford P. A. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49:1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schneiders T., Amyes S. G., Levy S. B. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47:2831–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sirot D., Labia R., Morand A., Courvalin P. 1987. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel β-lactamase. J. Antimicrob. Chemother. 20:323–334 [DOI] [PubMed] [Google Scholar]
- 40. Sulavik M. C., Dazer M., Miller P. F. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179:1857–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Visalli M. A., Murphy E., Projan S. J., Bradford P. A. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob. Agents Chemother. 47:665–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Webber M. A., Talukder A., Piddock L. J. 2005. Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob. Agents Chemother. 49:4390–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu J., Dunham W. R., Weiss B. 1995. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative response regulon in Escherichia coli. J. Biol. Chem. 270:10323–10327 [DOI] [PubMed] [Google Scholar]
- 44. Wu K. M., et al. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 191:4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yigit H. Q., et al. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]