Abstract
The synthesis of 2′-azido-5-cyano-2′-deoxyuridine, N3CNdU (1), from trityl-protected 2′-amino-2′-deoxyuridine was accomplished in four steps with a 12.5% overall yield. The IR absorption positions and profiles of the azide and nitrile group of N3CNdU were investigated in 14 different solvents and water/DMSO solvent mixtures. The azide probe was superior to the nitrile probe in terms of its extinction coefficient, which is 2–4 times larger. However, the nitrile IR absorbance profile is generally less complicated by accidental Fermi resonance. The IR frequencies of both probes undergo a substantial red shift upon going from water to aprotic solvents such as THF or DMSO. DFT calculations supported the hypothesis that the molecular origin of the higher observed frequency in water is primarily due to hydrogen bonds between the probes and water molecules.
Keywords: Infrared spectroscopy, azide, nitrile, nucleosides, DNA, density functional theory
Introduction
The use of vibrational probes to study biomolecules has a long and rich history. In most of the early work the probe was a ligand bound to hemoproteins. 1 For example, four decades ago McCoy and Caughey used infrared spectroscopy to investigate the binding of ligands such as azide, cyanide, and thiocyanate to metmyoglobin and methemoglobin. 2 More recently, Suydam and Boxer3 examined a series of vibrational probes for proteins and concluded that the most useful ones have the following properties: (1) an IR absorption band that is narrow, intense, and in a clear region of the spectrum; (2) sensitive to changes in local environment; (3) small size to minimize structural perturbations; and (4) chemical stability. It is therefore no surprise that nitriles and azides have emerged as the most useful vibrational probes, since both meet these criteria. Within the past decade the use of nitriles to study non-hemoproteins has become prevalent, and more recently azides have also been used for this purpose.
In terms of nitriles, the unnatural amino acid (UAA) p-cyanophenylalanine (pCNPhe) has been inserted into proteins both synthetically4–12 and by in vivo nonsense suppression. 13–15 The nitrile of pCNPhe served as a vibrational probe to study the hydration and electrostatic environments of several protein systems, including the MLCK peptide–calmodulin complex, 4 the mastoparan-X peptide, 5, 9 myoglobin, 13 and ribonuclease S;12 the latter was also studied with the UAAs m-cyanophenylananine and S-cyanohomocysteine. The chemical conversion of cysteine thiols into thiocyanates or aryl nitriles has been utilized to introduce a vibrational probe into numerous peptides and proteins. 16–18 The vibrational Stark effect of a nitrile-containing enzyme inhibitor was effectively used to measure the electric field in the active site of human aldose reductase. 19 The UAA 5-cyanotryptophan has also been employed as an infrared probe of protein hydration status. 20
In terms of azides, the UAAs azidoalanine and azidohomoalanine have been synthetically incorporated into peptide fragments of the Aβ peptide and the N-terminal domain of the ribosomal protein L9 (NTL9), respectively, to serve as vibrational probes of protein structure. 21, 22 An engineered aminoacyl-tRNA synthetase/tRNA pair has been employed to incorporate the UAA p-azidophenylalanine (pN3Phe) into the G-protein coupled receptor rhodopsin to study the helix movements during light activation via infrared spectroscopy. 23, 24 Aromatic azides such as pN3Phe have an additional property of photoreactivity. The instability of this probe is a disadvantage for vibrational experiments but it can be utilized for photocrosslinking experiments. 25, 26 Azido-nicotinamide adenine dinucleotide was recently synthesized and shown to be a 2D IR probe of the active sites of NAD-dependent enzymes. 27 These same workers showed that the structurally related 3-azidopyridine was less suitable as a vibrational probe because of a complex absorption profile resulting from accidental Fermi resonance. 28
The use of nitrile and azide vibrational probes in nucleic acids has lagged behind the protein work. These functional groups were first incorporated into nucleic acids decades ago, but the idea to employ them as vibrational probes was first proposed in 2007 when the Stark tuning rates of a number of nitrile- and azido-nucleosides were published. 29 Krummel and Zanni30 have demonstrated that the vibrational coupling of 14N- and 15N-labeled 5-cyano-2′-deoxyuridines (CNdU) can be used to measure distances and angles in a DNA oligomer, while the effects of solvent, hydrogen bonded heterodimer formation, and temperature on the nitrile and azido IR absorbance bands of CNdU and 2′-azido-2′-deoxyuridine (N3dU), respectively, have also been reported. 31, 32 Here, we believe for the first time, both nitrile and azido functional groups have been incorporated into the same biomolecular monomer to facilitate a direct comparison of the vibrational characteristics of these important probes. 2′-Azido-5-cyano-2′-deoxyuridine (N3CNdU, 1) was synthesized and the IR absorption profiles of the azide and nitrile groups in multiple solvents and solvent mixtures are compared. This work builds upon Cho’s comparison of amino acids containing azide, nitrile, or thiocyante groups in three different solvents. 21 Future incorporation of N3CNdU into nucleic acid oligomers will allow simultaneous monitoring of both the sugar backbone region and the major groove at a specific nucleotide site through analysis of the azide and nitrile IR absorbance bands, respectively.
Results and Discussion
Synthetic Chemistry
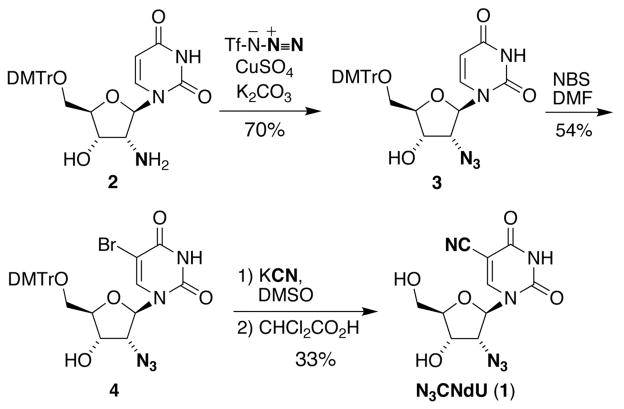
The synthesis of N3CNdU (1) was accomplished in four steps with a 12.5% overall yield, Scheme 1. Trityl-protected 2′-amino-2′-deoxyuridine 2 33 was subjected to a copper-catalyzed diazotransfer reaction using triflic azide to afford azide 3 in 70% yield. Aromatic bromination of 3 with NBS provided bromide 4 in moderate yield (54%). Cyanation with potassium cyanide followed by detritylation using dichloroacetic acid afforded a 33% yield of 1 for the two steps.
Scheme 1.
Infrared Spectroscopy
N3CNdU in Water
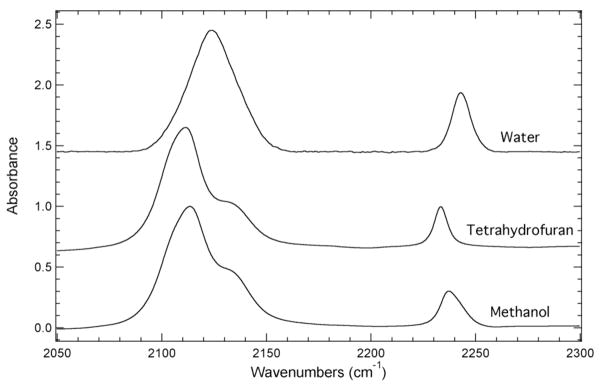
The dependence of the position, shape, and complexity of the IR absorbance bands corresponding to the azide asymmetric or the nitrile symmetric stretch of N3CNdU was explored in a number of solvents and solvent mixtures. The solvents and solvent mixtures were selected to present a wide range of local environments around the azide and nitrile groups of N3CNdU to compare and contrast the utility of these groups as vibrational probes. Figure 1 shows the normalized FTIR absorbance spectra of N3CNdU dissolved in water, tetrahydrofuran (THF), or methanol. The IR absorbance spectrum of N3CNdU in water shows two well-resolved bands at 2124.1 cm−1 and 2242.7 cm−1 corresponding to the azide asymmetric and the nitrile symmetric stretching frequency, respectively. The fwhm of the azide IR absorbance band is ~26 cm−1 compared to a fwhm of ~10 cm−1 for the nitrile IR absorbance band. These experimental results are consistent with previous studies of the single-modified nucleosides: N3dU32 and CNdU31. A third distinct difference between the bands is that the azide IR absorbance band is a factor of two larger than the nitrile IR absorbance band based upon the peak absorbance of each band. This difference increases to a factor of five when considering the relative integrated intensity of each band due to the significant difference in the fwhm.
Figure 1.
FTIR absorbance spectra of N3CNdU dissolved in water, tetrahydrofuran (THF), or methanol recorded at 293 K with a concentration of 50 mM. The maximum absorbance of each spectrum has been normalized to unity and the spectra have been offset for comparison.
N3CNdU in Tetrahydrofuran (THF)
The nitrile IR absorbance band of N3CNdU dissolved in the aprotic solvent THF is centered at 2233.5 cm−1. This position represents a 9.2 cm−1 red shift of the nitrile stretching frequency in THF relative to water. The higher frequency in water is the result of H-bonding between the nitrile group and water, which is absent in THF. This red shift is consistent with previous experimental studies of the solvent induced frequency shifts of nitrile-modified molecules such as acetonitrile, pCNPhe, thiocyanates, and CNdU. 4, 31, 34, 35
The impact of H-bonding with water on the nitrile stretching frequency of N3CNdU was further explored using DFT calculations at the B3PW91/6-31++G(d,p) level. The calculations resulted in two distinct geometry-optimized H-bonding configurations between the water molecule and the nitrile group of the model 5-cyanouracil (see Supporting Information for the geometry optimized structures). The first geometry was characterized by a C≡N···H angle of 153° between the nitrile group of 5-cyanouracil and the water molecule where a hydrogen atom of the water molecule interacted with the lone pair on the nitrogen atom of the nitrile group. This σ-H-bond resulted in a 8.6 cm−1 blue shift in the nitrile stretching frequency relative to isolated 5-cyanouracil. The C≡N···H angle in the second configuration was 90°, where a hydrogen atom of the water molecule interacted with the π-orbital of the nitrile group. The π-H-bond geometry resulted in a 8.4 cm−1 red shift in the nitrile group. Consequently, the nitrile stretching frequency is dependent on both the presence and geometry of H-bonding interactions with water. A balance of these H-bonding interactions results in the experimentally observed blue shift between the THF and aqueous solutions. The calculations were not in quantitative agreement with the experimental values since the calculations were performed in the gas phase, they do not include effects of anharmonicity, and limitations in the basis set. 36–45 However, these computational results are in agreement with previous theoretical studies exploring the impact of H-bonding interactions between the nitrile group of acetonitrile and water on the nitrile stretching frequency. 34, 36
The azide IR absorbance band of N3CNdU in THF displays a complex absorption profile with a maximum absorbance at 2111.5 cm−1. This band position represents a 12.6 cm−1 red shift relative to its position in water. The azide IR absorbance band for N3CNdU in THF, however, contains a pronounced high frequency shoulder resulting in an absorbance band comprised of at least two spectral components. The central frequency of the primary component of the azide IR absorbance band of N3CNdU in THF is 2109.8 cm−1 and the secondary component (the high frequency shoulder) is centered at 2135.2 cm−1. These positions are based upon a line shape analysis of the azide IR absorbance band utilizing two line shape functions described by Eqn. 1 (see Supporting Information). This added complexity is not surprising based upon previous experimental studies of the azide IR absorbance band of aryl azides such as 3-azidopyridine and pN3Phe. 23, 28, 46, 47
The observed complexity of the azide IR absorbance band of N3CNdU in THF is likely the result of either multiple conformations of N3CNdU in THF or accidental Fermi resonance involving the azide asymmetric stretch and a normally forbidden combination or overtone band of N3CNdU. The presence of two distinct conformations giving rise to the complex absorption profile is unlikely since the azide IR absorbance band for N3dU in THF does not show a pronounced high frequency shoulder (the band is only slightly asymmetric). 32, 48 Consequently, we interpret the 2109.8 cm−1 component to be the azide asymmetric stretching frequency and the secondary component at 2135.2 cm−1 to be the normally forbidden combination or overtone band participating in the Fermi resonance. The assignment of Fermi resonance is in agreement with a previous literature study, 28 although 2D IR studies of N3CNdU are needed for confirmation. 27, 28, 48–53
Similar to the nitrile IR absorbance band, the observed red shift in the azide IR absorbance band of N3CNdU in THF relative to water is due to specific solute-solvent interactions (H-bonding) between the azide group and water. The sensitivity of the azide asymmetric stretch to H-bonding with water was investigated by DFT calculations at the B3PW91/6-31++G(d,p) level. The calculations resulted in two geometry-optimized H-bonding configurations. The calculations were performed with the model 2-azido-1,2-dideoxyribose and one explicit water molecule (see Supporting Information for the geometry-optimized structures). The first configuration showed a σ-H-bond between the azide group and a water molecule characterized by a N=N···H angle of 150°. This σ-H-bond formed by an interaction between the hydrogen atom of the water molecule and the terminal nitrogen atom of the azide group resulted in a 7.4 cm−1 blue shift in the azide asymmetric stretching frequency relative to isolated 2-azido-1,2-dideoxyribose. The second H-bond geometry involved a π-H-bond between the water molecule and the azide group characterized by a N=N···H angle of 101°. This π-H-bond resulted in a 0.4 cm−1 red shift in the azide asymmetric stretching frequency relative to isolated 2-azido-1,2-dideoxyribose. These calculations are in agreement with previous theoretical studies probing the impact of H-bonding between the azide group of methyl azide and water. 54 The calculations here demonstrate the sensitivity of the azide asymmetric stretch to H-bonding with water, although the calculations are not in quantitative agreement with the experimental results due, in part, to limitations in the basis set. 41–45
N3CNdU in Methanol
The azide and nitrile IR absorbance bands of N3CNdU dissolved in methanol both display complex absorption profiles. The azide IR absorbance band has an absorbance maximum at 2113.5 cm−1 with a high frequency shoulder, which is between the band position observed in water and THF due to the potential of H-bonding between methanol and the azide group. Similar to the azide IR absorbance band of N3CNdU in THF, the azide IR absorbance band in methanol is comprised of at least two spectral components. The central frequency of the major component is 2111.9 cm−1 and the frequency of the minor component is 2135.6 cm−1 based upon line shape analysis of the experimental band to line shape functions described by Eqn. 1 (see Supporting Information). Once again, this complex absorption profile could be the result of different conformations of N3CNdU but is more likely due to accidental Fermi resonance, although 2D IR experiments are needed for a definitive determination. 27, 28, 48–53
The nitrile IR absorbance band of N3CNdU in methanol has an absorbance maximum at 2237.4 cm−1 and is comprised of two components positioned at 2236.3 cm−1 and 2241.1 cm−1 as determined by line shape analysis (see Supporting Information). The complex absorbance band is likely due to two distinct solvation states, 35, 49, 55 although Fermi resonance has been observed previously for nitrile absorbance bands.34, 56, 57 Based upon literature precedence, we attribute the high frequency component to a solvation state where the nitrogen lone pair of the nitrile group is involved in a σ-H-bond with the hydroxyl hydrogen atom of methanol. 49, 55 This assignment is supported by the similarity of the frequency with the observed position of the nitrile IR absorbance band in water. The low frequency component is due to a solvation state devoid of a σ-H-bond between the nitrile group and water. It is hypothesized that these two distinct solvation states give rise to the complex nitrile absorption profile in methanol. 49
N3CNdU in Water/DMSO Mixtures
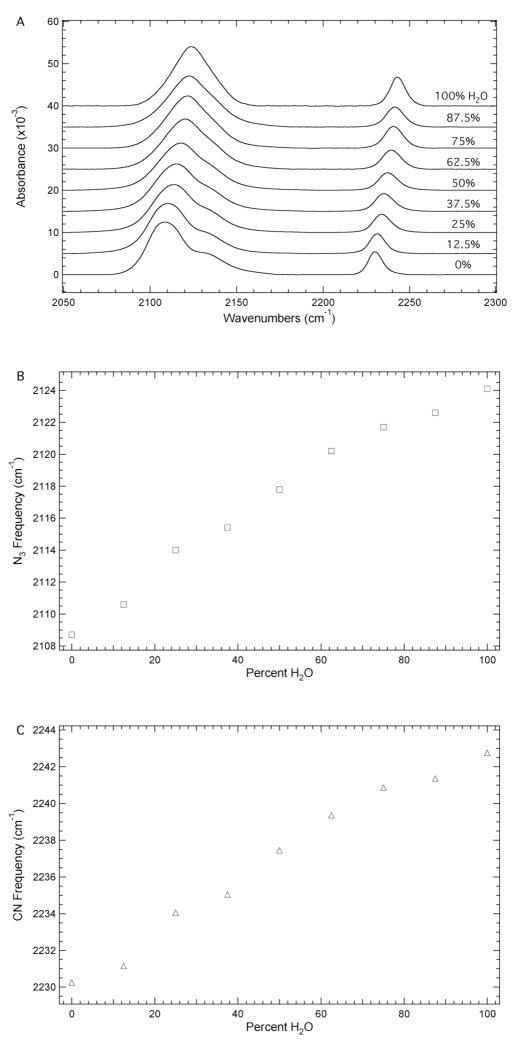
Figure 2A shows the dependence of the azide and nitrile IR absorbance bands on the systematic variation of the solvent system from water to DMSO in 12.5% increments. These solvent mixtures represent a wide range of local environments for the azide and nitrile groups of N3CNdU both in terms of solvent dielectric constants and the potential for specific solute-solvent interactions (H-bonding). The FTIR spectrum of N3CNdU in DMSO is similar to the spectrum in THF in that the azide IR absorbance band shows a complex profile consisting of at least two components while the nitrile IR absorbance profile is a single band. Again, the complexity of the azide IR absorbance band is likely due to accidental Fermi resonance. 28
Figure 2.
A. FTIR absorbance spectra of N3CNdU in water/DMSO mixtures ranging from 0 to 100% water in 12.5% increments recorded at 293 K with a concentration of 50 mM. The maximum absorbance of each spectrum has been normalized to unity and the spectra have been offset for comparison. B. Dependence of the position of maximum absorbance of the azide IR absorbance band on solvent composition. C. Dependence of the nitrile IR absorbance band position on solvent composition.
The frequency of maximum absorbance for the azide and nitrile IR absorbance bands of N3CNdU as a function of the solvent composition is plotted in Figures 2B and 2C, respectively. The positions of both bands decrease monotonically as the percent water is decreased in the water/DMSO solvent mixture. The position of the azide IR absorbance band decreases from 2124.4 to 2108.7 cm−1, while the nitrile IR absorbance band decreases from 2242.7 to 2230.2 cm−1 upon going from water to DMSO. The 15.7 cm−1 and 12.5 cm−1 red shifts are due to changes in H-bond interactions between the azide and nitrile groups with water, respectively. The azide IR absorbance band displays a larger peak extinction for each solvent mixture relative to the nitrile IR absorbance band while exhibiting a larger solvent induced frequency shift. The magnitude of the shift in response to solvent composition highlights the potential utility of azide and nitrile vibrational probes of local environments in nucleic acids.
Additional Solvent and Temperature Studies on N3CNdU
The IR absorption profiles of N3CNdU in ten additional solvents (2-propanol, 1-butanol, tert-butanol, 2,2,2-trifluoroethanol, acetone, nitromethane, formamide, N-methylformamide, N,N-dimethylformamide, and propylene carbonate) were investigated (see Supporting Information). The azide IR absorbance band is asymmetric in each of these solvents, but is less pronounced in 2,2,2-trifluoroethanol, nitromethane, and formamide. This complexity is presumably due to accidental Fermi resonance. The nitrile IR absorbance band also showed a complex absorption profile in 2-propanol, 1-butanol, tert-butanol, formamide, and N-methylformamide (see Supporting Information). In the alcoholic solvents the nitrile complexity is likely due to different solvation states corresponding to H-bonding and non-H-bonding configurations between the hydroxyl group of the solvent and the nitrile group of N3CNdU. The solvent induced frequency shifts of the vibrational probes did not correlate with solvent parameters such as solvent dielectric constant or solvent dipole moment, similar to previous studies of other nitrile or azide modified molecules. 21, 34 Additionally, both the azide and nitrile IR bands of N3CNdU showed a modest dependence on temperature in aqueous solution (see Supporting Information).
Conclusions
The three-atom azide and two-atom nitrile groups are effective vibrational probes of local environments as demonstrated by the solvent dependent positions of the azide asymmetric and nitrile symmetric stretching frequencies in N3CNdU. On the whole, the azide probe is superior to the nitrile probe because its extinction coefficient is 2–4 times larger. However, the nitrile IR absorbance profile is generally less complicated by accidental Fermi resonance although both the azide and nitrile IR absorbance profiles are effectively a single band in the biologically relevant aqueous solution. The IR frequencies of both probes exhibit a pronounced red shift upon going from water to DMSO (or THF). The frequencies of both observables were higher in water due to H-bonding with water, as confirmed by DFT calculations. As noted previously for 3-azidopyridine,28 the potential complex absorption profiles for the azide and nitrile probes must be considered when using them to study biomolecular structure and dynamics. These complexities28, 57, 58 could result in significant difficulties in the interpretation of the infrared signature of azide and nitrile groups in biological systems. A general experimental method to address this issue is currently under investigation.
The azide and nitrile groups of N3CNdU have the potential to simultaneously probe both the sugar backbone region and the major groove, respectively, of nucleic acids. The vibrational signatures of both groups are well resolved from each other permitting the solvent-dependent azide asymmetric and nitrile symmetric stretching frequencies to yield site-specific information about nucleic acid structure and dynamics. For instance, this minimally invasive modified nucleoside has the potential to aid in the investigation of conformation changes associated with DNA-protein interactions and RNA folding.
Supplementary Material
Acknowledgments
We are grateful to Carol Strausser for assistance in the editing of the manuscript, Lisa Mertzman for obtaining materials and supplies, and Beth Buckwalter for acquiring the NMR spectra. This work was supported by an F&M Snavely Summer Research stipend and a Snavely Research Award to XSG, the William M. and Lucille M. Hackman Scholars Program to BAC, a Mellon/CPC New Tasks/New Tools grant to EEF, and the NIH (R15GM093330) to SHB/EEF.
Footnotes
Electronic supplementary information (ESI) available: Full synthetic procedures; FTIR absorbance spectra of N3CNdU in 14 solvents; line shape analysis of the FTIR absorbance spectrum of N3CNdU in THF and methanol; variable temperature spectra of N3CNdU in water; DFT geometry-optimized structures.
References
- 1.Jung C. J Mol Recognit. 2000;13:325–351. doi: 10.1002/1099-1352(200011/12)13:6<325::AID-JMR507>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 2.McCoy S, Caughey WS. Biochemistry. 1970;9:2387–2393. doi: 10.1021/bi00814a001. [DOI] [PubMed] [Google Scholar]
- 3.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 4.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J Am Chem Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 5.Tucker MJ, Getahun Z, Nanda V, DeGrado WF, Gai F. J Am Chem Soc. 2004;126:5078–5079. doi: 10.1021/ja032015d. [DOI] [PubMed] [Google Scholar]
- 6.Aprilakis KN, Taskent H, Raleigh DP. Biochemistry. 2007;46:12308–12313. doi: 10.1021/bi7010674. [DOI] [PubMed] [Google Scholar]
- 7.Tucker MJ, Oyola R, Gai F. J Phys Chem B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 8.Tucker MJ, Oyola R, Gai F. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]
- 9.Tang J, Signarvic RS, DeGrado WF, Gai F. Biochemistry. 2007;46:13856–13863. doi: 10.1021/bi7018404. [DOI] [PubMed] [Google Scholar]
- 10.Marek P, Gupta R, Raleigh DP. Chembiochem. 2008;9:1372–1374. doi: 10.1002/cbic.200800052. [DOI] [PubMed] [Google Scholar]
- 11.Glasscock JM, Zhu YJ, Chowdhury P, Tang J, Gai F. Biochemistry. 2008;47:11070–11076. doi: 10.1021/bi8012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fafarman AT, Boxer SG. J Phys Chem B. 2010;114:13536–13544. doi: 10.1021/jp106406p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 14.Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- 15.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 16.Fafarman AT, Webb LJ, Chuang JI, Boxer SG. J Am Chem Soc. 2006;128:13356–13357. doi: 10.1021/ja0650403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon HA, Alfieri KN, Clark CAA, Londergan CH. J Phys Chem Lett. 2010;1:850–855. doi: 10.1021/jz1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo H, Culik RM, Korendovych IV, DeGrado WF, Gai F. Biochemistry. 2010;49:10354–10356. doi: 10.1021/bi101711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suydam IT, Snow CD, Pande VS, Boxer SG. Science. 2006;313:200–204. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]
- 20.Waegele MM, Tucker MJ, Gai F. Chem Phys Lett. 2009;478:249–253. doi: 10.1016/j.cplett.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh KI, Lee JH, Joo C, Han H, Cho M. J Phys Chem B. 2008;112:10352–10357. doi: 10.1021/jp801558k. [DOI] [PubMed] [Google Scholar]
- 22.Taskent-Sezgin H, Chung JA, Banerjee PS, Nagarajan S, Dyer RB, Carrico I, Raleigh DP. Angew Chem, Int Ed. 2010;49:7473–7475. doi: 10.1002/anie.201003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye SX, Huber T, Vogel R, Sakmar TP. Nat Chem Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye SX, Zaitseva E, Caltabiano G, Schertler GFX, Sakmar TP, Deupi X, Vogel R. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 25.Zhang KC, Diehl MR, Tirrell DA. J Am Chem Soc. 2005;127:10136–10137. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- 26.Carrico IS, Maskarinec SA, Heilshorn SC, Mock ML, Liu JC, Nowatzki PJ, Franck C, Ravichandran G, Tirrell DA. J Am Chem Soc. 2007;129:4874–4875. doi: 10.1021/ja070200b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta S, Cook RJ, Houtman JCD, Kohen A, Cheatum CM. Anal Biochem. 2010;407:241–246. doi: 10.1016/j.ab.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nydegger MW, Dutta S, Cheatum CM. J Chem Phys. 2010;133:134506. doi: 10.1063/1.3483688. [DOI] [PubMed] [Google Scholar]
- 29.Silverman LN, Pitzer ME, Ankomah PO, Boxer SG, Fenlon EE. J Phys Chem B. 2007;111:11611–11613. doi: 10.1021/jp0750912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krummel AT, Zanni MT. J Phys Chem B. 2008;112:1336–1338. doi: 10.1021/jp711558a. [DOI] [PubMed] [Google Scholar]
- 31.Watson MD, Gai XS, Gillies AT, Brewer SH, Fenlon EE. J Phys Chem B. 2008;112:13188–13192. doi: 10.1021/jp8067238. [DOI] [PubMed] [Google Scholar]
- 32.Gai XS, Fenlon EE, Brewer SH. J Phys Chem B. 2010;114:7958–7966. doi: 10.1021/jp101367s. [DOI] [PubMed] [Google Scholar]
- 33.McGee DPC, Vaughn-Settle A, Vargeese C, Zhai YS. J Org Chem. 1996;61:781–785. doi: 10.1021/jo9510548. [DOI] [PubMed] [Google Scholar]
- 34.Reimers JR, Hall LE. J Am Chem Soc. 1999;121:3730–3744. [Google Scholar]
- 35.Maienschein-Cline MG, Londergan CH. J Phys Chem A. 2007;111:10020–10025. doi: 10.1021/jp0761158. [DOI] [PubMed] [Google Scholar]
- 36.Choi JH, Oh KI, Lee H, Lee C, Cho M. J Chem Phys. 2008;128:134506. doi: 10.1063/1.2844787. [DOI] [PubMed] [Google Scholar]
- 37.Lindquist BA, Corcelli SA. J Phys Chem B. 2008;112:6301–6303. doi: 10.1021/jp802039e. [DOI] [PubMed] [Google Scholar]
- 38.Lindquist BA, Haws RT, Corcelli SA. J Phys Chem B. 2008;112:13991–14001. doi: 10.1021/jp804900u. [DOI] [PubMed] [Google Scholar]
- 39.Oh KI, Choi JH, Lee JH, Han JB, Lee H, Cho M. J Chem Phys. 2008;128:154504. doi: 10.1063/1.2904558. [DOI] [PubMed] [Google Scholar]
- 40.Waegele MM, Gai F. J Phys Chem Lett. 2010;1:781–786. doi: 10.1021/jz900429z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boese AD, Martin JML, Klopper W. J Phys Chem A. 2007;111:11122–11133. doi: 10.1021/jp072431a. [DOI] [PubMed] [Google Scholar]
- 42.Lee JS. J Chem Phys. 2007;127:085104. doi: 10.1063/1.2761881. [DOI] [PubMed] [Google Scholar]
- 43.Riley KE, Hobza P. J Phys Chem A. 2007;111:8257–8263. doi: 10.1021/jp073358r. [DOI] [PubMed] [Google Scholar]
- 44.Santra B, Michaelides A, Scheffler M. J Chem Phys. 2007;127:184104. doi: 10.1063/1.2790009. [DOI] [PubMed] [Google Scholar]
- 45.Scott JN, Nucci NV, Vanderkooi JM. J Phys Chem A. 2008;112:10939–10948. doi: 10.1021/jp8058239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lieber E, Rao CNR, Thomas AE, Oftedahl E, Minnis R, Nambury CVN. Spectrochim Acta. 1963;19:1135–1144. [Google Scholar]
- 47.Dyall LK, Kemp JE. Aust J Chem. 1967;20:1395–1402. [Google Scholar]
- 48.Tucker MJ, Gai XS, Fenlon EE, Brewer SH, Hochstrasser RM. Phys Chem Chem Phys. 2011;13:2237–2241. doi: 10.1039/c0cp01625j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YS, Hochstrasser RM. J Phys Chem B. 2009;113:8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon J, Yang S, Choi JH, Cho M. Acc Chem Res. 2009;42:1280–1289. doi: 10.1021/ar900014e. [DOI] [PubMed] [Google Scholar]
- 51.Ganim Z, Chung HS, Smith AW, Deflores LP, Jones KC, Tokmakoff A. Acc Chem Res. 2008;41:432–441. doi: 10.1021/ar700188n. [DOI] [PubMed] [Google Scholar]
- 52.Shim SH, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Proc Natl Acad Sci USA. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maekawa H, De Poli M, Toniolo C, Ge NH. J Am Chem Soc. 2009;131:2042–2043. doi: 10.1021/ja807572f. [DOI] [PubMed] [Google Scholar]
- 54.Choi JH, Oh KI, Cho MH. J Chem Phys. 2008;129:174512. doi: 10.1063/1.3001915. [DOI] [PubMed] [Google Scholar]
- 55.Lindquist BA, Furse KE, Corcelli SA. Phys Chem Chem Phys. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]
- 56.Andrews SS, Boxer SG. J Phys Chem A. 2000;104:11853–11863. [Google Scholar]
- 57.Tucker MJ, Kim YS, Hochstrasser RM. Chem Phys Lett. 2009;470:80–84. doi: 10.1016/j.cplett.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinnaman CS, Cremeens ME, Romesberg FE, Corcelli SA. J Am Chem Soc. 2006;128:13334–13335. doi: 10.1021/ja064468z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.