We show that GUN4-porphyrin complexes help to channel protoporphyrin IX into chlorophyll biosynthesis by binding to the ChlH subunit of Mg-chelatase with a higher affinity than unliganded GUN4 on Arabidopsis chloroplast membranes. GUN4 and ChlH used distinct mechanisms to associate with chloroplast membranes, and mutant alleles of GUN4 and Mg-chelatase subunit genes cause sensitivity to intense light.
Abstract
The GENOMES UNCOUPLED4 (GUN4) protein stimulates chlorophyll biosynthesis by activating Mg-chelatase, the enzyme that commits protoporphyrin IX to chlorophyll biosynthesis. This stimulation depends on GUN4 binding the ChlH subunit of Mg-chelatase and the porphyrin substrate and product of Mg-chelatase. After binding porphyrins, GUN4 associates more stably with chloroplast membranes and was proposed to promote interactions between ChlH and chloroplast membranes—the site of Mg-chelatase activity. GUN4 was also proposed to attenuate the production of reactive oxygen species (ROS) by binding and shielding light-exposed porphyrins from collisions with O2. To test these proposals, we first engineered Arabidopsis thaliana plants that express only porphyrin binding–deficient forms of GUN4. Using these transgenic plants and particular mutants, we found that the porphyrin binding activity of GUN4 and Mg-chelatase contribute to the accumulation of chlorophyll, GUN4, and Mg-chelatase subunits. Also, we found that the porphyrin binding activity of GUN4 and Mg-chelatase affect the associations of GUN4 and ChlH with chloroplast membranes and have various effects on the expression of ROS-inducible genes. Based on our findings, we conclude that ChlH and GUN4 use distinct mechanisms to associate with chloroplast membranes and that mutant alleles of GUN4 and Mg-chelatase genes cause sensitivity to intense light by a mechanism that is potentially complex.
INTRODUCTION
Arabidopsisis thaliana GENOMES UNCOUPLED4 (GUN4) is a chloroplast-localized protein of 22 kD that was identified in a screen for mutant alleles that cause defects in plastid-to-nucleus signaling. GUN4 is encoded by a single-copy nuclear gene. The null mutant gun4-2 does not accumulate chlorophyll when grown in fluence rates that are optimal for chlorophyll accumulation in wild-type seedlings, but it can accumulate low levels of chlorophyll when grown in low fluence rates that are suboptimal for the wild type (Larkin et al., 2003; Peter and Grimm, 2009). Thus, unlike chlorophyll biosynthetic enzymes, GUN4 is not absolutely required for the accumulation of chlorophyll. GUN4 is conserved among organisms that perform oxygenic photosynthesis but is absent in Rhodobacter species, which perform anoxygenic photosynthesis (Larkin et al., 2003). In certain species, GUN4 is encoded by more than one gene. For instance, the genome of Synechocystis sp PCC 6803 (hereafter referred to as Synechocystis), encodes three relatives of GUN4 (Larkin et al., 2003). Wilde et al. (2004) knocked out two of these genes: the Synechocystis relative of GUN4 that encodes the protein with the highest sequence similarity to GUN4 (i.e., sll0558) and a more distant relative of GUN4 (i.e., sll1380). They found that sll0558 (hereafter referred to as SynGUN4) is required for the accumulation of chlorophyll, whereas the more distant relative of GUN4 (i.e., sll1380) is not required.
GUN4 promotes the accumulation of chlorophyll at least in part by stimulating Mg-chelatase (Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005); Mg-chelatase commits porphyrins to chlorophyll biosynthesis by catalyzing the insertion of Mg2+ into protoporphyrin IX (PPIX), yielding Mg-protoporphyrin IX (Mg-PPIX) (Willows, 2003; Masuda, 2008; Mochizuki et al., 2010). Mg-chelatase activity does not absolutely depend on GUN4 in vitro (Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005), which is consistent with chlorophyll accumulation not absolutely depending on GUN4 in vivo (Larkin et al., 2003; Peter and Grimm, 2009). Mg-chelatase activity does absolutely require three subunits in vitro and in vivo. These three subunits are conserved from prokaryotes to plants and are commonly referred to as BchH or ChlH, BchD or ChlD, and BchI or ChlI. In Arabidopsis, these subunits are 140, 79, and 40 kD, respectively. ChlH is the porphyrin binding subunit and is likely the Mg2+ binding subunit of Mg-chelatase. ChlI and ChlD are related to AAA-type ATPases and form an oligomer that interacts with ChlH and drives the ATP-dependent insertion of Mg2+ into PPIX (Willows et al., 2003; Masuda, 2008; Mochizuki et al., 2010).
GUN4 and cyanobacterial relatives of GUN4 stimulate their respective Mg-chelatases by a mechanism that depends on GUN4 binding the porphyrin substrate (PPIX) and the product (Mg-PPIX) of Mg-chelatase (Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005). This stimulatory mechanism also appears to depend on GUN4 binding to ChlH. Two lines of evidence suggest that GUN4 also binds ChlH: (1) ChlH is the only protein that copurifies with GUN4 from solubilized Arabidopsis thylakoid membranes on immunoaffinity columns that were constructed with affinity-purified anti-GUN4 antibodies, as shown by silver-stained SDS gels and mass spectrometry (Larkin et al., 2003); and (2) ChlH copurifies with an epitope-tagged SynGUN4, as shown by immunoblotting (Sobotka et al., 2008).
The porphyrin binding activity of GUN4 was first quantified for its cyanobacterial relatives. These studies used deuteroporphyrin IX (DPIX) and Mg-deuteroporphyrin IX (Mg-DPIX); DPIX and Mg-DPIX are significantly more water soluble than PPIX and Mg-PPX because they lack two vinyl groups found in PPIX and Mg-PPIX. Recently, a modification to a porphyrin binding assay allowed for the quantification of GUN4 binding to PPIX, Mg-PPIX, and various other natural porphyrins. GUN4 was found to bind Mg-PPIX with significantly higher affinity than all other porphyrins tested (Adhikari et al., 2009). These findings are consistent with the Mg-PPIX binding activity of GUN4 contributing significantly to its Mg-chelatase stimulatory activity, as was previously concluded (Verdecia et al., 2005).
A proteinaceous cofactor that regulates an enzyme by binding the enzyme, a substrate, and a product is unique. This type of enzymology may help protect plants from reactive oxygen species (ROS) that are produced by collisions between O2 and porphyrins that are exposed to bright light. GUN4 or GUN4-ChlH complexes were proposed to envelop and thereby shield PPIX and Mg-PPIX from collisions with O2 that might yield ROS (Larkin et al., 2003). ROS inactivate Mg-chelatase, inhibit chlorophyll biosynthesis (Willows et al., 2003; Aarti et al., 2006), and promote both photobleaching and cell death (Duke et al., 1991; Galvez-Valdivieso and Mullineaux, 2010). Such a shielding activity is potentially significant because PPIX and Mg-PPIX are not rapidly and completely used for chlorophyll biosynthesis but accumulate to readily detectable levels in vivo (Falbel and Staehelin, 1994; Pöpperl et al., 1998; Papenbrock et al., 1999; Mohapatra and Tripathy, 2007; Mochizuki et al., 2008; Moulin et al., 2008). PPIX and Mg-PPIX begin accumulating to readily detectable levels at dawn. Their levels decline later in the day to low levels or are undetectable at night (Falbel and Staehelin, 1994; Pöpperl et al., 1998; Papenbrock et al., 1999).
GUN4 and ChlH are found in both soluble and membrane-containing fractions when purified chloroplasts are lysed and fractionated. Inducing a rise in chloroplastic PPIX and Mg-PPIX promotes interactions between pea (Pisum sativum) chloroplast membranes and in vitro–translated and imported GUN4, pea GUN4, and pea ChlH. Inducing a rise in PPIX and Mg-PPIX also increases the amount of Mg-chelatase activity associated with pea chloroplast membranes (Adhikari et al., 2009). These data are consistent with GUN4 and/or GUN4-ChlH complexes binding and shielding the PPIX and Mg-PPIX that accumulate in chloroplast membranes from collisions with O2 that might yield ROS. These data are also consistent with GUN4 helping to channel porphyrins into chlorophyll biosynthesis by binding chlorophyll precursors and ChlH on chloroplast membranes, promoting interactions between ChlH and chloroplast membranes, and activating Mg-chelatase. In the following, we describe experiments with Mg-chelatase subunit gene mutants and stably transformed Arabidopsis plants that express only the porphyrin binding-deficient forms of GUN4 in order to test these ideas. We found that ChlH and GUN4 used distinct mechanisms to associate with chloroplast membranes. We also found that although mutant alleles of GUN4 and Mg-chelatase genes cause sensitivity to intense light by a potentially complex mechanism, we did not obtain evidence that the porphyrin binding activity of GUN4 attenuates ROS production.
RESULTS
gun4 Mutants Are Chlorophyll Deficient
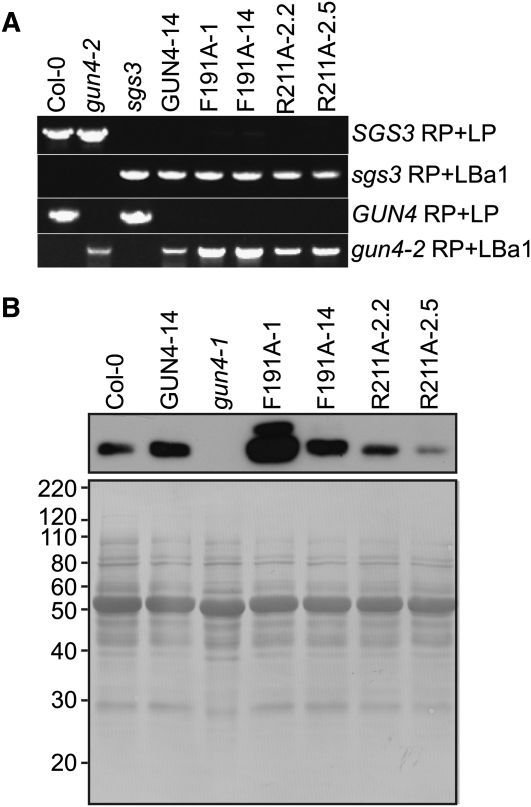
To test the in vivo significance of the porphyrin binding activities that were previously quantified for GUN4 in vitro, we stably transformed Arabidopsis plants with transgenes in which the native GUN4 promoter drives expression of wild-type GUN4 (Larkin et al., 2003) or with gun4 alleles that cause Arg-to-Ala (R211A) or Phe-to-Ala (F191A) substitutions in the amino acid sequence of the GUN4 protein. The F191A and R211A substitutions cause an ~2-fold increase in KdDPIX and a 3- to 5-fold increase in KdMg-DPIX (Adhikari et al., 2009). Changing residues that are homologous to Phe-191 and Arg-211 to Ala in SynGUN4 lowers the affinity of SynGUN4 for DPIX and Mg-DPIX and reduces the Mg-chelatase stimulatory activity of SynGUN4 in vitro (Davison et al., 2005; Verdecia et al., 2005). We transformed Arabidopsis plants that are heterozygous for gun4-2, a T-DNA insertion allele that is a null (Larkin et al., 2003), and homozygous for a T-DNA insertion allele of SUPPRESSOR OF GENE SILENCING3 (SGS3; N.D. Adhikari, unpublished data) that confers resistance to transgene-induced silencing (Butaye et al., 2004). We included this sgs3 allele because GUN4-expressing transgenes were previously shown to induce robust cosuppression of GUN4 in Arabidopsis (Larkin et al., 2003). Next, we identified stably transformed Arabidopsis plants that contain single GUN4- and gun4-expressing transgenes by scoring antibiotic resistance in segregating populations (N.D. Adhikari, unpublished data). We then isolated lines that are homozygous for the individual GUN4-expressing transgenes, gun4-2, and sgs3 (Figure 1A).
Figure 1.
Analysis of Arabidopsis Plants That Were Stably Transformed with GUN4-Derived Transgenes.
(A) Genotyping of the sgs3 and gun4-2 T-DNA insertion alleles in Arabidopsis plants that were stably transformed with GUN4-derived transgenes. Transgenic plants were homozygous for gun4-2, sgs3, the transgene indicated above lanes 4 to 8. Genomic DNA was extracted from the wild type (Col-0), the indicated mutant, or the indicated stably transformed line. Plants were screened for wild-type and T-DNA insertion alleles by PCR-based genotyping with oligonucleotides that can amplify PCR products from SGS3 (SGS3 RP + LP), GUN4 (GUN4 RP +LP), or particular T-DNA insertion alleles (sgs3 RP + LBa1 or gun4-2 RP + LBa1). PCR products were analyzed by electrophoresis in agarose gels followed by staining with ethidium bromide.
(B) Analysis of GUN4 protein levels in Arabidopsis plants that are stably transformed with GUN4-derived transgenes. Transgenic plants were homozygous for gun4-2, sgs3, and the indicated transgene. Whole seedling extracts were prepared from the indicated mutant or the indicated stably transformed line grown in 100 μmol m−2 s−1 broad-spectrum white light. Aliquots of these whole seedling extracts that contained 10 μg of protein were analyzed by immunoblotting using anti-GUN4 antibodies (top panel) to detect the 22-kD band that corresponds to GUN4. After immunoblotting, the polyvinylidene fluoride membrane was stained with Coomassie blue (bottom panel). Mass standards are indicated at the left in kilodaltons.
The previously described mutant alleles for GUN4 in Arabidopsis are severe loss-of-function alleles that cause barely detectable levels of protein to accumulate or are protein nulls (Larkin et al., 2003; Peter and Grimm, 2009). By contrast, immunoblotting analysis of whole seedling extracts prepared from 7-d-old transgenic seedlings indicated that the transgenic lines GUN4-14, F191A-14, and R211A-2.2 express similar but slightly higher levels of GUN4, R211A, and F191A proteins, respectively, relative to the wild type (Figure 1B). We found that other transgenic lines prepared by the same procedures express much higher (F191A-1) or much lower (R211A-2.5) levels of either F191A or R211A than the wild type (Figure 1B). We attribute these differences to the insertion of these transgenes into different positions within the genome.
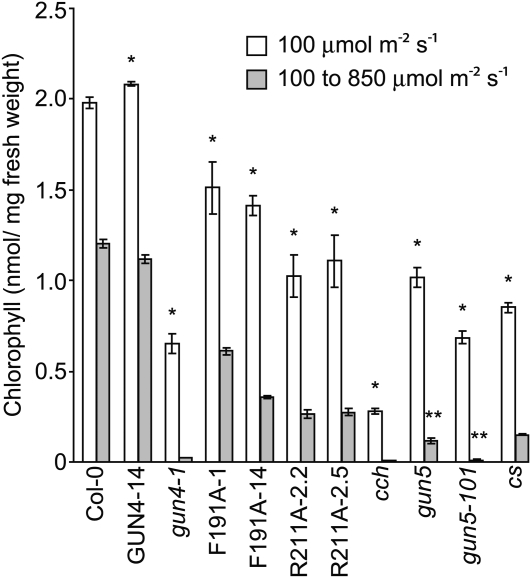
Based on previous analyses of GUN4 and SynGUN4 (Verdecia et al., 2005; Adhikari et al., 2009), we predicted that lines expressing F191A and R211A would contain reduced Mg-chelatase activity and therefore would exhibit chlorophyll deficiencies relative to the wild type. Indeed, we found that, like the chlorophyll-deficient gun4-1 (Vinti et al., 2000; Mochizuki et al., 2001; Larkin et al., 2003) that causes a Leu-to-Phe (L88F) substitution (Larkin et al., 2003), the F191A- and R211A-expressing lines accumulate 23 to 48% less chlorophyll per mg fresh weight than the wild type when these seedlings were grown for 7 d in 100 μmol m−2 s−1 white light (Figure 2; see Supplemental Figures 1A and 1B online). Under these conditions, gun4-1 accumulated 67% less chlorophyll per mg fresh weight than the wild type (Figure 2; see Supplemental Figures 1A and 1B online), which is consistent with previous analyses of gun4-1 (Vinti et al., 2000; Mochizuki et al., 2001).
Figure 2.
Analysis of Chlorophyll Levels in gun4, chlH/gun5, and cs Mutants Grown under Various Fluence Rates.
Wild type (Col-0) and the indicated mutants and transgenic lines were grown in continuous 100 μmol m−2 s−1 white light for 7 d (white bars) or for 3 d in continuous 100 μmol m−2 s−1 white light and then 4 d in continuous 850 μmol m−2 s−1 white light (gray bars). Chlorophyll was extracted from at least three biological replicates for each mutant or line in each condition. Error bars indicate se. The statistical significance of the difference between the chlorophyll levels of the wild type (Col-0) and a particular mutant or transgenic line was calculated with an unpaired t test. A single asterisk indicates a statistically significant difference in chlorophyll levels between Col-0 and a particular mutant or transgenic line (P = 0.0001 to 0.03). Using an unpaired t test, we calculated the statistical significance of the decrease in chlorophyll levels following the fluence rate shift from 100 μmol m−2 s−1 to 850 μmol m−2 s−1 white light between (1) gun5-101 and gun4-1 and (2) between gun5 and the following mutants and transgenic lines: cs, R211A-2.2, and R211A-2.5. A double asterisk indicates a statistically significant difference (P = 0.003 to 0.04).
Chlorophyll Deficiency in gun4 Mutants Is Enhanced by Intense Light
Mutants that are impaired in their ability to synthesize chlorophyll were previously reported to exhibit more severe chlorophyll deficiencies after fluence rates were increased (Falbel et al., 1996). To test whether these F191A- and R211A-expressing lines exhibit light-sensitive phenotypes similar to other chlorophyll-deficient mutants, we transferred them to a higher fluence rate. We transferred 3-d-old seedlings from 100 μmol m−2 s−1 to 850 μmol m−2 s−1 white light and grew them for an additional 4 d. The transgenic line that expresses wild-type GUN4 (i.e., GUN4-14) accumulates levels of chlorophyll very similar to those of the wild type in both light conditions tested (Figure 2). The wild type and GUN4-14 grown in 850 μmol m−2 s−1 white light contain 54 to 62% as much chlorophyll as the same seedlings grown in 100 μmol m−2 s−1 white light (Figure 2).
We found that high-intensity light enhances chlorophyll deficiencies in gun4-1 and the F191A- and R211A-expressing lines relative to the wild type. When 3-d-old seedlings were transferred from 100 μmol m−2 s−1 to 850 μmol m−2 s−1 and grown for an additional 4 d, the transgenic lines contained 25 to 54% of the chlorophyll as found in the wild type, and gun4-1 contained only 4% of the chlorophyll relative to the wild type (Figure 2; see Supplemental Figures 2A and 2B online). With the exception of F191A-1, the decrease in chlorophyll was greater in these transgenic lines and gun4-1 than in the wild type following the fluence rate shift (see Supplemental Table 1 online). Although F191A-1 accumulates nearly 2-fold more chlorophyll than F191-14 when seedlings are grown in 850 μmol m−2 s−1 white light, chlorophyll accumulates to the same levels in R211A-2.2 and R211A-2.5 when they are grown in 850 μmol m−2 s−1 white light (Figure 2). Thus, variations in the levels of F191A and R211A protein would appear to affect the accumulation of chlorophyll only in lines such as F191A-1, which accumulates the F191A protein at much higher levels than the GUN4 protein in the wild type (Figure 1A).
Chlorophyll Deficiency in chlH and chlI Mutants Is Enhanced by Intense Light
Next, we tested whether available Arabidopsis Mg-chelatase subunit gene mutants exhibited light-sensitive phenotypes similar to gun4-1, R211A, and F191A. For these experiments, we tested gun5 and cch mutants, which are missense alleles of the ChlH gene in Arabidopsis (Mochizuki et al., 2001). We hereafter refer to this gene as ChlH/GUN5 to avoid confusion. We also tested the Arabidopsis gun5-101 mutant, which we isolated from a new gun mutant screen (Ruckle et al., 2007). gun5-101 is a missense allele that causes a Phe-to-Leu (P450L) substitution in the derived amino acid sequence of ChlH/GUN5. We cloned gun5-101 by positional cloning (see Supplemental Figures 3A and 3B online). gun5, cch, and gun5-101 accumulate 49 to 86% less chlorophyll than the wild type when seedlings are grown in 100 μmol m−2 s−1 for 7 d (Figure 2; see Supplemental Figures 1A and 1B online). gun5 and gun5-101 accumulate 90 and 99% less chlorophyll, respectively, than the wild type, and chlorophyll is not detectable in cch when seedlings are grown in 100 μmol m−2 s−1 for 3 d and then transferred to 850 μmol m−2 s−1 for 4 d (Figure 2; see Supplemental Figures 2A and 2B online). We also tested the light sensitivity of the cs mutant, which is strikingly deficient in chlorophyll because of a defect in the ChlI subunit of Mg-chelatase (Koncz et al., 1990; Mochizuki et al., 2001; Adhikari et al., 2009). The cs mutant contains 57% less chlorophyll than the wild type when seedlings are grown in 100 μmol m−2 s−1 for 7 d (Figure 2; see Supplemental Figures 1A and 1B online) and 88% less chlorophyll than the wild type when 3-d-old seedlings are transferred from 100 μmol m−2 s−1 to 850 μmol m−2 s−1 and grown for an additional 4 d (Figure 2; see Supplemental Figures 2A and 2B online). Thus, the decrease in chlorophyll content is greater in gun5, gun5-101, cch, and cs than in the wild type following the fluence rate shift (see Supplemental Table 1 online). Among all of these mutants and transgenic lines, the most chlorophyll-deficient seedlings in 100 μmol m−2 s−1 white light exhibited the greatest chlorophyll deficiency after they were transferred to 850 μmol m−2 s−1 (Figure 2; see Supplemental Table 1 online). Also, we found that the mutant alleles of the ChlH/GUN5 and ChlI genes that we tested cause greater light sensitivity than the mutant alleles of the GUN4 genes that we tested. Specifically, gun5-101 accumulates the same level of chlorophyll as gun4-1 in 100 μmol m−2 s−1 but more than 59% less than gun4-1 in 850 μmol m−2 s−1. gun5 and cs accumulate similar levels of chlorophyll as both R211A lines in 100 μmol m−2 s−1 but 41 to 56% less chlorophyll than both R211A lines in 850 μmol m−2 s−1 (Figure 2).
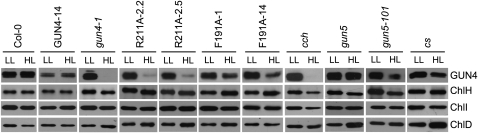
To test for possible mechanisms that might cause these light-sensitive phenotypes, we used immunobloting to monitor the levels of GUN4 and Mg-chelatase subunit levels in response to light intensity. We observed a reduction in GUN4 protein levels in R211A, F191A, gun4-1, cch, and gun5-101 relative to the wild type after seedlings were transferred from 100 μmol m−2 s−1 to 850 μmol m−2 s−1 white light (Figure 3). By contrast, transferring gun5 to 850 μmol m−2 s−1 did not similarly affect the accumulation of the GUN4 protein (Figure 3). The cs allele causes chlorophyll deficiencies that are similar to these R211A and F191A lines, gun4-1, cch, and gun5-101 (Figure 2). If the cs allele significantly affects GUN4 protein levels, this effect is much less severe than the effects of the R211A and F191A lines, gun4-1, cch, and gun5-101 (Figure 3). These data indicate that particular amino acid substitutions in GUN4 and ChlH can reduce GUN4 protein levels when seedlings are grown in 850 μmol m−2 s−1 white light. These effects appear specific to GUN4; ChlH/GUN5, ChlI, and ChlD levels were either unaffected or only slightly reduced following this fluence rate shift (Figure 3).
Figure 3.
Analysis of GUN4 and Mg-Chelatase Subunit Levels in 100 μmol m−2 s−1 and 850 μmol m−2 s−1 White Light.
Seedlings were grown either in 100 μmol m−2 s−1 white light for 7 d (LL) or in 100 μmol m−2 s−1 for 3 d and then transferred 850 μmol m−2 s−1 white light for 4 d (HL) as indicated in Figure 2. Whole seedling extracts were prepared from the indicated mutant or the indicated stably transformed line. Aliquots of these whole seedling extracts that contained 10 μg of protein were analyzed by immunoblotting using the following antibodies: anti-GUN4, anti-ChlH, anti-ChlI, or anti-ChlD. When faint bands were observed in extracts from HL-treated seedlings, exposures were adjusted so that these faint bands were observable.
GUN4 and ChlH/GUN5 Associate with Chloroplast Membranes
When chloroplasts are lysed and fractionated into soluble fractions and pellet fractions that previously were shown to contain chloroplast membranes (Perry et al., 1991), more GUN4 and ChlH/GUN5 protein accumulates in the membrane-containing pellet fraction when an increase is induced in chloroplastic porphyrin levels (Adhikari et al., 2009). ChlH/GUN5 can form aggregates (Sirijovski et al., 2006) that could potentially accumulate in the pellet fraction without binding membranes. However, when subjected to the conditions and procedures used here, GUN4 and Mg-chelatase were previously shown to accumulate in the pellet fraction only when chloroplast membranes are present (Adhikari et al., 2009). Additionally, GUN4 and ChlH/GUN5 cosediment with chloroplast envelope membranes near the center of discontinuous Suc gradients, rather than accumulating only in the pellet fractions of these Suc gradients (Nakayama et al., 1998; Larkin et al., 2003). Based on these data, we conclude that GUN4 and ChlH/GUN5 accumulate in the membrane-containing pellet fraction because they interact with chloroplast membranes and not because they precipitate during the chloroplast lysis and fractionation procedure used here.
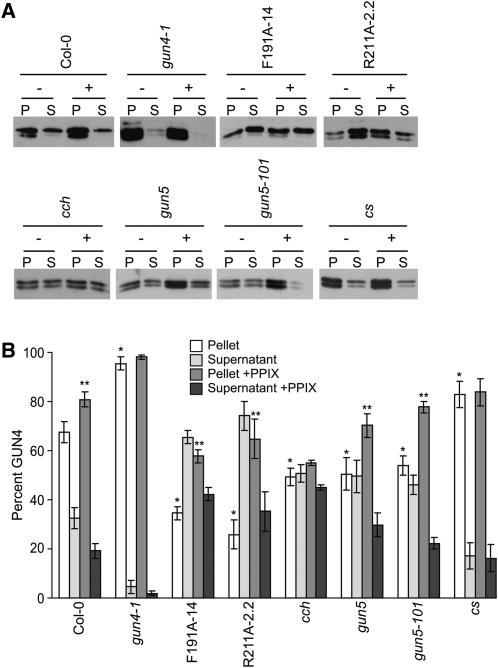
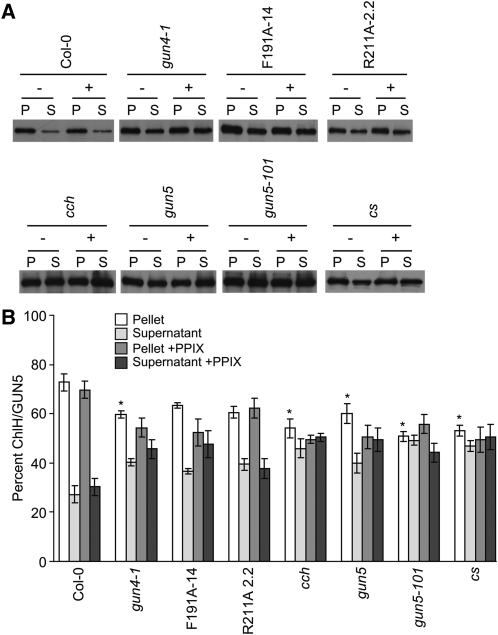
Considering our previous finding that porphyrin binding promotes the association of GUN4 with chloroplast membranes (Adhikari et al., 2009), we expected that the porphyrin binding-deficient versions of GUN4 (i.e., F191A and R211A) would not associate with chloroplast membranes as stably as GUN4. We also tested whether interactions between GUN4 and chloroplast membranes might increase in gun4-1. Amino acid substitutions that are homologous to the L88F substitution caused by gun4-1 increase the porphyrin binding affinities of Thermosynechococcus elongatus GUN4 and Synechocystis GUN4 (Davison et al., 2005). The gun4-1 allele also causes a severe reduction in GUN4 protein levels in Arabidopsis (Larkin et al., 2003) and causes the GUN4 protein to accumulate in the insoluble fraction when expressed in Escherichia coli, in contrast with the GUN4 protein from the wild type (Adhikari et al., 2009). We found that when purified chloroplasts were lysed and fractionated, a significantly lower percentage of GUN4 was associated with the membrane-containing pellet fractions derived from R211A-2.2 and F191A-14 (26 to 34% in the pellet fraction) than in the membrane-containing pellet fractions derived from the wild type (67% in the pellet fraction) (Figures 4A and 4B). A greater proportion of GUN4 appeared in the supernatant fractions derived from R211A-2.2 and F191A-14 than in the wild type (Figures 4A and 4B). We also found that a significantly higher percentage of GUN4 associated with the membrane-containing pellet fractions derived from gun4-1 (93% in the pellet fraction) than in the membrane-containing pellet fractions derived from the wild type (67% in the pellet fraction) (Figures 4A and 4B). A lower proportion of GUN4 appeared in the supernatant fractions derived from gun4-1 than in supernatant fractions derived from the wild type (Figures 4A and 4B).
Figure 4.
Distribution of GUN4 in Lysed and Fractionated Chloroplasts That Were Either Fed or Not Fed with PPIX.
(A) Immunoblot analysis of lysed and fractionated chloroplasts using anti-GUN4 antibodies. Chloroplasts were purified from gun4-1, F191A-14, R211A-2.2, cch, gun5, gun5-101, and cs. Purified intact chloroplasts (200 μg) were either fed (+) or not fed (−) with 20 μM PPIX. Chloroplasts were then fractionated into soluble (S) and membrane-containing pellet (P) fractions of equal volume. Equal volumes were analyzed by SDS-PAGE and immunoblotting with anti-GUN4 antibodies. Representative immunoblots are shown.
(B) Proportions of GUN4 in soluble and pellet fractions. The chemiluminescence that was emitted from the immunoreactive bands described in (A) was quantified. The percentage of GUN4 in the pellet (white bars) and supernatant (light-gray bars) fractions derived from chloroplasts that were not fed PPIX and the percentage of GUN4 in the pellet (medium-gray bars) and supernatant (dark-gray bars) fractions derived from chloroplasts that were fed PPIX are indicated for wild type (Col-0) and each mutant and transgenic line. Results from at least five independent experiments for Col-0 and four independent experiments for all mutants and transgenic lines are shown. Error bars indicate se. A single asterisk indicates a statistically significant difference in the levels of GUN4 in the membrane-containing pellet fractions between Col-0 and the gun4 mutants gun4-1, F191A-14, and R211A-2.2 (P < 0.0005) and between Col-0 and the chlH/gun5 mutants cch (P < 0.01), gun5, gun5-101, and cs (P < 0.04). A double asterisk indicates a statistically significant difference in the percent GUN4 in the membrane-containing pellet fractions derived from chloroplasts that were fed PPIX and not fed PPIX and were purified from Col-0 (P = 0.009), F191A-14 (P = 0.002), R211A-2.2 (P = 0.002), gun5 (P = 0.04), and gun5-101 (P = 0.007).
Although previous findings indicate that GUN4 associates with ChlH and that both proteins interact with chloroplast membranes (Nakayama et al., 1998; Larkin et al., 2003; Adhikari et al., 2009), whether interaction between these two proteins is required for their association with chloroplast membranes was not previously established. To test whether the interactions between GUN4 and chloroplast membranes depend on ChlH/GUN5, we purified chloroplasts from gun5, gun5-101, and cch. We then lysed them, fractionated them into soluble and membrane-containing pellet fractions, and analyzed these fractions by immunoblotting with anti-GUN4 antibodies. We found that a significantly lower percentage of GUN4 associated with the membrane-containing pellet fractions derived from gun5, gun5-101, and cch (49 to 54% in the pellet fraction) than in the membrane-containing pellet fractions that were derived from the wild type (67% in the pellet fraction; Figures 4A and 4B). We found a higher percentage of GUN4 in the supernatant fractions derived from gun5, gun5-101, and cch than from the wild type (Figures 4A and 4B).
To test whether other defects in other Mg-chelatase subunits besides ChlH/GUN5 might affect interactions between GUN4 and chloroplast membranes, we performed a similar analysis with cs. When we purified chloroplasts from cs and then lysed and fractionated them into soluble and membrane-containing pellet fractions, we found a small but significant increase in the percentage of GUN4 associated with the membrane-containing pellet fraction (83% in the pellet fraction) relative to the wild type (67% in the pellet fraction) and a decrease in the proportion of GUN4 in the supernatant fractions derived from cs relative to the wild type (Figures 4A and 4B).
To test whether interactions between ChlH/GUN5 and chloroplast membranes are influenced by GUN4 or Mg-chelatase activity, we reanalyzed these same fractions by immunoblotting with anti-ChlH/GUN5 antibodies. A lower percentage of ChlH/GUN5 associated with the chloroplast membrane–containing pellet fractions (51 to 65%) relative to the wild type (73% in the pellet fraction) (Figures 5A and 5B) in all of the mutants and transgenic lines tested. A greater percentage of ChlH/GUN5 accumulated in the soluble fractions derived from all of the mutants and transgenic lines tested. In contrast with the other mutants and transgenic lines, the decrease in the proportion of ChlH/GUN5 associated with the chloroplast membrane–associated pellet fractions derived from F191A-14 and R211A-2.2 was not statistically significant (Figures 5A and 5B).
Figure 5.
Distribution of ChlH/GUN5 in Lysed and Fractionated Chloroplasts That Were Either Fed or Not fed with PPIX.
(A) Analysis of lysed and fractionated chloroplasts by immunoblotting with anti-ChlH/GUN5 antibodies. The same fractions described in Figure 4 were analyzed by immunoblotting with anti-ChlH/GUN5 antibodies. As described for Figure 4, 200 μg of purified intact chloroplasts were either fed (+) or not fed (−) with 20 μM PPIX. These chloroplasts were then fractionated into soluble (S) and membrane-containing pellet (P) fractions of equal volume. Equal volumes were analyzed by SDS-PAGE and immunoblotting.
(B) Proportions of ChlH/GUN5 in soluble and membrane fractions. The percentage of ChlH/GUN5 in the pellet (white bars) and supernatant (light-gray bars) fractions derived from chloroplasts that were not fed PPIX and the percentage of ChlH/GUN5 in the pellet (medium-gray bars) and supernatant (dark-gray bars) fractions derived from chloroplasts that were fed PPIX are indicated for the wild type (Col-0) and each mutant and transgenic line. Results from seven independent experiments are shown for Col-0, five independent experiments are shown for cs, and four independent experiments are shown for all other mutants and transgenic lines. Error bars indicate se. The asterisk indicates a statistically significant difference in the levels of ChlH/GUN5 in the membrane-containing pellet fractions between Col-0 and gun4-1 (P = 0.02), between Col-0 and cch (P = 0.008), and between gun5 (P = 0.04), gun5-101, and cs (P < 0.002).
In summary, we found similar decreases in the amount of GUN4 and ChlH/GUN5 that associates with chloroplast membranes in gun5, gun5-101, and cch relative to the wild type (Figures 4 and 5; see Supplemental Table 2 online) and striking differences in the amount of GUN4 and ChlH/GUN5 that associates with chloroplast membranes in gun4-1, F191A-14, R211A-2.2, and cs relative to the wild type. We found a higher percentage of GUN4 and lower percentage of ChlH/GUN5 associated with the chloroplast membrane–containing pellet fraction in both gun4-1 and cs relative to the wild type (Figures 4 and 5; see Supplemental Table 2 online). In chloroplasts purified from F191A-14 and R211A-2.2, we observed a statistically significant decrease of ~30 to 40% in the GUN4 that associates with the chloroplast membrane–containing pellet fraction, but we did not observe statistically significant decreases in the percentage of ChlH/GUN5 that associates with the chloroplast membrane–containing pellet fraction (Figures 4 and 5; see Supplemental Table 2 online).
Porphyrin Binding Increases the Affinity of GUN4 for Chloroplast Membranes
The distribution of pea GUN4 and in vitro–translated and imported GUN4 are similar in fractionated pea chloroplasts to those reported here for fractionated Arabidopsis chloroplasts. Inducing an increase in the porphyrin levels of purified pea chloroplasts promotes interactions between pea chloroplast membranes and both pea GUN4 and in vitro–translated and imported GUN4. By contrast, in vitro–translated and imported R211A or F191A do not associate with pea chloroplast membranes, regardless of whether porphyrin levels are increased (Adhikari et al., 2009). The inability of elevated porphyrin levels to promote interactions between pea chloroplast membranes and either R211A or F191A is consistent with the following: (1) R211A and F191A disrupting interactions between GUN4 and chloroplast membranes by not only affecting porphyrin binding but also by inhibiting some other function of GUN4 besides porphyrin binding, and (2) a technical limitation of the pea system such as the vast molar excess of pea GUN4 competing more effectively with in vitro–translated and imported R211A and F191A than with in vitro–translated and imported wild-type GUN4. To distinguish between these possibilities, we induced an increase in the porphyrin levels of chloroplasts that were purified from gun4 and gun5/chlH mutants by feeding these chloroplasts with PPIX, as previously described by Adhikari et al. (2009). PPIX feeding caused a significant increase in the association of GUN4 with chloroplast membranes of wild-type chloroplasts (Figures 4A and 4B; see Supplemental Table 3 online). This increase in the association of GUN4 with chloroplast membranes from Arabidopsis was similar to increases previously reported with chloroplast membranes from pea (Adhikari et al., 2009). PPIX feeding did not significantly increase the percentage of GUN4 in the chloroplast membrane–containing pellet fractions derived from gun4-1 and cs (Figures 4A and 4B; see Supplemental Table 3 online). Increases in PPIX levels may not further promote interactions between GUN4 and chloroplast membranes in gun4-1 and cs because the bulk of GUN4 already stably associates with the membrane-containing pellet fractions of chloroplasts purified from gun4-1 and cs prior to PPIX feeding (Figures 4A and 4B; see Supplemental Table 3 online). By contrast, feeding PPIX to chloroplasts purified from the F191A-14 and R211A-2.2 caused a significant 20 to 40% increase in the GUN4 retained in the membrane-containing pellet fraction (Figures 4A and 4B; see Supplemental Table 3 online). Our conclusion, based on these findings, is that porphyrin binding increases the affinity of GUN4 for chloroplast membranes. We conclude that the R211A and F191A substitutions attenuate interactions between the GUN4 protein and chloroplast membranes of Arabidopsis by decreasing the affinity of GUN4 for porphyrins, thereby decreasing the proportion of porphyrin-bound GUN4. We conclude that inducing a rise in chloroplastic porphyrin levels promotes interactions between GUN4 and chloroplast membranes of Arabidopsis by increasing the proportion of porphyrin-bound GUN4, regardless of whether the amino acid sequence is wild type or contains the F191A and R211A substitutions. We also conclude that in Adhikari et al. (2009), the in vitro–translated and imported R211A or F191A probably do not associate with pea chloroplast membranes regardless of whether porphyrin levels are increased because of a technical limitation of the pea system, as described above and as previously suggested by Adhikari et al. (2009).
We also observed that PPIX feeding of chloroplasts purified from gun5 and gun5-101 (but not cch) caused a significant 20% increase in the GUN4 associated with the membrane-containing pellet fractions (Figures 4A and 4B; see Supplemental Table 3 online). In summary, we found that amino acid substitutions that attenuate the porphyrin binding activity of GUN4 and the activity of ChlH/GUN5 promote the dissociation of GUN4 from chloroplast membranes when chloroplasts are lysed and fractionated. Also, inducing an increase in PPIX levels can partially reverse these membrane dissociation effects, thereby increasing the proportion of GUN4 in the membrane-containing pellet fraction. The only exception was that PPIX feeding did not increase the proportion of GUN4 in the membrane-containing pellet fraction derived from cch. These data persuade us that ChlH/GUN5 helps tether GUN4 to chloroplast membranes when chloroplasts are lysed and fractionated.
Inducing a rise in chloroplastic porphyrin levels can promote interactions between pea ChlH/GUN5 and pea chloroplast membranes, although not to the same degree as with pea GUN4 and pea chloroplast membranes (Adhikari et al., 2009). To test whether the porphyrin binding promotes interactions between ChlH/GUN5 as it does with GUN4, we analyzed these same fractions by immunoblotting with anti-ChlH/GUN5 antibodies. We found that in contrast with GUN4, PPIX feeding does not affect the association between GUN5/ChlH and chloroplast membranes in the wild type or in any of these mutants and transgenic lines (Figures 5A and 5B).
Some but Not All ROS-Inducible Genes Are Expressed at Higher Levels in F191A-14 and R211A-2.2 Than in the Wild Type
The chlorophyll-deficient phenotypes of chlorophyll biosynthesis mutants are dependent on fluence rates. In bright light, deficiencies in chlorophyll, chlorophyll binding proteins, and grana thylakoids are enhanced relative to the wild type (Allen et al., 1988; Falbel et al., 1996). Mutant alleles that attenuate Mg-chelatase activity were proposed to enhance sensitivity to high-intensity light at least in part by causing PPIX to overaccumulate. This overaccumulated PPIX was proposed to diffuse away from the site of biosynthesis and to induce the production of singlet oxygen (1O2) in illuminated chloroplast membranes (Falbel and Staehelin, 1994; Falbel et al., 1996). Differential production of ROS from the light reactions of photosynthesis (Niyogi, 1999; Li et al., 2009) in these mutants might also contribute to their light sensitivities. To test this possibility and to distinguish between ROS production from misregulated chlorophyll biosynthesis and photosynthesis, we first grew the wild type, mutants, and transgenic lines in a low fluence rate (10 μmol m−2 s−1) white light. The levels of chlorophyll, levels and compositions of the photosystems, and thylakoid ultrastructures of particular chlorophyll-deficient mutants and the wild type are more similar when plants are grown under low fluence rates (Allen et al., 1988; Falbel et al., 1996). Consistent with these previous findings, the chlorophyll levels in these mutants and transgenic lines tested range from ~70 to 80% of the wild type or are essentially the same as the wild type when seedlings are grown in 10 μmol m−2 s−1 white light (see Supplemental Figures 4A to 4C online).
Based on these data, we expected that the light reactions of photosynthesis might produce similar ROS in all of these mutants, transgenic lines, and the wild type when they are grown in 10 μmol m−2 s−1 white light. To test this idea, we assessed photosynthetic properties in intact leaves by measuring saturation pulse chlorophyll a fluorescence yield changes in F191A-14, R211A-2.2, and the wild type. We measured the maximal (Fv/Fm) and steady state (ΦII) photosystem II quantum yields, total nonphotochemical quenching of excitation energy (NPQ), energy-dependent exciton quenching (qE), and long-lived inhibitory quenching (qI) in seedlings grown in 10 μmol m−2 s−1 continuous white light (see Supplemental Figures 5A to 5D online). Consistent with the light reactions of photosynthesis functioning similarly in F191A-14, R211A-2.2, and the wild type when they are grown in 10 μmol m−2 s−1 white light, we found no significant differences in these photosynthetic parameters among F191A-14, R211A-2.2, and the wild type using an unpaired t test (P > 0.05).
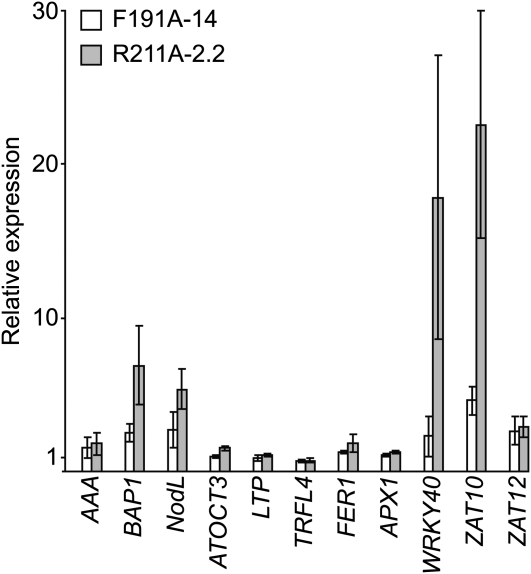
To test whether high fluence rates induce a greater production of 1O2 and other ROS in F191A-14 and R211A-2.2 relative to the wild type, we grew them in 10 μmol m−2 s−1 for 7 d and then transferred them to 850 μmol m−2 s−1 white light. We extracted RNA from seedlings that were collected immediately before (0 h) and at 0.5, 1, and 3 h after this fluence rate shift. We then quantified the expression of five genes whose expression is specifically induced by the 1O2 that is produced in the flu mutant or by Rose Bengal treatments. flu alleles cause the overaccumulation of protochlorophyllide, a chlorophyll precursor. Protochlorophyllide and Rose Bengal can function as photosensitizers by transferring energy to O2 yielding 1O2. These 1O2-inducible genes encode an AAA-ATPase (AAA), BON ASSOCIATION PROTEIN1 (BAP1), a nodulin-like protein (NodL), an organic cation/carnitine transporter (ATOCT3), a member of the protease inhibitor/seed storage/lipid transfer protein family protein (LPT), and a protein that is similar to a telomere binding protein (TRFL4) (op den Camp et al., 2003; Gadjev et al., 2006; Baruah et al., 2009a, 2009b). We also quantified the expression of two genes whose expression is specifically induced by superoxide (O2•–) and hydrogen peroxide (H2O2). These O2•–- and H2O2-inducible genes encode ascorbate peroxidase 1 (APX1) and ferritin1 (FER1) (Petit et al., 2001; Rizhsky et al., 2004). We also quantified the expression of three genes whose expression is induced by a number of different types of ROS, including 1O2. These ROS-inducible genes encode WRKY40, ZAT10, and ZAT12 (Gadjev et al., 2006).
At 0 h, we found that the expression of BAP1, NodL, WRKY40, and ZAT10 was 5- to 20-fold higher in R211A-2.2 relative to the wild type and that some of these genes are expressed at 2- to 5-fold higher levels in F191A-14 relative to the wild type at 0 h (Figure 6). By contrast, we found that AAA, ATOCT3, LTP, TRFL4, FER1, APX1, and ZAT12 were not present at elevated levels in F191A-14 and R211A-2.2 relative to the wild type (Figure 6). Consistent with previous work (Gadjev et al., 2006; Baruah et al., 2009a, 2009b), we found that the transcripts derived from most of these genes accumulate in the wild type after the fluence rate shift with various kinetics (see Supplemental Figure 6 online). We found that the expression of these genes in F191A-14 and R211A-2.2 resembled the wild type after the 10 to 850 μmol m−2 s−1 fluence rate shift (see Supplemental Figure 6 online).
Figure 6.
Analysis of ROS-Regulated Gene Expression in F191A and R211A Grown in 10 μmol m−2 s−1 White Light.
The wild type (Col-0), F191A-14, and R211A-2.2 were grown for 7 d in 10 μmol m−2 s−1 white light. Levels of transcripts indicated were quantified by means of qRT-PCR. Expression in F191A-14 (white bars) and R211A-2.2 (gray bars) is reported relative to Col-0, which is assigned a value of 1. Four biological replicates were analyzed for each line. Error bars represent se.
Photoperiodic Light Induces the Expression of Some but Not All ROS-Inducible Genes in R211A-2.2, cch, and cs
Considering the data presented in Figure 6, we conclude that either (1) porphyrin-derived ROS is regulating the expression of ROS-inducible genes by a mechanism that is at least partially distinct from the ROS that is derived from overaccumulated protochlorophyllide in flu, or (2) ROS is not regulating the expression of these ROS-inducible genes in F191A-14 and R211A-2.2 when seedlings are grown in 10 μmol m−2 s−1 white light. To test whether porphyrin-derived ROS might induce the expression of these ROS-inducible genes in dim light, we monitored their expression in continuous and photoperiodic light. There is a burst of PPIX and Mg-PPIX biosynthesis at dawn that causes PPIX and Mg-PPIX to accumulate to readily detectable levels (Falbel and Staehelin, 1994; Pöpperl et al., 1998; Papenbrock et al., 1999). This diurnal accumulation of porphyrins can induce photooxidative stress in mutants with defects in porphyrin metabolism (Meskauskiene et al., 2001; Huq et al., 2004). We propose that GUN4 or a GUN4-ChlH/GUN5 complex binds pools of PPIX and Mg-PPIX that accumulate in photoperiodic light to shield them from collisions with O2. If porphyrin-derived ROS is inducing the expression of these genes, photoperiodic light should induce higher levels of these ROS-inducible genes than continuous light in gun4 and Mg-chelatase subunit gene mutants.
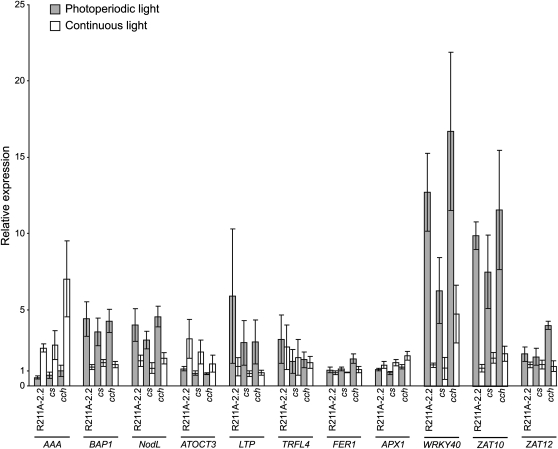
To test this idea, we quantified ROS-inducible gene expression in wild-type, R211A-2.2, cch, and cs seedlings that were grown in 2 μmol m−2 s−1 continuous white light and in a photoperiod that contained 12 h of 2 μmol m−2 s−1 white light followed by 12 h of darkness. Including cch and cs in this experiment allows us to also test whether GUN4 and/or ChlH/GUN5 might perform porphyrin binding functions that are distinct from their chlorophyll biosynthetic functions. If the pools of GUN4-bound and/or GUN4-ChlH/GUN5-bound porphyrins that accumulate during the diurnal cycle are distinct from the pools of GUN4-bound and/or GUN4-ChlH/GUN5-bound porphyrins that are associated with the chlorophyll biosynthetic pathway, then mutants and transgenic lines that attenuate GUN4 and ChlH/GUN5 activity, such as R211A and cch, should accumulate more ROS than mutants such as cs that attenuates the activity of ChlI, a Mg-chelatase subunit that does not bind porphyrins and only contributes to chlorophyll biosynthesis.
We grew wild-type, R211A-2.2, cch, and cs seedlings in 2 μmol m−2 s−1 continuous and photoperiodic white light for 7 d. We then quantified Fv/Fm, ΦII, NPQ, qE, and qI in the wild-type, R211A-2.2, cch, and cs seedlings that were grown in both conditions (see Supplemental Figures 7 and 8 online). Using an unpaired t test, we found that none of the differences that we observed in these photosynthetic parameters were statistically significant (P > 0.05) except that cs exhibited a small but statistically significant reduction in ΦII relative to the wild type when grown in 2 μmol m−2 s−1 photoperiodic white light (P < 0.05). Based on these data, we conclude that the ROS that is produced by the light reactions of photosynthesis is very similar or, in most cases, essentially the same as the wild type when R211A-2.2, cch, and cs are grown in 2 μmol m−2 s−1 continuous white light and 2 μmol m−2 s−1 photoperiodic white light. We found that the expression of the BAP1 and NodL is induced from 2- to 4-fold higher levels in R211A-2.2, cs, and cch relative to the wild type when seedlings are grown in photoperiodic relative to continuous light (Figure 7). We also found that the expression of WRKY40 and ZAT10 was induced from 6- to 17-fold in R211A-2.2, cs, and cch relative to the wild type in photoperiodic relative to continuous light (Figure 7). The expression of AAA, ATOCT3, LTP, TRFL4, FER1, APX1, and ZAT12 was more similar in R211A-2.2, cs, cch, and the wild type in photoperiodic and continuous light (Figure 7). Additionally, transcripts from BAP1, NodL, WRKY40, and ZAT10 did not accumulate to higher levels in R211A and cch than they did in cs (Figure 7).
Figure 7.
Analysis of ROS-Inducible Gene Expression in Photoperiodic and Continuous Light.
Seedlings were grown for 7 d in 12 h of 2 μmol m−2 s−1 white light followed by 12 h darkness (gray bars) or in continuous 2 μmol m−2 s−1 white light (white bars). All seedlings were harvested 1 h after dawn. The expression of ROS-inducible genes was quantified by means of qRT-PCR as described in Figure 6. Expression is reported relative to Col-0 in continuous light, which was assigned a value of 1. Four biological replicates were analyzed for each line in each condition. Error bars represent se.
The expression of AAA was higher when seedlings were grown in continuous light than in photoperiodic light (Figure 7). This elevated expression in continuous light may result from signals that are distinct from ROS-based signals such as those produced by the circadian clock. Consistent with this interpretation, AAA is expressed at 3-fold higher levels in wild-type seedlings grown in continuous relative to photoperiodic light (Figure 7; see Supplemental Figure 9 online). By contrast, genes whose expression is induced when R211A-2.2, cs, and cch are grown in photoperiodic light relative to continuous light (i.e., BAP1, NodL, WRKY40, and ZAT10; Figure 7) are expressed at essentially the same level in wild-type seedlings regardless of whether wild-type seedlings are grown in continuous or photoperiodic light (Figure 7; see Supplemental Figure 9 online). This analysis indicates that the R211A-2.2 transgene and the cs and cch alleles induce the expression of BAP1, NodL, WRKY40, and ZAT10 when seedlings are grown in photoperiodic light.
A Mutant Allele of EXECUTER1 Does Not Suppress gun4-1
Our finding that only two of the 1O2-inducible genes that we tested were induced by malfunctioning GUN4 and Mg-chelatase led us to suspect that the flu mutant might produce more ROS than gun4 and Mg-chelatase subunit gene mutants. An alternative explanation is that malfunctioning Mg-chelatase might regulate nuclear gene expression by a mechanism that is not triggered by ROS. If GUN4 and Mg-chelatase subunit gene mutants regulate ROS-inducible gene expression by a mechanism that is distinct from flu mutants, then we would expect that suppressors of flu might not suppress mutant alleles of GUN4. Loss-of-function alleles of EXECUTER1 (EX1) suppress the ROS-regulated gene expression and chlorophyll-deficient phenotypes of flu (Wagner et al., 2004; Lee et al., 2007). To test whether ex1 alleles might also suppress the mutant alleles of GUN4, we monitored chlorophyll levels in the wild type, gun4-1, and a gun4-1 ex1 double mutant. We chose to test for suppression of the chlorophyll-deficient phenotype rather than the ROS-inducible gene expression phenotype because our quantification of ROS-inducible gene expression (Figures 6 and 7) does not clearly indicate that the expression of these particular genes is induced by elevated chloroplastic ROS in our mutants and transgenic lines. The ex1 allele that we used has a T-DNA insertion in an exon of the EX1 gene and was derived from Salk_022735 (see Supplemental Figure 10A online). Analysis of EX1 expression in these mutants is consistent with this T-DNA insertion causing at least a partial loss of function or possibly knocking out EX1 function (see Supplemental Figure 10B online).
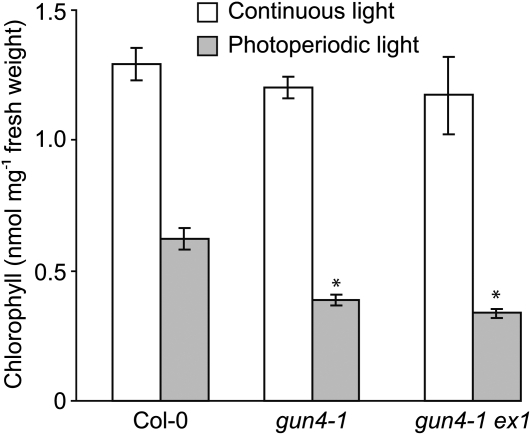
Consistent with previous results (Mochizuki et al., 2001; Larkin et al., 2003), gun4-1 accumulates significantly less chlorophyll than the wild type when seedlings are grown in 100 μmol m−2 s−1 white light, regardless of whether the light is continuous or photoperiodic. Similar levels of chlorophyll accumulate in gun4-1 and in gun4-1 ex1 (see Supplemental Figure 11 online). gun4 mutants were previously shown to accumulate more chlorophyll in low fluence rates of white light (Larkin et al., 2003; Peter and Grimm, 2009). Also, gun4 mutants were previously shown to accumulate more chlorophyll in continuous 10 μmol m−2 s−1 white light than in the same fluence rate of photoperiodic white light (Peter and Grimm, 2009). Consistent with this previous work, we found that the wild type, gun4-1, and gun4-1 ex1 accumulate essentially the same amount of chlorophyll when grown in continuous 10 μmol m−2 s−1 white light but that gun4-1 accumulates nearly 50% less chlorophyll than the wild type when grown in the same fluence rate of photoperiodic white light (Figure 8; see Supplemental Figure 12 online). Consistent with EX1 not contributing to the enhanced chlorophyll-deficient phenotype caused by gun4-1 in photoperiodic light, we found that gun4-1 and gun4-1 ex1 accumulate similar levels of chlorophyll when seedlings are grown in 10 μmol m−2 s−1 photoperiodic white light (Figure 8).
Figure 8.
Quantitative Analysis of Chlorophyll Levels in the Wild Type (Col-0), gun4-1, and gun4-1 ex1.
Wild-type (Col-0), gun4-1, and gun4-1 ex1 seedlings were grown for 14 d in a photoperiod in which 12 h of 10 μmol m−2 s−1 white light was followed by 12 h of darkness (gray bars) or in continuous 10 μmol m−2 s−1 white light (white bars). Four biological replicates were collected for each line in each condition. The statistical significance of the difference between the levels of chlorophyll in the wild type (Col-0) and a particular mutant was calculated with an unpaired t test. Error bars indicate se. The asterisk indicates a statistically significant difference in the levels of chlorophyll between the wild type (Col-0) and gun4-1 (P = 0.002) and between the wild type and gun4-1 ex1 (P = 0006) grown in photoperiodic light.
DISCUSSION
Mg-chelatase is an unusual enzyme in that it requires a regulatory protein that binds one of its subunits (ChlH/GUN5), one of its substrates (PPIX), and one of its products (Mg-PPIX) for robust activity. This proteinaceous cofactor, GUN4, was proposed to have evolved in part to attenuate the production of ROS by shielding PPIX and Mg-PPIX from collisions with O2 (Larkin et al., 2003; Verdecia et al., 2005). A crystal structure guided the engineering of a collection of site-directed mutants that cause 45 single amino acid substitutions in SynGUN4 (Verdecia et al., 2005). Analyses of these amino acid substitutions in porphyrin binding assays, Mg-chelatase assays, and other kinetic measures (Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005) indicate that the porphyrin binding activity of SynGUN4 underpins its Mg-chelatase stimulatory activity. Analyses of the association of GUN4 and pea GUN4 with pea chloroplast membranes following induced increases in chloroplastic porphyrin levels is consistent with GUN4 helping to channel PPIX and Mg-PPIX into chlorophyll biosynthesis by binding PPIX, Mg-PPIX, and stimulating Mg-chelatase on chloroplast membranes (Adhikari et al., 2009). The work of Adhikari et al. (2009) is also consistent with pea GUN4 possibly promoting interactions between pea ChlH and chloroplast membranes. However, Adhikari et al. (2009) could not test this idea because the pea system does not provide mutant alleles of GUN4 and Mg-chelatase subunit genes.
In this report, we describe the engineering of transgenic plants that express only gun4 alleles that significantly inhibit the porphyrin binding activity of GUN4 in vitro but do not affect the accumulation of GUN4 protein in vivo. The previously available gun4 alleles were limited to protein nulls or severe loss-of-function alleles that cause barely detectable levels of GUN4 protein to accumulate (Larkin et al., 2003; Peter and Grimm, 2009). Using these transgenic plants, a new chlH/gun5 allele, and previously available gun4, chlH/gun5, and chlI alleles, we tested whether the porphyrin binding activity of GUN4 that was previously demonstrated in vitro and Mg-chelatase activity contribute significantly to chlorophyll biosynthesis, interactions between chloroplast membranes and both GUN4 and ChlH/GUN5, and photoprotection in vivo. Our analysis of transgenic plants indicates that GUN4 binds porphyrins in vivo and is consistent with GUN4 helping to channel porphyrins into chlorophyll biosynthesis by binding both porphyrins and ChlH/GUN5 on chloroplast membranes. We also found that GUN4 and ChlH/GUN5 bind to chloroplast membranes using different mechanisms. Our analysis of ROS-regulated gene expression and chlorophyll accumulation in the gun4-1 ex1 double mutant did not lead to evidence that the porphyrin binding activity of GUN4 helps attenuate ROS production or that gun4 and flu alleles use the same mechanism to induce chlorophyll deficiencies.
GUN4 Helps Channel Porphyrins into Chlorophyll Biosynthesis by Binding to ChlH/GUN5 on Chloroplast Membranes
When purified chloroplasts were lysed and fractionated, we observed greater proportions of F191A and R211A (the porphyrin binding-deficient versions of GUN4) in soluble fractions and lower proportions in membrane-containing pellet fractions than GUN4.
Similarly, when chloroplasts were purified from chlH/gun5 mutants and then lysed and fractionated, we observed greater proportions of GUN4 in soluble fractions and lower proportions in the membrane-containing pellet fractions. These data indicate that the porphyrin binding activity of GUN4 and the activity of ChlH/GUN5 promote interactions between GUN4 and chloroplast membranes. To test whether the reduced membrane association of the F191A and R211A proteins results from their attenuated porphyrin binding activity or from the attenuation of some other function of GUN4, we increased chloroplastic PPIX levels by feeding PPIX to purified chloroplasts. In the wild type, F191A-14, R211A-2.2, gun5, and gun5-101, we found that feeding PPIX to purified chloroplasts promotes the association of GUN4 with chloroplast membranes. Based on these findings, we conclude that GUN4-porphyrin complexes indeed have a higher affinity for chloroplast membranes than unliganded GUN4. This effect of PPIX feeding promoting the association of GUN4 with chloroplast membranes does not occur in cch. This result is difficult to explain because the effect of the P642L substitution on the activity of ChlH/GUN5 is severe and potentially pleiotropic (Mochizuki et al., 2001; Davison and Hunter, 2011). Although the amino acid substitutions caused by gun5 and cch may not cause identical effects in Arabidopsis and Synechocystis, our findings are consistent with the amino acid substitutions caused by cch more severely attenuating Mg-chelatase than gun5 in both Arabidopsis and Synechocystis (Mochizuki et al., 2001; Davison and Hunter, 2011) and with SynGUN4 more effectively stimulating the Synechocystis Mg-chelatase that is inactivated by the amino acid substitution caused by gun5 than the amino acid substitution caused by cch (Davison and Hunter, 2011). The simplest and most likely explanation of these data is that GUN4-porphyrin complexes bind ChlH/GUN5 on chloroplast membranes with a higher affinity than unliganded GUN4, the gun5 and gun5-101 alleles attenuate these interactions, and the cch allele may diminish the ability of ChlH/GUN5 to distinguish between porphyrin-bound and unliganded GUN4.
In contrast with F191A-14, R211A-2.2, gun5, and gun5-101, we observed that a higher percentage of GUN4 associates with the chloroplast membrane–containing pellet fractions derived from gun4-1 and cs than those derived from the wild type. GUN4 accumulates to barely detectable levels in gun4-1 (Larkin et al., 2003). Also, the L88F substitution caused by gun4-1 causes a 6- to 15-fold increase in the affinity of cyanobacterial GUN4 proteins for porphyrins (Davison et al., 2005). These two effects of the gun4-1 allele may cause a greater proportion of GUN4 to associate with chloroplast membranes by causing a greater proportion of GUN4 to associate with porphyrins. Similar mechanisms may explain the cs allele promoting an increase in the proportion of GUN4 that associates with chloroplast membranes. Mg-chelatase mutants accumulate PPIX when grown in photoperiodic light (Falbel and Staehelin, 1994; Zhang et al., 2011). Thus, the photoperiodic light used here might elevate PPIX in cs, thereby causing a greater proportion of GUN4 to associate with chloroplast membranes by causing a greater proportion of GUN4 to associate with porphyrins. However, we did not analyze the PPIX levels in cs. We may not safely assume that PPIX accumulates in cs because in contrast with the results of Falbel and Staehelin (1994) and Zhang et al. (2011), downregulating Mg-chelatase activity by expressing antisense RNA does not cause significant changes in PPIX levels (Papenbrock et al., 2000a, 2000b). Furthermore, cs is different from gun4-1 in that it does not accumulate lower levels of GUN4 than the wild type (see Supplemental Figure 13 online). Thus, although accumulation of PPIX in cs may promote the association of GUN4 with the membrane-containing pellet fraction, we cannot rule out that the cs allele may promote the association of GUN4 with ChlH/GUN5 on chloroplast membranes by a currently unknown mechanism.
The association of GUN4 with chloroplast membranes is attenuated as leaves age. The lysis and fractionation of the 10-d-old Arabidopsis seedlings used here and the 6- to 8-d-old pea seedlings (Adhikari et al., 2009) yield one-third of GUN4 in the soluble fraction and two-thirds in the membrane-containing pellet fraction. By contrast, lysis and fractionation of chloroplasts purified from fully expanded rosette leaves of Arabidopsis using the same buffer conditions and MgCl2 concentrations used here yields the bulk of GUN4 in the soluble fraction (Larkin et al., 2003). As young leaves become mature, the expression of genes that encode chlorophyll biosynthetic enzymes wanes (Matsumoto et al., 2004). The data in this report, Larkin et al. (2003), and Adhikari et al. (2009) are consistent with the channeling activity of GUN4 also decreasing with age.
The high viscosity of the chloroplast stroma (Köhler et al., 2000) probably prevents GUN4 from rapidly redistributing between the chloroplast stroma and chloroplast membranes in vivo. Additionally, although chloroplasts were fed PPIX, Mg-chelatase might convert a fraction of this PPIX to Mg-PPIX. GUN4 binds both of these porphyrins (Adhikari et al., 2009). Thus, the simplest interpretation of our data regarding the association of GUN4 with chloroplast membranes derived from fractionated chloroplasts is (1) that interactions between GUN4 and chloroplast membranes largely depend on GUN4 binding active ChlH/GUN5 and (2) that porphyrin-bound GUN4 more stably interacts with ChlH/GUN5 than unliganded GUN4. Thus, when chloroplasts are lysed and fractionated, the relatively higher stability of complexes that contain porphyrin-bound GUN4 and active ChlH/GUN5 yields a higher proportion of GUN4 associated with membrane-containing pellet fractions. The data reported here and previously published data (Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005; Sobotka et al., 2008; Adhikari et al., 2009) support a role for GUN4 in the channeling of porphyrins into chlorophyll biosynthesis, especially in young leaves, by a mechanism that includes binding both porphyrins and ChlH/GUN5 on chloroplast membranes and by stimulating the Mg-chelatase activity that associates with chloroplast membranes. Our finding that gun5, gun5-101, and cch cause a greater proportion of GUN4 to accumulate in soluble fractions derived from lysed chloroplasts and that the cs allele does not cause a greater proportion of GUN4 to accumulate in soluble fractions derived from lysed chloroplasts provides evidence that GUN4 specifically binds ChlH/GUN5 on chloroplast membranes in vivo. However, we cannot rule out the possibility that more complex mechanisms such as unliganded GUN4 and GUN4-porphyrin complexes bind chloroplast membranes by also binding to membrane proteins that contribute to chlorophyll biosynthesis, such as protoporphyrinogen IX oxidase (Matringe et al., 1992).
GUN4 and ChlH/GUN5 Use Different Mechanisms to Bind Chloroplast Membranes
The availability of loss-of-function alleles for GUN4, ChlH/GUN5, and ChlI allowed us to test whether GUN4 and ChlH/GUN5 use similar or distinct mechanisms to associate with chloroplast membranes. Our data are consistent with GUN4 and ChlH/GUN5 using distinct mechanisms to associate with chloroplast membranes. Although loss-of-function alleles for ChlH/GUN5 similarly reduce the association of both GUN4 and ChlH/GUN5 with chloroplast membranes, loss-of-function alleles for ChlI and the F191A and R211A substitutions in GUN4 cause completely different effects on the association of GUN4 and ChlH/GUN5 with chloroplast membranes. The differences in the proportions of membrane-associated ChlH/GUN5 between the wild type and both F191A-14 and R211A-2.2 were the smallest differences between the wild type and any of the mutants tested and were not statistically significant. By contrast, the largest decreases in the proportions of membrane-associated GUN4 between the wild type and all of the mutants and transgenic lines were observed in F191A-14 and R211A-2.2. Additionally, feeding PPIX to purified chloroplasts had no effect on the association of ChlH/GUN5 and chloroplast membranes in the wild type and in all of the mutants tested but promoted the association of GUN4 with chloroplast membranes. Based on these findings, we conclude that GUN4 uses a mechanism that largely depends on binding both porphyrins and active ChlH/GUN5 to associate with chloroplast membranes. We also conclude that ChlH/GUN5 binds chloroplast membranes using a mechanism that is influenced by Mg-chelatase activity and depends less on the porphyrin binding activity of GUN4 than does GUN4.
Approximately 50% of pea ChlH associates with the membrane-containing pellet fraction when purified pea chloroplasts are lysed and fractionated. Inducing a rise in porphyrin levels causes ~70% of pea ChlH/GUN5 to associate with the membrane-containing pellet fractions (Adhikari et al., 2009). By contrast, we report here that the same methods used by Adhikari et al. (2009) yield 70% of ChlH/GUN5 in membrane-containing pellet fractions derived from wild-type Arabidopsis and that feeding PPIX to Arabidopsis chloroplasts does not further promote the association of ChlH/GUN5 with membrane-containing pellet fractions. We propose that ChlH/GUN5 uses similar mechanisms to associate with chloroplast membranes in Arabidopsis and pea and that PPIX feeding cannot promote the association of ChlH/GUN5 with chloroplast membranes, so that more than 70% of ChlH/GUN5 associates with the membrane-containing pellet fraction derived from both pea and wild-type Arabidopsis when analyzed with the methods used here and by Adhikari et al. (2009).
A Complex Mechanism Likely Explains the Light-Sensitive Phenotypes of Chlorophyll-Deficient Mutants
The GUN4 protein decreased to low levels or was undetectable in gun4-1, F191A-14, R211A-2.2, gun5-101, and cch, but not in gun5, cs, and the wild type, following a fluence rate shift from 100 to 850 μmol m−2 s−1 white light. Based on previous kinetic analyses of Synechocystis Mg-chelatase (Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005), we expect that this reduction in GUN4 protein levels significantly attenuates Mg-chelatase activity. However, these decreases in GUN4 protein levels are not the sole cause of the enhanced chlorophyll deficiencies of these mutants following this fluence rate shift from 100 to 850 μmol m−2 s−1 because F191A-14, R211A-2.2, gun5, and cs are similarly chlorophyll deficient following this fluence rate shift.
The reduced levels of GUN4 protein following a fluence rate shift from 100 to 850 μmol m−2 s−1 white light may result from an enhanced turnover of GUN4 protein and/or reduced expression of the GUN4 gene. This reduction of GUN4 protein levels was specific to GUN4; similar reductions were not observed for the ChlH/GUN5, ChlI, or ChlD subunits of Mg-chelatase. Consistent with these findings, Peter and Grimm (2009) did not observe a reduction in ChlH levels in a gun4 mutant. Peter and Grimm (2009) proposed that GUN4 can contribute to the posttranslational regulation of tetrapyrrole metabolism in part by binding excess Mg-porphyrins and that, upon binding excess Mg-porphyrins, GUN4 may become targeted for degradation. Our findings indicate that GUN4 protein levels are reduced (1) when porphyrin binding activity is attenuated, when ChlH/GUN5 activity is attenuated, or when GUN4 less stably associates with chloroplast membranes; and (2) plants are simultaneously exposed to 850 μmol m−2 s−1 white light. Some of these conditions are expected to increase the proportion of unliganded GUN4. Whether this degradation of GUN4 is specific to particular mutants or whether degradation of GUN4 in wild-type plants can occur in conditions that wild-type plants experience in nature is important to test.
We observed that transferring wild-type seedlings from 100 μmol m−2 s−1 to 850 μmol m−2 s−1 white light causes a decrease in chlorophyll levels to 59 to 68% of levels in wild-type seedlings that continue to grow in 100 μmol m−2 s−1 white light. We expect that the Suc in the growth medium could contribute to this decrease in chlorophyll content in wild-type seedlings. Although we included Suc in the growth medium to promote uniform growth and viability of these chlorophyll-deficient mutants, especially in 850 μmol m−2 s−1 white light (N.D. Adhikari, unpublished data), carbohydrates can attenuate chloroplast function (To et al., 2003; Stettler et al., 2009). We suggest that perhaps the combination of Suc and 850 μmol m−2 s−1 white light cause chlorophyll levels to decrease in the wild type. Nonetheless, intense light causes enhanced chlorophyll deficiency relative to the wild type in chlorophyll biosynthesis mutants even when sucrose is not included in the growth medium (Falbel et al., 1996). Similar to other chlorophyll biosynthesis mutants, gun4-1, cs, the chlH/gun5 mutants, and the transgenic lines expressing F191A and R211A are more chlorophyll deficient than the wild type when they are grown in high-intensity white light. Also consistent with previous work, we found that fluence rate increases induce the expression of several ROS-inducible genes. ROS can attenuate the production of chlorophyll precursors (Aarti et al., 2006, 2007; Stenbaek et al., 2008) and trigger plastid-to-nucleus signaling that induces nuclear gene expression and promotes chlorophyll deficiency (Wagner et al., 2004; Lee et al., 2007). Our analysis of ROS-inducible gene expression is consistent with the wild type, F191A-14, and R211A-2.2, producing similar levels of ROS following the fluence rate shift from 100 to 850 μmol m−2 s−1. Based on these data, we conclude that increased levels of chloroplastic ROS may contribute to the chlorophyll deficiencies of the wild type, F191A-14, and R211A-2.2 when these plants are exposed to intense light but that F191A-14 and R211A-2.2 do not accumulate more ROS than the wild type following this fluence rate shift. Thus, elevated ROS production relative to the wild type would not appear to cause the enhanced light sensitivity of these chlorophyll-deficient mutants following the fluence rate shift from 100 to 850 μmol m−2 s−1. Our conclusion, based on these data, is that deficiencies in the porphyrin binding activity of GUN4 do not necessarily cause ROS production following the fluence rate shift from 100 to 850 μmol m−2 s−1. However, we cannot rule out the possibility that these ROS-inducible genes are expressed at maximal levels in the wild type following the fluence-rate shift. If these genes are expressed at maximal levels following the fluence rate shift, then F191A-14 and R211A-2.2 may actually contain more ROS than the wild type after the fluence rate shift. Another possibility is that the F191A-14 and the R211A-2.2 might produce the same amount of ROS as the wild type following the fluence rate shift and that these amino acid substitutions increase the sensitivity of the chlorophyll biosynthetic pathway to inhibition by ROS. Consistent with this interpretation, the production of ROS in response to the exposure of plants to intense light inhibits chlorophyll biosynthesis and causes PPIX to accumulate (Aarti et al., 2006).
Inadequate Mg-chelatase activity may partially explain the enhanced light sensitivity of these mutants. Chlorophyll turnover can increase when plants are exposed to high-intensity light (Feierabend and Dehne, 1996; Beisel et al., 2010). The attenuated rate of Mg-PPIX biosynthesis in these mutants may prevent the adequate replacement of the chlorophyll that is turned over in 850 μmol m−2 s−1 white light. Indeed, we did observe that the mutants that are more severely chlorophyll deficient in 100 μmol m−2 s−1 exhibit a greater enhancement of their chlorophyll deficiency following transfer to 850 μmol m−2 s−1. Nonetheless, the correlation between chlorophyll deficiency in 100 μmol m−2 s−1 and enhanced chlorophyll deficiency in 850 μmol m−2 s−1 white light is not perfect. We found that in 850 μmol m−2 s−1 white light, chlorophyll levels were ~50% lower in gun5, gun5-101, and cs than in gun4 mutants and transgenic lines that had similar chlorophyll levels when grown in 100 μmol m−2 s−1. One possible explanation for these data is that the alleles of ChlH/GUN5 and ChlI that were used in this study may enhance the ROS sensitivity of the chlorophyll biosynthetic pathway that was previously reported (Aarti et al., 2006) to a greater degree than the alleles of GUN4.
Mutant Alleles of GUN4 and Mg-Chelatase Subunit Genes Do Not Cause Major Effects on ROS-Inducible Gene Expression
We found that the expression of only two of the five genes (i.e., BAP1 and NodL) whose expression is induced by 1O2 in flu mutants is also induced in R211A-2.2, cs, and cch relative to the wild type. We found that two genes whose expression is specifically induced by O2•–H2O2 are not induced in R211A-2.2, cs, and cch relative to the wild type. We found increases in the expression of two of the three genes whose expression is induced by a number of different types of ROS in R211A-2.2, cs, and cch relative to the wild type. The simplest interpretation of these data is that R211A, cs, and cch induce the expression of these genes by means of a mechanism that is distinct from the 1O2-based mechanism that is triggered by the overaccumulation of protochlorophyllide in flu mutants. Consistent with this interpretation, although loss-of-function alleles of EX1 can suppress the chlorophyll-deficient phenotypes of flu (Wagner et al., 2004), we found that a loss-of-function allele of EX1 does not suppress the chlorophyll-deficient phenotype of gun4-1.
A screen for mutant alleles that cause defects in the plastid regulation of Lhcb gene expression yielded loss-of-function alleles of GUN4, ChlH/GUN5, and other genes that contribute to tetrapyrrole biosynthesis in the chloroplast (Mochizuki et al., 2001; Larkin et al., 2003). An analysis of these mutants indicates that all chlorophyll biosynthesis mutants do not also have defects in plastid-regulated gene expression and that mutants with more severe chlorophyll-deficient phenotypes do not exhibit more severe plastid-regulated gene expression phenotypes. These findings implicated Mg-chelatase, Mg-PPIX biosynthesis, or chloroplastic tetrapyrrole metabolism in the plastid-to-nucleus signaling that regulates photosynthesis-related gene expression (Mochizuki et al., 2001). Data generated using distinct experimental approaches provide evidence that Mg-chelatase, Mg-PPIX biosynthesis, or chloroplastic tetrapyrrole metabolism can affect gene expression in Arabidopsis and other plants (Oster et al., 1996; Papenbrock et al., 2000b; Vinti et al., 2000; La Rocca et al., 2001, 2007; McCormac and Terry, 2002; Strand et al., 2003; Alawady and Grimm, 2005; Gadjieva et al., 2005; Ankele et al., 2007; Zhang et al., 2011), Chlamydomonas reinhardtii (Johanningmeier and Howell, 1984; Kropat et al., 1997, 2000; Falciatore et al., 2005; Vasileuskaya et al., 2005; von Gromoff et al., 2008), and Synechocystis (Osanai et al., 2009). Mg-PPIX also regulates DNA replication in Cyanidioschyzon merolae and tobacco (Nicotiana tabacum) BY-2 cell cultures (Kobayashi et al., 2009). A simple mechanism such as the reduced accumulation of Mg-PPIX per se or the reduced production of ROS from Mg-PPIX regulating photosynthesis-related nuclear gene expression in plants is not consistent with the available data (Voigt et al., 2010; Mochizuki et al., 2010; Zhang et al., 2011). If our analyses of ROS-inducible gene expression were consistent with GUN4 and ChlH/GUN5 binding distinct pools of porphyrins (e.g., one pool that associates with chlorophyll biosynthesis and a distinct pool that is not associated with chlorophyll biosynthesis), this finding might contribute to our knowledge of plastid-to-nucleus signaling mechanisms. However, our analysis of ROS-regulated gene expression does not contribute to our understanding of plastid-to-nucleus signaling mechanism. We did find that both of our F191A and R211A expressing transgenic lines are gun mutants and that their gun phenotypes are similar to gun4-1 (see Supplemental Figure 14 online), a mutant with a more severe chlorophyll-deficient phenotype. Additionally, we found that although cs has a chlorophyll-deficient phenotype that is similar to both of our F191A- and R211A-expressing transgenic lines, cs is not a gun mutant (see Supplemental Figure 14 online), which is consistent with previous work (Mochizuki et al., 2001). These data add to the body of evidence that is inconsistent with a model in which a simple mechanism such as the reduced accumulation of Mg-PPIX per se or the reduced production of ROS from Mg-PPIX causes the misregulation of photosynthesis-related gene expression in tetrapyrrole biosynthesis mutants of Arabidopsis.
In summary, our main finding is that the porphyrin binding activity of GUN4 and the activity of ChlH/GUN5 promote interactions between GUN4 and chloroplast membranes. Thus, GUN4 promotes chlorophyll biosynthesis not only by stimulating Mg-chelatase activity but also by binding both porphyrins and ChlH/GUN5 on chloroplast membranes. By contrast, we found that the porphyrin binding activity of GUN4 is not similarly important for interactions between ChlH/GUN5 and chloroplast membranes. We also found that the mechanism that causes the light-sensitive phenotypes of several chlorophyll biosynthesis mutants is potentially complex and does not appear to depend only on reduced levels of the GUN4 protein, attenuated chlorophyll biosynthesis, or enhanced production of ROS relative to the wild type.
METHODS
Plant Materials
The wild type and all mutants were Arabidopsis thaliana plants of the Columbia-0 (Col-0) ecotype. gun5, cch, gun4-1, and gun4-2 were previously described (Mochizuki et al., 2001; Larkin et al., 2003). All Salk T-DNA insertion lines were obtained from the ABRC (Ohio State University). Salk_039005 has a T-DNA insertion in SGS3 and is hereafter referred to as sgs3. gun4-2 and sgs3 were crossed and the F2 progeny that were homozygous for the sgs3 and heterozygous for the gun4-2 T-DNA insertion alleles were identified using PCR-based genotyping with strategies recommended by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/). Primers 5′-ACACAATCATTGCTTTCCTGTGACGGTTC-3′ (RP) and 5′-ACACAGTGATGGTAGATTCGGATACAGC-3′ (LP) were used to score GUN4, and 5′-ACTCTCAAGGTTCTCTCCCCC-3′ (RP) and GAGCTGTTCCAAAGCCTCATC-3′ (LP) were used to score SGS3. The LBa1 oligonucleotide (O’Malley et al., 2007) and the indicated gene-specific oligonucleotides were used to score T-DNA insertion alleles. These sgs3 gun4-2 mutants were transformed with transgenes in which the native promoter drives expression of the wild-type GUN4 protein or GUN4 proteins in which the F191A and R211A amino acid substitutions were engineered by site-directed mutagenesis. GUN4 proteins containing these amino acid substitutions are hereafter referred to as F191A and R211A. Site-directed mutants were prepared using the QuickChange XL site-directed mutagenesis kit (Stratagene) as previously described (Adhikari et al., 2009). All mutations were confirmed by sequencing at the Research Technology Support Facility (Michigan State University). These transgenes included a genomic fragment that contained the complete GUN4 coding sequence as well as 845 bp upstream and 164 bp downstream of the complete GUN4 coding sequence inserted into pPZP221, as previously described (Larkin et al., 2003). Two lines each for F191A and R211A were judged to contain single transgenes by scoring for the gentamicin resistance gene that is linked to the GUN4 genes during segregation analysis. Using PCR-based genotyping, we determined that these lines are homozygous for the sgs3 and the gun4-2 alleles. We propagated these primary transformants and obtained lines that are homozygous GUN4-, F191A-, and R211A-expressing transgenes, gun4-2, and sgs3.
We isolated gun5-101 from a previously described gun mutant screen (Ruckle et al., 2007). This mutant allele was identified by positional cloning after gun5-101 mutants were crossed to a Landsberg erecta line. F2 progeny that exhibited a pale phenotype were used to map gun5-101 with simple sequence length polymorphism and cleaved-amplified polymorphic sequence markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) that were designed using the Cereon Arabidopsis Polymorphism Collection (Jander et al., 2002) (see Supplemental Table 4 online) and previously described procedures (Weigel and Glazebrook, 2002). To sequence the gun5-101 allele, the GUN5 coding sequence was amplified by means of Platinum Pfx DNA polymerase (Invitrogen) using GUN5-specific oligonucleotides in at least 10 aliquots that were subsequently pooled, purified from agarose gels using the QIAquick gel extraction kit (Qiagen), and sequenced with gene-specific oligonucleotides by the Research Technology Support Facility (Michigan State University).
The gun4-1 ex1 double mutants were prepared by crossing gun4-1 and Salk_022735, which has a T-DNA insertion in the sixth exon of EX1. Salk_022735 is hereafter referred to as ex1. The ex1 allele was scored using PCR-based genotyping with 5′-TGAGTAGATAGCAAAGTGTGATTTCTCTACG-3′ and 5′-ATCCTTAAGCATCTGCAACGTGAATTTCTAG-3′ as described above for gun4-2 and sgs3. We determined the approximate levels of EX1 expression in the ex1 mutant relative to the wild type by RT-PCR as described by Ruckle et al. (2007) using 5′-TCGTCTCCCCGATCCCACCGC-3′ and 5′-TCCAGGTGAAGTCAAAAACCTACCAGG-3′ to estimate the relative expression of exons 1 to 6 (i.e., sequences that are upstream of the T-DNA insertion) and using 5′-ATCGCAGTGAAATTTGTGATAGGCGATATTG-3′ and 5′-ATCAATGCGACGAAATTTTGTCAAACCCG-3′ to estimate the relative expression of exons 6 and 7 (i.e., sequences that are downstream of the T-DNA insertion). The gun4-1 allele was scored by its striking chlorophyll-deficient phenotype (Vinti et al., 2000; Mochizuki et al., 2001; Larkin et al., 2003).
Growth Conditions
Seeds for all lines were surface sterilized by incubation first in 70% ethanol, 0.5% Triton X-100 solution for 10 min on a tube mixer and then in 95% ethanol for 10 min on a tube mixer, followed by air drying on filter paper soaked in 95% ethanol in a laminar flow hood. Seeds were plated on Linsmaier and Skoog medium containing 1% Suc and 0.5% phytoblend (Caisson Laboratories). To test for gun phenotypes, seeds were plated on Linsmaier and Skoog media that contained 2% Suc and either 5 μM norflurazon or no norflurazon as previously recommended (Susek et al., 1993). Seeds were stratified for 4 d at 4°C and grown for the indicated number of days at 21°C. With the exception of the high-intensity light experiments, plants were grown in the indicated fluence rate of white light from broad-spectrum fluorescent tube lamps. For high-intensity light (850 μmol m−2 s−1) experiments, white light was obtained from a combination of high-pressure sodium and metal halide lamps. Light was filtered through neutral density filters (Roscolux 397; Rosco Laboratories) to obtain particular fluence rates. For high-intensity light experiments, plates were placed in envelopes made by taping neutral density filters to 3MM paper (Whatman International). For experiments with lower fluence rates of light, neutral density filters were clamped on the transparent lids of plastic boxes with black-painted sides and bottoms and with multiple layers of black electrical tape sealing cracks at the top of the boxes as previously recommended (Fankhauser and Casal, 2004). Using a thermometer, we determined that the temperatures inside these envelopes and boxes were identical to the air temperatures of the growth chambers (i.e., 21°C) during these fluence rate shift experiments. In the growth chambers and in the envelopes and boxes covered with neutral density filters, fluence rates were measured with an LI-250A photometer using a PAR sensor (LI-COR Biosciences).
Chlorophyll a Fluorescence Measurements
Chlorophyll a fluorescence yield was measured in cotyledons as previously described (Livingston et al., 2010) using a nonfocusing optics flash spectrophotometer/fluorometer modified to measure signal from ~2-mm-diameter samples. Fluorescence yields were corrected for background signals in the absence of a cotyledon. Each cotyledon was subjected to a series of four increasing light intensities: 5, 9, 18, and 37 μmol m−2 s−1. Steady state photosynthetic measurements were made after 14 min of continuous illumination at each light intensity to ensure cotyledons reached steady state. Maximal (Fv/Fm) and steady state (ΦII) photosystem II quantum yields were calculated as previously described (Genty et al., 1989). Total NPQ, energy-dependent exciton quenching (qE), and long-lived inhibitory quenching (qI) were measured using previously described methods (Baker et al., 2007). The extent of qE was calculated from Fm’ measured just before the actinic light was turned off and Fm” measured after an 8-min dark recovery. Photosynthetic parameters were averaged from quadruplicate measurements. The unpaired t test was used to measure the statistical significance of the differences between the wild type and the mutants.
Quantitative Analysis of Chlorophyll Levels
For quantitative analysis of chlorophyll levels, seedlings were harvested under a very dim-green light, flash frozen in liquid nitrogen, and stored at −80°C. Chlorophyll a and b was extracted using N, N’-dimethylformamide and quantified exactly as previously described (Porra et al., 1989) except that biological replicates were placed in 1.5-mL microfuge tubes that contained a single 3-mm very-high-density zirconium oxide bead (Glen Mills) and homogenized with a Retsch TissueLyser (Qiagen) set at the maximum frequency for 5 min.
Purification, PPIX Feeding, Fractionation, and Analysis of Intact Chloroplasts
Intact chloroplasts were purified from Arabidopsis seedlings and the indicated Arabidopsis mutants and transgenic lines grown in 125 μmol m−2 s−1 white light with a diurnal cycle of 12 h of light and 12 h of dark. To facilitate harvesting, sterile 125-mm-diameter circles of grade 3 filter paper (Whatman International) were placed on the growth media in Petri dishes as described above. Surface-sterilized seeds were then evenly distributed on the filter paper–covered media. Seedlings were grown as described above. Chloroplasts were purified from 40 g of 10-d-old Arabidopsis seedlings using procedures that were previously described (Kubis et al., 2008). The only modification to Kubis et al. (2008) was that intact chloroplasts were separated from crude chloroplast fractions (i.e., fractions that contain intact chloroplasts, broken chloroplasts, and other debris) using 30% Percoll cushions rather than linear Percoll gradients to facilitate the simultaneous isolation of intact chloroplasts from the wild type and a number of different mutants and transgenic lines. Briefly, after homogenizing seedlings, filtering homogenates, and centrifuging the filtered homogenates as recommended by Kubis et al. (2008), crude chloroplast fractions were resuspended in 2 mL of 50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol, 5 mM MgCl2, and 5 mM EDTA. Each resuspended crude chloroplast fraction was applied to 10 mL of 30% (v/v) Percoll (Sigma-Aldrich), 50 mM HEPES-KOH, pH 8.0, and 330 mM sorbitol. Intact chloroplasts were isolated by centrifuging these Percoll cushions at 3000 rpm in an Eppendorf 5810R centrifuge with an A-4-81 swinging-bucket rotor for 5 min at 4°C. Intact chloroplasts were recovered from the bottom of the Percoll cushion, washed once by resuspending them in 5.0 mL of 50 mM HEPES-KOH, pH 8.0, and 330 mM sorbitol, and then centrifuging them using an Eppendorf 5810R centrifuge with an A-4-81 swing-bucket rotor at 1500 rpm for 5 min at 4°C. These pelleted, intact chloroplasts were resuspended in 1.0 mL 50 mM HEPES-KOH, pH 8.0, and 330 mM sorbitol. Yields of intact chloroplasts were determined by quantifying total protein rather than chlorophyll because of the striking variation in chlorophyll levels that were observed among the wild type and the various transgenic lines and mutants when they were grown in 125 μmol m−2 s−1 white light. Protein was quantified using the Bio-Rad protein assay and a BSA standard.
The method for feeding PPIX to purified intact chloroplasts was very similar to the method described by Adhikari et al. (2009). Briefly, purified chloroplasts were suspended in 50 mM HEPES-KOH, pH 8.0, and 330 mM sorbitol to a final protein concentration of 1 mg/mL. PPIX was added to a final concentration of 20 μM. Then, chloroplasts were incubated at 26°C for 15 min in the dark. Chloroplast lysis and fractionation was exactly as described by Adhikari et al. (2009). Briefly, after PPIX feeding, chloroplasts were purified using a 30% Percoll cushion. Pelleted, intact chloroplasts were resuspended in 200 μL of lysis buffer (25 mM HEPES-KOH, pH 8.0, and 4 mM MgCl2), incubated on ice for 20 min, and then centrifuged at 16,000g for 5 min. Pellet fractions prepared using this method contain both envelope and thylakoid membranes (Perry et al., 1991). Soluble and pellet fractions were diluted to the same volume using SDS-PAGE loading buffer and analyzed using SDS-PAGE and immunoblotting with affinity purified anti-GUN4, -ChlH/GUN5, -ChlI, and -ChlD antibodies as previously described (Larkin et al., 2003; Adhikari et al., 2009). Quantitation of immunoblots was performed by quantifying the chemiluminescence produced by immunoreactive bands using the Versadoc MP 4000 (Bio-Rad) as described by Adhikari et al. (2009). The statistical significance of the difference between GUN4 and ChlH/GUN5 levels in membrane-containing pellet fractions of the wild type (Col-0) and a particular mutant or transgenic line was calculated with an unpaired t test. The statistical significance of GUN4 and GUN5/ChlH levels in the membrane-containing pellet fractions derived from chloroplasts that were fed PPIX and chloroplasts purified from the same mutant or transgenic line that were not fed with PPIX were calculated with a paired t test
Analysis of Protein from Whole Seedlings
We extracted protein from whole seedlings by first powdering the seedlings with liquid N2. The powdered seedlings were mixed into 3 volumes of homogenization buffer: 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 20% glycerol, 1 mM DTT, 4 mM Pefabloc SC (Roche Applied Science), and 33 μL protease inhibitor cocktail for plant cell and tissue extracts (Sigma-Aldrich) per mL homogenization buffer. This suspension was clarified at 16,000g at 4°C for 10 min. The protein concentration of the clarified supernatants was quantified using Coomassie Plus (Bradford) protein assay (Thermo Fisher Scientific) and a BSA standard. These whole seedling extracts were analyzed by immunoblotting as described for chloroplast fractions.
Analysis of RNA
For quantitative RT-PCR (qRT-PCR) analyses, we extracted total RNA using the RNeasy plant mini kit (Qiagen). We synthesized cDNA using Superscript III (Invitrogen). The oligonucleotides used for qRT-PCR analysis of AAA, BAP1, FER1, APX1, WRKY40, ZAT10, ZAT12, and UBQ10 expression were as previously reported (Czechowski et al., 2005; Walley et al., 2007; Giraud et al., 2008; Baruah et al., 2009b). The oligonucleotides used for qRT-PCR analysis of NodL, ATOCT3, LPT, and TRFL4 expression were 5′-GAGCTTGTTCAGGGTGTTATCG-3′ and 5′-AGCTCTAGCTGAACCTTGTCAAAC-3′, 5′-TCCCACGTGGCACTGTCTT-3′ and 5′-AGATGTCGGAGGCTGAAGGA-3′, 5′-CCTGCGTTCGCACAGTACAT-3′ and 5′-TGCAAGCAAGGAGCACTTTG-3′, and 5′-GCCCAAGTGGCTGGAAGAT-3′ and 5′-CAACGAGCCGATAACAATTCG-3′, respectively. The oligonucleotides used for qRT-PCR analysis of Lhcb1.4 (At2g34430) expression were 5′-GCCTTCGCTACCAACTTCGTC-3′ and 5′-AACCGGATACACACAACTCGATC-3′. We assayed transcript levels using the Fast SYBR Green Master Mix and the ABI 7500 Fast Real-time PCR system (Applied Biosystems). Relative mRNA levels were calculated using the comparative CT method (Pfaffl, 2001) and normalized to UBQ10 expression.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database and the GenBank/EMBL data libraries under the following accession numbers: GUN4, At3g59400, NM_115802; SGS33, At5g23570, NM_122263; ChlH/GUN5, At5g13630, NM_121366; ChlD, At1g08520, NM_100725; ChlI-1, At4g18480, NM_117962; ChlI-2, At5g45930, NM_123961; AAA, At3g28580, NM_113778; BAP1, At3g61190, NM_115983; NodL, At5g64870, NM_125885; ATOCT3, At1g16390, NM_101505; LPT, At1g66850, NM_105356; TRFL4, At3g53790, NM_115239; FER1, At5g01600, NM_120238; APX1, At1g07890, NM_202057; WRKY40, At1g80840, NM_106732; ZAT10, At1g27730, NM_102538; and ZAT12, At5g59820, NM_125374. T-DNA insertion lines used were ex1 (Salk_022735) and sgs3 (Salk_039005).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Representative Seedlings Grown in 100 μmol m−2 s−1 White Light for 7 d.
Supplemental Figure 2. Representative Seedlings Grown in 100 μmol m−2 s−1 White Light for 3 d and Then Shifted to 850 μmol m−2 s−1 White Light for 4 d.
Supplemental Figure 3. Positional Cloning and Sequence Analysis of gun5-101.
Supplemental Figure 4. Analysis of Chlorophyll Levels in gun4, chlH/gun5, and cs Mutants Grown in 10 μmol m−2 s−1 White Light.
Supplemental Figure 5. Photosynthetic Parameters of Seedlings Grown in 10 μmol m−2 s−1 Continuous White Light.
Supplemental Figure 6. Induced Expression of ROS-Regulated Genes Following a Fluence Rate Shift.
Supplemental Figure 7. Photosynthetic Parameters of Seedlings Grown in 2 μmol m−2 s−1 Continuous White Light.
Supplemental Figure 8. Photosynthetic Parameters of Seedlings Grown in 2 μmol m−2 s−1 Photoperiodic White Light.
Supplemental Figure 9. Expression of ROS-Inducible Genes in Wild-Type Seedlings Grown in Continuous and Photoperiodic Light.
Supplemental Figure 10. Characterization of an ex1 Allele from Salk_022735.
Supplemental Figure 11. Quantitative Analysis of Chlorophyll Levels in the Wild Type (Col-0), gun4-1, and gun4-1 ex1 Grown in 100 μmol m−2 s−1 White Light.
Supplemental Figure 12. Representative gun4-1 and gun4-1 ex1 Seedlings Grown in 10 μmol m−2 s−1 White Light.
Supplemental Figure 13. Analysis of GUN4 Protein Levels in the Wild Type and cs That Were Grown in 100 μmol m−2 s−1 and 850 μmol m−2 s−1 White Light.
Supplemental Figure 14. Analysis of Lhcb1.4 Expression in Norflurazon-Treated Wild Type, gun4-1, F191A-14, R211A-2.2, cch, gun5, gun5-101, and cs.
Supplemental Table 1. Decrease in Chlorophyll Following a Transfer to 850 μmol m−2 s−1 White Light.
Supplemental Table 2. Differences in the Percentage of GUN4 and ChlH/GUN5 Associated with Chloroplast Membranes Purified from the Indicated Mutants and Transgenic Lines Relative to the Wild Type.
Supplemental Table 3. Increases in the Percentage of GUN4 in Chloroplast Membrane–Containing Pellet Fractions Following PPIX Feeding.
Supplemental Table 4. Simple Sequence Length Polymorphism and CAPS Markers Used for Mapping gun5-101.
Acknowledgments
We thank Michael E. Ruckle for providing the oligonucleotides that were used to quantify Lhcb1.4 expression. We also thank Karen Bird for editorial assistance in the preparation of this manuscript. This work was supported by the National Science Foundation (Grants IOB 0517841 and IOS-1021755) and by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Grant DE-FG02-91ER20021).
References
- Aarti D., Tanaka R., Ito H., Tanaka A. (2007). High light inhibits chlorophyll biosynthesis at the level of 5-aminolevulinate synthesis during de-etiolation in cucumber (Cucumis sativus) cotyledons. Photochem. Photobiol. 83: 171–176 [DOI] [PubMed] [Google Scholar]
- Aarti D.P., Tanaka R., Tanaka A. (2006). Effects of oxidative stress on chlorophyll biosynthesis in cucumber (Cucumis sativus) cotyledons. Physiol. Plant. 128: 186–197 [Google Scholar]
- Adhikari N.D., Orler R., Chory J., Froehlich J.E., Larkin R.M. (2009). Porphyrins promote the association of GENOMES UNCOUPLED 4 and a Mg-chelatase subunit with chloroplast membranes. J. Biol. Chem. 284: 24783–24796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawady A.E., Grimm B. (2005). Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 41: 282–290 [DOI] [PubMed] [Google Scholar]
- Allen K.D., Duysen M.E., Staehelin L.A. (1988). Biogenesis of thylakoid membranes is controlled by light intensity in the conditional chlorophyll b-deficient CD3 mutant of wheat. J. Cell Biol. 107: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankele E., Kindgren P., Pesquet E., Strand A. (2007). In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19: 1964–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.R., Harbinson J., Kramer D.M. (2007). Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ. 30: 1107–1125 [DOI] [PubMed] [Google Scholar]
- Baruah A., Simková K., Apel K., Laloi C. (2009a). Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol. Biol. 70: 547–563 [DOI] [PubMed] [Google Scholar]
- Baruah A., Simková K., Hincha D.K., Apel K., Laloi C. (2009b). Modulation of O-mediated retrograde signaling by the PLEIOTROPIC RESPONSE LOCUS 1 (PRL1) protein, a central integrator of stress and energy signaling. Plant J. 60: 22–32 [DOI] [PubMed] [Google Scholar]
- Beisel K.G., Jahnke S., Hofmann D., Köppchen S., Schurr U., Matsubara S. (2010). Continuous turnover of carotenes and chlorophyll a in mature leaves of Arabidopsis revealed by 14CO2 pulse-chase labeling. Plant Physiol. 152: 2188–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.J., Ecker J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Butaye K.M., Goderis I.J., Wouters P.F., Pues J.M., Delauré S.L., Broekaert W.F., Depicker A., Cammue B.P., De Bolle M.F. (2004). Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39: 440–449 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison P.A., Hunter C.N. (2011). Abolition of magnesium chelatase activity by the gun5 mutation and reversal by Gun4. FEBS Lett. 585: 183–186 [DOI] [PubMed] [Google Scholar]
- Davison P.A., Schubert H.L., Reid J.D., Iorg C.D., Heroux A., Hill C.P., Hunter C.N. (2005). Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 44: 7603–7612 [DOI] [PubMed] [Google Scholar]
- Duke S.O., Becerril J.M., Sherman T.D., Lehnen L.P., Matsumoto H. (1991). Photosensitizing porphyrins as herbicides. ACS Symp. Ser. 449: 371–386 [Google Scholar]
- Falbel T.G., Meehl J.B., Staehelin L.A. (1996). Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol. 112: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel T.G., Staehelin L.A. (1994). Characterization of a family of chlorophyll-deficient wheat (Triticum) and barley (Hordeum vulgare) mutants with defects in the magnesium-insertion step of chlorophyll biosynthesis. Plant Physiol. 104: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A., Merendino L., Barneche F., Ceol M., Meskauskiene R., Apel K., Rochaix J.D. (2005). The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev. 19: 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Casal J.J. (2004). Phenotypic characterization of a photomorphogenic mutant. Plant J. 39: 747–760 [DOI] [PubMed] [Google Scholar]
- Feierabend J., Dehne S. (1996). Fate of the porphyrin cofactors during the light-dependent turnover of catalase and of the photosystem II reaction-center protein D1 in mature rye leaves. Planta 198: 413–422 [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjieva R., Axelsson E., Olsson U., Hansson M. (2005). Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol. Biochem. 43: 901–908 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G., Mullineaux P.M. (2010). The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant. 138: 430–439 [DOI] [PubMed] [Google Scholar]
- Genty B., Briantais J.-M., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92 [Google Scholar]
- Giraud E., Ho L.H., Clifton R., Carroll A., Estavillo G., Tan Y.F., Howell K.A., Ivanova A., Pogson B.J., Millar A.H., Whelan J. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanningmeier U., Howell S.H. (1984). Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardi. Possible involvement of chlorophyll synthesis precursors. J. Biol. Chem. 259: 13541–13549 [PubMed] [Google Scholar]
- Kobayashi Y., Kanesaki Y., Tanaka A., Kuroiwa H., Kuroiwa T., Tanaka K. (2009). Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl. Acad. Sci. USA 106: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler R.H., Schwille P., Webb W.W., Hanson M.R. (2000). Active protein transport through plastid tubules: Velocity quantified by fluorescence correlation spectroscopy. J. Cell Sci. 113: 3921–3930 [DOI] [PubMed] [Google Scholar]
- Koncz C., Mayerhofer R., Koncz-Kalman Z., Nawrath C., Reiss B., Redei G.P., Schell J. (1990). Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 9: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kropat J., Oster U., Rüdiger W., Beck C.F. (1997). Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. USA 94: 14168–14172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Oster U., Rüdiger W., Beck C.F. (2000). Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24: 523–531 [DOI] [PubMed] [Google Scholar]
- Kubis S.E., Lilley K.S., Jarvis P. (2008). Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol. Biol. 425: 171–186 [DOI] [PubMed] [Google Scholar]
- La Rocca N., Rascio N., Oster U., Rüdiger W. (2001). Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213: 101–108 [DOI] [PubMed] [Google Scholar]
- La Rocca N., Rascio N., Oster U., Rüdiger W. (2007). Inhibition of lycopene cyclase results in accumulation of chlorophyll precursors. Planta 225: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Larkin R.M., Alonso J.M., Ecker J.R., Chory J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lee K.P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Livingston A.K., Cruz J.A., Kohzuma K., Dhingra A., Kramer D.M. (2010). An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 22: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T. (2008). Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 96: 121–143 [DOI] [PubMed] [Google Scholar]
- Matringe M., Camadro J.M., Block M.A., Joyard J., Scalla R., Labbe P., Douce R. (1992). Localization within chloroplasts of protoporphyrinogen oxidase, the target enzyme for diphenylether-like herbicides. J. Biol. Chem. 267: 4646–4651 [PubMed] [Google Scholar]
- Matsumoto F., Obayashi T., Sasaki-Sekimoto Y., Ohta H., Takamiya K., Masuda T. (2004). Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 135: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac A.C., Terry M.J. (2002). Loss of nuclear gene expression during the phytochrome A-mediated far-red block of greening response. Plant Physiol. 130: 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan J.A., Larkin R., Nagatani A., Chory J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A.G., Tanaka A., Terry M.J. (2010). The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15: 488–498 [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra A., Tripathy B.C. (2007). Differential distribution of chlorophyll biosynthetic intermediates in stroma, envelope and thylakoid membranes in Beta vulgaris. Photosynth. Res. 94: 401–410 [DOI] [PubMed] [Google Scholar]
- Moulin M., McCormac A.C., Terry M.J., Smith A.G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Masuda T., Bando T., Yamagata H., Ohta H., Takamiya K. (1998). Cloning and expression of the soybean chlH gene encoding a subunit of Mg-chelatase and localization of the Mg2+ concentration-dependent ChlH protein within the chloroplast. Plant Cell Physiol. 39: 275–284 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (1999). PHOTOPROTECTION REVISITED: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 333–359 [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Alonso J.M., Kim C.J., Leisse T.J., Ecker J.R. (2007). An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2: 2910–2917 [DOI] [PubMed] [Google Scholar]
- op den Camp R.G., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., Wagner D., Hideg E., Göbel C., Feussner I., Nater M., Apel K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai T., Imashimizu M., Seki A., Sato S., Tabata S., Imamura S., Asayama M., Ikeuchi M., Tanaka K. (2009). ChlH, the H subunit of the Mg-chelatase, is an anti-sigma factor for SigE in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 106: 6860–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster U., Brunner H., Rüdiger W. (1996). The greening process in cress seedlings. V. Possible interference of chlorophyll precursors, accumulated after thujaplicin treatment, with light-regulated expression of Lhc genes. J. Photochem. Photobiol. B 36: 255–261 [Google Scholar]
- Papenbrock J., Mock H.-P., Kruse E., Grimm B. (1999). Expression studies in tetrapyrrole biosynthesis: Inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208: 264–273 [Google Scholar]
- Papenbrock J., Mock H.P., Tanaka R., Kruse E., Grimm B. (2000b). Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J., Pfündel E., Mock H.P., Grimm B. (2000a). Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 22: 155–164 [DOI] [PubMed] [Google Scholar]
- Perry S.E., Li H.M., Keegstra K. (1991). In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol. 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Peter E., Grimm B. (2009). GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2: 1198–1210 [DOI] [PubMed] [Google Scholar]
- Petit J.M., Briat J.F., Lobréaux S. (2001). Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J. 359: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöpperl G., Oster U., Rüdiger W. (1998). Light-dependent increase in chlorophyll precursors during th day-night cycle in tobacco and barley seedlings. J. Plant Physiol. 153: 40–45 [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 957: 384–394 [Google Scholar]
- Rizhsky L., Davletova S., Liang H., Mittler R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Ruckle M.E., DeMarco S.M., Larkin R.M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirijovski N., Olsson U., Lundqvist J., Al-Karadaghi S., Willows R.D., Hansson M. (2006). ATPase activity associated with the magnesium chelatase H-subunit of the chlorophyll biosynthetic pathway is an artefact. Biochem. J. 400: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotka R., Dühring U., Komenda J., Peter E., Gardian Z., Tichy M., Grimm B., Wilde A. (2008). Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J. Biol. Chem. 283: 25794–25802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbaek A., Hansson A., Wulff R.P., Hansson M., Dietz K.J., Jensen P.E. (2008). NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 582: 2773–2778 [DOI] [PubMed] [Google Scholar]
- Stettler M., Eicke S., Mettler T., Messerli G., Hörtensteiner S., Zeeman S.C. (2009). Blocking the metabolism of starch breakdown products in Arabidopsis leaves triggers chloroplast degradation. Mol. Plant 2: 1233–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand Å., Asami T., Alonso J., Ecker J.R., Chory J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Susek R.E., Ausubel F.M., Chory J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- To J.P.C., Reiter W.-D., Gibson S.I. (2003). Chloroplast biogenesis by Arabidopsis seedlings is impaired in the presence of exogneous glucose. Physiol. Plant. 118: 456–463 [Google Scholar]
- Vasileuskaya Z., Oster U., Beck C.F. (2005). Mg-protoporphyrin IX and heme control HEMA, the gene encoding the first specific step of tetrapyrrole biosynthesis, in Chlamydomonas reinhardtii. Eukaryot. Cell 4: 1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia M.A., Larkin R.M., Ferrer J.L., Riek R., Chory J., Noel J.P. (2005). Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biol. 3: e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinti G., Hills A., Campbell S., Bowyer J.R., Mochizuki N., Chory J., López-Juez E. (2000). Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J. 24: 883–894 [DOI] [PubMed] [Google Scholar]
- Voigt C., Oster U., Börnke F., Jahns P., Dietz K.J., Leister D., Kleine T. (2010). In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol. Plant. 138: 503–519 [DOI] [PubMed] [Google Scholar]
- von Gromoff E.D., Alawady A., Meinecke L., Grimm B., Beck C.F. (2008). Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20: 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Walley J.W., Coughlan S., Hudson M.E., Covington M.F., Kaspi R., Banu G., Harmer S.L., Dehesh K. (2007). Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Wilde A., Mikolajczyk S., Alawady A., Lokstein H., Grimm B. (2004). The gun4 gene is essential for cyanobacterial porphyrin metabolism. FEBS Lett. 571: 119–123 [DOI] [PubMed] [Google Scholar]
- Willows R.D. (2003). Biosynthesis of chlorophylls from protoporphyrin IX. Nat. Prod. Rep. 20: 327–341 [DOI] [PubMed] [Google Scholar]
- Willows R.D., Lake V., Roberts T.H., Beale S.I. (2003). Inactivation of Mg chelatase during transition from anaerobic to aerobic growth in Rhodobacter capsulatus. J. Bacteriol. 185: 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.W., Yuan S., Feng H., Xu F., Cheng J., Shang J., Zhang D.W., Lin H.H. (2011). Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs - New evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 168: 714–721 [DOI] [PubMed] [Google Scholar]