By means of a combination of experimental and modeling approaches applied to the hyperbranching mutant étoile, cell–cell communication, likely mediated by novel transmembrane proteins that share similarities with metazoan Notch receptors, was shown to account for the establishment of filament patterning and cell differentiation in the filamentous brown alga Ectocarpus siliculosus.
Abstract
Brown algae are multicellular marine organisms evolutionarily distant from both metazoans and land plants. The molecular or cellular mechanisms that govern the developmental patterning in brown algae are poorly characterized. Here, we report the first morphogenetic mutant, étoile (etl), produced in the brown algal model Ectocarpus siliculosus. Genetic, cellular, and morphometric analyses showed that a single recessive locus, ETL, regulates cell differentiation: etl cells display thickening of the extracellular matrix (ECM), and the elongated, apical, and actively dividing E cells are underrepresented. As a result of this defect, the overrepresentation of round, branch-initiating R cells in the etl mutant leads to the rapid induction of the branching process at the expense of the uniaxial growth in the primary filament. Computational modeling allowed the simulation of the etl mutant phenotype by including a modified response to the neighborhood information in the division rules used to specify wild-type development. Microarray experiments supported the hypothesis of a defect in cell–cell communication, as primarily Lin-Notch-domain transmembrane proteins, which share similarities with metazoan Notch proteins involved in binary cell differentiation were repressed in etl. Thus, our study highlights the role of the ECM and of novel transmembrane proteins in cell–cell communication during the establishment of the developmental pattern in this brown alga.
INTRODUCTION
Brown algae are multicellular organisms that live along the coasts all over the world under tropical, temperate, and arctic climates. Their sizes are diverse, ranging from microscopic filaments (Laminaria gametophytes) to up to 50-meter-long giant kelps (Macrocystis sp). They also display a wide range of morphologies, spanning from uniseriate filaments to blades with conic, plain, or digitated flat shapes.
Brown algae belong to the heterokont phylum, also comprising oomycetes and diatoms (Baldauf, 2008). It is estimated that this phylum emerged more than one billion years ago from a unicellular eukaryotic ancestor common to all the eukaryotic lineages known today (Yoon et al., 2004). Therefore, the very distant phylogenetic relationship between brown algae and other multicellular organisms, namely, the archaeplastida (land plants and red and green algae) and the opisthokonta (metazoan and fungi), raises the possibility that these plants developed distinct cellular mechanisms to achieve multicellular development.
The brown algae Fucus and Pelvetia were previously used to study the establishment of plant zygote polarity during the first divisions of the embryo (Hable and Kropf, 2005; reviewed in Kropf, 1992). During the first divisions, the cytoskeleton and associated proteins are highly implicated in the establishment of cell polarity, in addition to a Rho family GTPase (Fowler et al., 2004). At later embryonic stages, cell wall–mediated intercellular communication processes seem to participate in the determination and maintenance of cell fate in the fucoid embryo (Berger et al., 1994; Bouget et al., 1998), but the nature of the underlying molecular components remains unknown.
Despite their use as models for embryo polarization, mature thalli of fucoid algae cannot be cultured in vitro and are therefore not appropriate models to study postembryonic development. Furthermore, genomic and genetic tools are not available. By contrast, the emerging brown alga model Ectocarpus siliculosus (Peters et al., 2004; Charrier et al., 2008) displays both a body and a reproductive cycle amenable to morphogenetic and genetic studies (Peters et al., 2008). In addition, its genome is sequenced (Cock et al., 2010) and proteomic (Ritter et al., 2010) and transcriptomic tools such as microarrays (Dittami et al., 2009) and real-time PCR normalization genes (Le Bail et al., 2008b) are available.
The early developmental pattern of wild-type E. siliculosus sporophytes was previously described (Le Bail et al., 2008a). Despite a high level of morphological plasticity, several constants were observed. After zygote germination, E. siliculosus sporophytes grow a uniaxial prostrate filament composed of two cell types, E (for elongated) and R (for round). E cells are always located at apical and subapical positions, and R cells are grouped in the center of the filament. Filament growth is ensured essentially by apical E cell division and elongation, while R cells are produced by progressive subapical E cell differentiation. Subsequent axillary branching occurs mainly in the central part of the filament. This overall prostrate filamentous body then differentiates erect filaments, which will later carry the sporangia. Previous computer modeling showed that the developmental patterning that takes place at the early filament stages could be accounted for by local positional information based on the recognition of neighboring cells (Billoud et al., 2008). The progression of the developmental program (branching and shift to the reproductive phase) depends on the phytohormone auxin, which suggests an overall regulation of the developmental pattern based on auxin-mediated positional information (Le Bail et al., 2010). Analyses of morphologically affected mutants were then initiated to further decipher the mechanisms involved in the regulation of the morphogenesis in brown algae. Here, we report a morphogenetic mutant of a brown alga, étoile (etl), in E. siliculosus. We provide evidence that cell differentiation is altered in this mutant, resulting in an overall modification of its early developmental pattern, and point to a probable defect in the interplay between the extracellular matrix (ECM) and novel transmembrane proteins containing a Notch domain, whose presence and role previously have only been identified in animals.
RESULTS
Morphogenesis in the Brown Alga E. siliculosus Is Regulated by the Recessive Single Locus ETL
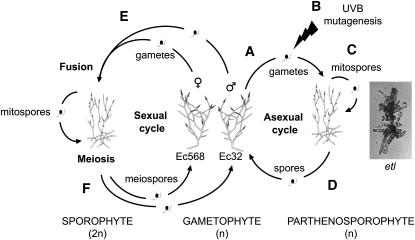
The life cycle of E. siliculosus is biphasic, heteromorphic, and dioecious (Figure 1). Both male and female unfertilized gametes are able to generate haploid sporophytes called parthenosporophytes (reviewed in Charrier et al., 2008), which are particularly amenable to mutant selection. Gametes from the male strain Ec32 were irradiated by UV-B, and after 2 weeks, the mutant etl was isolated. It displayed a hyperbranched phenotype, which remained stable for at least six parthenogenetic generations. etl gametophytes displayed no morphological variation compared with wild-type gametophytes.
Figure 1.
Mutagenesis and Genetic Characterization of the Mutant etl.
The life cycle of E. siliculosus can be both sexual (left) and asexual (right).
(A) Unfertilized gametes can generate haploid sporophytes called parthenosporophytes.
(B) Gametes from the strain Ec32 were irradiated with UV-B, and the etl mutant was isolated after 2 weeks of growth.
(C) The phenotype was stable for at least six parthenogenetic generations produced via germination of mitospores released from plurilocular sporangia.
(D) etl spores were isolated from unilocular sporangia to generate gametophytes, which displayed no morphological variation compared with wild-type gametophytes.
(E) Male etl gametes were crossed with female gametes of the strain Ec568 and the F1 generation displayed a wild-type phenotype.
(F) Unilocular sporangia were isolated from the F1, and the genotype of the spores was identified from the phenotype of the corresponding parthenosporophyte.
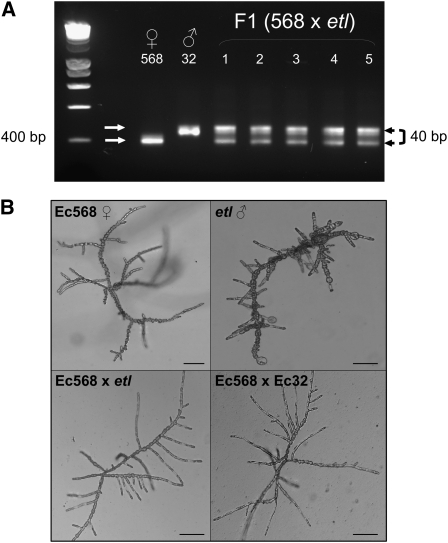
The mutant etl was then crossed with a female wild-type strain (Ec568). The heterozygous state of the F1 was tested by PCR amplification of a microsatellite marker, which exhibits length polymorphism between the two strains (Figure 2A). The 10 zygotes obtained from this cross all showed a wild-type phenotype, indicating that the etl mutation is recessive (Figure 2B).
Figure 2.
The etl Locus Is Recessive.
The male mutant etl (genetic background Ec32) was crossed with the female strain Ec568.
(A) Ten F1 individuals were tested by PCR for the presence of the polymorphic microsatellite marker 427 (40 bp difference; Heesch et al., 2010) between the two strains (only five shown).
(B) The 10 F1 individuals displayed a phenotype similar to that of the progeny of the control cross Ec568 × Ec32, showing the recessive character of the etl mutation. Bars = 100 μm.
To analyze the segregation of the etl mutation in the subsequent generations, 12 unilocular sporangia, each of them the site of a single meiotic event, were isolated from the heterozygote diploid sporophyte (Figure 1F). For each of them, 26 to 74 gametophytes were subisolated, and the morphology of the corresponding parthenosporophytes grown from the germination of unfertilized gametes (parthenogenesis) was analyzed. Altogether, 263 gametophytes produced parthenosporophytes with a mutant phenotype, and 275 gametophytes produced parthenosporophytes with a wild-type phenotype (Table 1). The results obtained were consistent with a 1:1 segregation ratio and a Mendelian inheritance of a single locus mutation (χ2 test, df = 90, χ2 = 96, P value = 0.313).
Table 1.
Segregation Analysis of Gametophytes Derived from Individual Unilocular Sporangia from One F1 (Ec568 × etl) Sporophyte
| Unilocular Sporangium | No. of etl Gametophytes | No. of Wild-Type Gametophytes |
| 1 | 13* | 13* |
| 2 | 32* | 42* |
| 3 | 14* | 22* |
| 4 | 14* | 23* |
| 5 | 20* | 14* |
| 6 | 30 | 16 |
| 7 | 23* | 13* |
| 8 | 34* | 28* |
| 9 | 14* | 18* |
| 10 | 19 | 37 |
| 11 | 40 | 20 |
| 12 | 22* | 17* |
| Total | 263* | 275* |
Segregation analysis was performed from 12 meiotic events (one event per unilocular sporangium). Twenty-six to 60 etl or wild-type gametophytes were isolated from each sporangium and grown to produce parthenosporophytes, on which the phenotype was observed. χ2 tests show that the population does not significantly deviate from a 50% (wild type)/50% (etl) segregation, indicating that a single mutated locus is responsible for the etl phenotype. Asterisk indicates that the value does not deviate significantly from 1:1 (χ2 test, P > 0.05).
Additional crosses were performed to further analyze the inheritance of the etl mutation. The sex of nine mutant gametophytes derived from the F1 was determined by attempting to cross them with male Ec32 and female Ec568 gametophytes. Four mutant gametophytes could be crossed only with the male Ec32 strain and five only with the female Ec568 strain, demonstrating that the etl mutation segregated independently of the sex locus.
Higher Branching Activity Increases etl Overall Growth Rate
Wild-type filaments of young E. siliculosus sporophytes (prostrate filaments) first grow as a linear primary filament (PF; Figure 3A) before lateral branching occurs mainly in the center of the filament giving rise to secondary filaments (SFs; Figure 3B) that grow parallel to the surface at the early stages (Figure 3C). By contrast, etl growth was characterized by an early arrest in apical growth and an earlier emergence of numerous branches all along the PF (Figures 3D and 3E), ultimately producing a bushy organism growing in all spatial dimensions (Figure 3F). An overall growth curve showing the number of cells as a function of time demonstrated that etl kept on growing faster than the wild-type filament 3 d after germination of the mitospores (Figure 3G). Detailed analyses of both the position and the time of emergence of the SFs can account for this increase in the growth rate. Indeed, while in wild-type filaments branching occurs preferentially in the central part of the filament, in etl, branches emerge with similar probabilities all along the filament (Figure 3H). In addition, observations of the developmental stage at which SF emergence occurs showed that branches develop earlier in etl than in the wild type. First and second SFs emerge at the 11- and 14-cell stages, respectively, in the wild type, while they emerge at the 5- and 8-cell stages in etl (Figures 3I and 3J). Finally, the emergence of branches was also more frequent in the etl mutant than in the wild type, with the PF growing on average 2.2 cells between the first and the second emergences, compared with 3.6 cells in the wild type.
Figure 3.
Hyperbranching Phenotype of the etl Mutant.
(A) to (F) Morphology of wild-type (A, B, C) and etl (D, E, F) sporophytes. Sporophytic wild-type PFs are composed of two cell types, namely, the elongated E cells located at the apical parts and the round R cells located in the center (A). Ten days after germination, SFs differentiate, mainly in the center and on R cells (Le Bail et al., 2008a) (B), thereby generating a loosely filamentous organism after 20 d (C). In etl sporophytes, cell types are less distinct (D), and the arrest of apical division and the more prolific branching (E) leads to a bushy, compact organism (F).
(G) Changes in cell number in the whole organism over time. For each observed individual, the time series was adjusted so that the first division occurred at t = 0. The number of observed wild-type (WT) and etl filaments was 54 and 35, respectively. Mean values ± sd.
(H) Position of the first nonaxial division, giving rise to the first SF (SF I). The y axis represents the relative deviation from the values expected under the assumption of a uniform distribution of SF.
(I) and (J) Developmental stage (number of cells in the primary filament) when the first (top) and second (bottom) nonaxial division occurs, giving rise to the first and second SFs (SF I [I] and SF II [J]), respectively. Means (represented as a vertical bar) and sd are indicated under the x axis. n = 50 individuals for both (I) and (J).
Auxin, which was shown to modulate wild-type E. siliculosus branching pattern (Le Bail et al., 2010), had no noticeable effect on etl development.
etl Regulates Cell Differentiation
At the early stages of development, two cell types can be differentiated in filaments of wild-type sporophytes: R cells with a length/width ratio ≤ 1.5 and E cells with a ratio > 1.5 (Figure 3A; Le Bail et al., 2008a). In etl, cell size and shape were significantly altered compared with the wild type, with E cells being chunkier and R cells being bigger (Figure 4A; P < 0.01). While in the wild type the two cell types were present in the same proportion within the filament (50% each; Le Bail et al., 2008a), in etl 90% of the cells were R (P < 0.01 for each stage; Figure 4B). Interestingly, in spite of this difference, the evolution in the cell type proportion was similar in the wild type and in etl during the entire experiment (up to the 20-cell stage). In both cases, the cell type proportion increased progressively from the 3-cell stage to the 12-cell stage, where it reached a plateau. These data indicate that etl alters the cell type proportion, without altering its evolution and its stabilization as the filament grows.
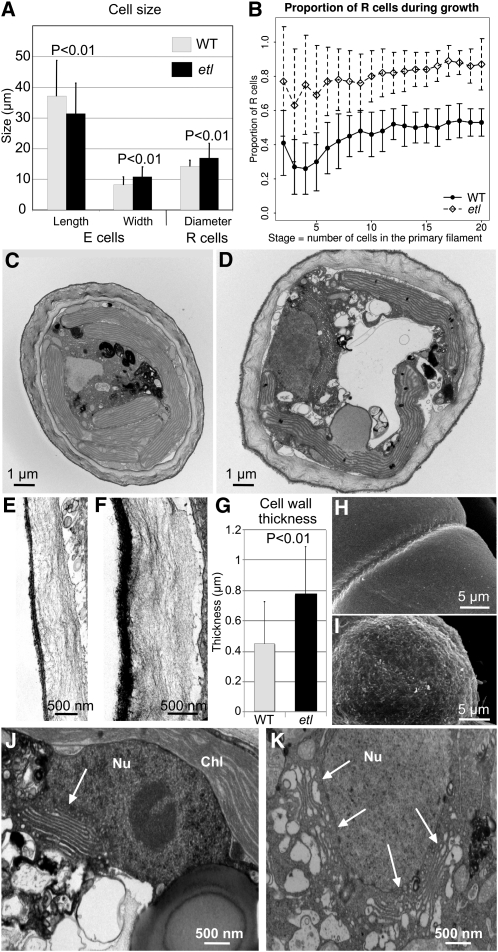
Figure 4.
Modification in Cell Size, Proportion, and Ultrastructure in etl.
(A) Cell size. In etl, E cells are shorter (31 μm versus 37 μm in the wild type [WT]) and wider (11 μm versus 8 μm in the wild type; t test, P < 0.01) and R cells are larger (17 μm diameter) than in the wild type (14 μm; P < 0.01). Means and sd are indicated (error bars) (n > 100).
(B) Proportion of R cells at each stage. Means and sd are indicated (error bars). n > 100 for each stage in both the wild type and etl.
(C) and (D) General morphology of the wild-type (C) and etl (D) cells observed by TEM.
(E) and (F) Details of the ECM in the wild-type (E) and etl (F) cells showing a similar organization with two layers: a thin outer black layer and a thick inner gray layer displaying an alignment of cellulose fibers.
(G) Thickness of the ECM. Several measurements were performed on >80 wild-type and etl cells observed by TEM. The mean ± sd of the smallest thickness per cell are presented (t test, P < 0.01).
(H) and (I) Cell surface viewed with a scanning electron microscope in wild-type (H) and etl (I) cells.
(J) and (K) Details of wild-type (J) and etl (K) cell ultrastructure by TEM. Nu, nucleus; Chl, chloroplast. The arrows indicate Golgi bodies. Note that this illustrates the most extreme and opposite phenotypes observed in the two organisms.
Furthermore, etl displayed a modification of several structural features of the cell. Transmission electron microscopy (TEM) showed that, in addition to the general diameter increase observed in etl cells, the ECM looked thicker compared with the wild type (Figures 4C and 4D; details in Figures 4E and 4F). Measurements on TEM sections (n > 80) confirmed that the ECM was significantly thicker in etl than in the wild type (t test, P value < 0.001; Figure 4G). Moreover, when observed by scanning electron microscopy, etl cells displayed a less regular and a rougher surface than wild-type cells (Figures 4H and 4I). However, the organization of the ECM did not seem to be modified in etl cells, as cellulose fibers were still observed parallel to the cell surface, and wild-type and etl ECM materials had a similar density (Figures 4E and 4F). In addition to a thicker ECM, etl cells displayed a slightly modified Golgi apparatus: it appeared larger and often encircled the cell nucleus (4% of etl cells displayed a discrete Golgi compared with 24% in the wild type, n = 30; Figures 4J and 4K). Furthermore, a higher proportion of etl cells displayed a low number of cisternae (25% of etl cells contained less than three cisternae per Golgi, compared with 10% in the wild type; Figures 4J and 4K). At their extremities, cisternae displayed large white vesicles at a higher proportion in etl cells (56%) than in wild-type cells (40%; Figure 4K), and this was accompanied by an abundant cis-Golgi. Therefore, the higher abundance of Golgi stacks with a reduced number of cisternae bearing big white budding vesicles indicate that the Golgi apparatus is more active in etl cells than in the wild type. This probably accounts for the thicker ECM, as the main components of the brown algal ECM (i.e., alginate and fucoidan polysaccharides) are transported to the ECM via Golgi-derived vesicles (Nagasato and Motomura, 2009).
In addition to changes in the proportion of R and E cells, the position of these cell types along the filament was altered. Figure 5 shows their distribution in >100 wild-type or etl primary filaments grown in similar conditions and observed from the 2-cell stage to the 10-cell stage. Because of the high level of morphological plasticity in this alga, wild-type filaments displayed a large range of combinations. However, a preference for filaments with R cells located in the center and always adjacent to each other was observed in the wild type (Le Bail et al., 2008a). In etl, the filament population was different. While some combinations were shared by the wild-type and etl populations, many others were specific to etl. In contrast with the wild type, etl R cells were distributed along the entire filament, and in rare but significant cases, R cells were separated by E cells. Altogether, these observations show that, in addition to the higher proportion of R cells and the modification of some of their subcellular features, tissue patterning in etl was impaired, as both the position of R cells along the filament and their clustering were affected.
Figure 5.
Filament Patterning in E. siliculosus Wild Type and etl.
Different filament organizations, according to the position of R cells (circle) and E cells (rectangle), were observed at each cell stage, from the 2-cell (left) to the 10-cell (right) stage. Some combinations were observed in both wild-type (WT) and etl populations (purple), while others occurred exclusively in wild-type (blue) or etl (red) populations. The respective frequency (%) of each combination in either of the two populations is indicated on the left side of the corresponding drawing with the same color code. n > 100 individuals at each stage.
Computational Simulation of etl Development Suggests Impairment in Local Positional Information
To generate a hypothesis of the mechanisms perturbed in etl, we modeled its development attempting to reproduce the cell-type distribution observed at each stage of filament growth (from the 2- to the 10-cell stages). Both the types of distribution and their frequency of occurrence were taken into account (Figure 5). The early developmental pattern of E. siliculosus wild-type sporophyte filaments was previously modeled using the cellular automaton ECTOMAT (Billoud et al., 2008). ECTOMAT uses five transition rules, based on the cellular processes taking place during the development of the alga in vivo (taking into account both cell division and cell differentiation rules: I, division of an R cell with at least one free side; E, E cell differentiation from an R cell; A, apical division of an E cell; R, R cell differentiation from an E cell; and D, simultaneous division and differentiation of an E cell into two R cells; Billoud et al., 2008; Figures 6A and 6B). These processes were modeled as stochastic transitions between cell states, which make use of information about the cellular neighborhood of the cell undergoing the transition. For example, rules R and D occur only if the considered cell is an E cell and if it has one R cell on one side and another cell (of any type) on the other side. Likewise, rule A depicts the apical division and is modeled as a transformation of an E cell into two E cells, provided that the initial E cell is bordered by one cell and an empty space. As these cellular transitions were also observed in the etl mutant, we investigated whether etl development patterning could be accounted for by changing only the probability of occurrence of these transition rules. For both wild-type and etl modeling, the probabilities were optimized for the distribution of R cells counted at each filament stage and were then used to simulate the generation of the population observed in Figure 5 (both filament pattern and frequency of occurrence at each filament stage). Figure 6B displays the best probabilities obtained for both wild-type and etl filament populations. It shows that, while the optimization heuristic method managed to simulate the wild-type population (low indicator of discrepancy = 40.6), it failed to obtain a set of probabilities able to simulate the observed etl population (high indicator of discrepancy = 250.4). This means that the transition rules used to simulate the development of wild-type E. siliculosus were not adequate and that the automaton rules themselves needed to be changed.
Figure 6.
Computational Modeling of the Early Developmental Pattern in the Wild Type and etl.
(A) Transition rules. Filaments are schematized by circles (R cells) and rectangles (E cells). The different events of cell differentiation are indicated by arrows. A letter, corresponding to the transition rule, is given above the arrow. The symbols accompanying the letter designate any required cells adjacent to that undergoing the transition: * for an empty neighborhood, ° for an R cell, and Δ for either an R or an E cell. No symbol means that the transition occurs with any type of neighboring cell. Black arrows and letters represent the rules used to model the development of the wild-type filaments; red dashed arrows and red letters are those added to model the development of etl filaments.
(B) Optimized probabilities of occurrence used to simulate the developmental pattern of wild-type (WT) and etl filaments. Each starting configuration (left) is defined by a cell type for the central cell (in brown) and its immediate neighbors (open gray shapes). The possible states are E (elongated, a rectangle), R (round, a circle), and 0 (empty, a star). Superimposed symbols mean that either state is allowed. The transitions (arrows) affect only the central cell; they are called I for initial, E for elongate, A for apical, P for partition, V for variegate, R for round, D for double, and S for split. All transitions are stochastic (i.e., they occur with a given probability). For each transition rule, the optimized probabilities enabling the best simulation at each developmental stage are shown on the right part, for both the wild type and the etl mutant. In each case, the simulation was optimized with either five or eight rules. An indicator of discrepancy between the simulated population and the observed population is given (see Methods for details).
(C) Simulation of a virtual population of etl filaments at the 10-cell stage. The simulation was conducted with the optimized probabilities given in (B). All patterns observed in vivo (Figure 5) were generated by the simulation. The frequency of each combination, either generated by the simulation (black) or observed in vivo (gray), is shown. Patterns with a simulated frequency of <1% are grouped in others. The linear correlation coefficient between the frequencies (r) and the P value of the Pearson test (p) is indicated.
In vivo observations revealed that, compared with the wild-type organism, etl undergoes additional cellular transitions during its early development. Figure 6A shows that the three additional rules (dashed lines in red) all contribute to the generation of more R cells, in agreement with the previous description of etl filaments. These rules were incorporated in the ECTOMAT model, and the probabilities of the now eight transition rules were optimized as described above (Figure 6B). This approach allowed the successful simulation of the in vivo etl population at all the developmental stages (low indicator of discrepancy = 3.5). As an example, Figure 6C provides a comparison of the observed and the simulated populations at the 10-cell stage (correlation coefficient = 0.89). When these eight transition rules were used to similarly simulate the development of the wild-type population (correlation coefficient = 0.85), the optimal probabilities of the three additional rules S, P, and V remained close to 0 (Figure 6B). This shows that the gain in adequacy for etl modeling brought by these new rules did not result from the increase in the number of free parameters. Instead, it supports the conclusion that these cellular transitions are specific to etl and do not participate in the development patterning of wild-type filaments, in agreement with our in vivo observations.
These results allowed us to speculate on the cellular mechanisms altered in etl. In addition to generating more R cells, the additional rules implemented a modification in the type of immediate neighboring cells required to promote a given transition in a cell (Figure 6A). Rule P imposed a different and weaker constraint on the nature of the neighboring cells than rule D, as only an empty space bordering the differentiating cell was required. Rule S, whose probability of occurrence was among the highest (Figure 6B), was unique as it did not impose any type of constraint on the neighboring cells. Altogether, computational simulations demonstrated that etl could be modeled using both additional transitions occurring during the initial developmental patterns and modified local constraints, which were weaker (rule P and V), different (rule P), or nonexistent (rule S). This result indicates that etl may be impaired in the recognition of neighboring cells, a process which is likely to participate in the establishment of local positional information required for proper filament patterning in the wild type.
etl Underexpresses Lin-Notch Domain Transmembrane Proteins
To identify the target genes of this mutation, we analyzed the transcriptome of etl using a large-scale microarray approach (Dittami et al., 2009). In addition to the wild-type organism, we included as a technical reference another E. siliculosus morphological mutant, called mutant A. After normalization of the data (see Supplemental Data Set 1 online), a statistical analysis was performed using significance analysis of microarrays (false discovery rate = 0.05). In etl, 24 ESTs displayed a modified level. This low number was not due to technical limitation of sensitivity or too stringent analysis conditions, as 943 ESTs were shown to be up- or downregulated in mutant A compared with the wild type (see Supplemental Data Set 2 online). Confirmation by quantitative RT-PCR (qRT-qPCR) allowed the identification of only two upregulated ESTs and five downregulated ESTs. The upregulated ESTs belong to a genomic region corresponding to a putative transposable element (Cock et al., 2010) related to a retrotransposon-like family member from zebra fish (Danio rerio; XP001339559.3 BLAST E value = 10−34). This upregulation was not correlated to an increase in the transposition activity of this element, as shown by the qPCR measurement of its genome copy number (see Supplemental Figure 1 online). The five downregulated genes, named ERG #1-5 (for etl-regulated gene), whose sequences were not modified in etl compared with those in the wild-type organisms, contained specific protein domains. ERG5 contained a short-chain dehydrogenase domain. The four other proteins featured a signal peptide and one or several predicted transmembrane domains (Figure 7). In addition, Lin-Notch domains (Interpro IPR000800, also named Notch domain or LNR) were present in up to three copies in three of the ERG sequences (ERG2, ERG3, and ERG4). In E. siliculosus, at least 14 predicted proteins with Lin-Notch domains (E value < 10−5) are present (see Supplemental Data Set 3 online), while at the same significance level, none have been found in the genomes of the monocellular diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum, which constitute the phylogenetically most closely related organisms with available genome sequences.
Figure 7.
ERG Proteins Are Targets of the ETL Locus.
Microarray experiments were conducted on four independent biological replicates corresponding to either wild-type or etl samples. Significance analysis of microarrays (Tusher et al., 2001) identified 19 significantly downregulated genes in etl, of which only five were confirmed by qRT-PCR.
(A) Map of the five ERG proteins. The genome sequences of the ERG genes are available at http://bioinformatics.psb.ugent.be/webtools/bogas/ with the accession numbers indicated in parentheses. Protein domains are indicated with a color code. The number of amino acids of the corresponding protein is indicated at the right-hand side of the gene map.
(B) Expression level. Transcript levels were measured by qRT-PCR and normalized according to Le Bail et al. (2008b). Data are expressed as the log2 of the ratio with the wild-type (WT) organism. n = 4 biological replicates; error bars are sd.
The low number of ESTs showing a modified expression level in etl remains surprising, considering the pleiotropic phenotype of this mutant. Therefore, it seems likely that the phenotypic alterations reflect additional molecular phenomena, which cannot be detected by transcriptome analysis. Indeed, in addition to transcriptional regulation of gene expression, various processes are able to account for phenotypic variation. Hundreds of plant genes are regulated by microRNAs (Zhang et al., 2010), which can cleave the mRNA at the recognition site or block mRNA translation (reviewed in Voinnet, 2009), thereby reducing protein production, while maintaining the corresponding transcripts at nearly normal levels (Brodersen et al., 2008). Therefore, we cannot exclude the possibility that additional genes are misexpressed in etl, which cannot be detected by the microarray approach.
DISCUSSION
Most of the previous studies on brown algal development were performed on fucoid zygote polarization (Fucus sp or Silvetia sp). They allowed the identification of molecular actors involved in this initial process of development (for an update, see Hable and Kropf, 2005). Interestingly, Berger et al. (1994) showed that, later in the embryonic development, a component of the cell wall was determining the cell fate. Unfortunately, because of the experimental limitations of fucoid models, this component remains unidentified more than 15 years later. This study used another model, E. siliculosus, which is more amenable to the investigation of thallus development. Previous studies on its wild-type sporophyte filament have shown that, at the early stages, the apical E cells both elongate and divide away from the center of the filament, ensuring most of its growth, while the resulting subapical E cells progressively differentiate into R cells (Le Bail et al., 2008a). The balance between these two activities (i.e., cell elongation and division on one hand and cell differentiation on the other hand) might be at least partially regulated by auxin, as this phytohormone is accumulated in the E cells where it inhibits both E-to-R cell differentiation and branching (Le Bail et al., 2010).
In this study, we characterized a morphogenetic mutant of a brown alga. The single locus mutation etl is responsible for a hyperbranching activity with an earlier and more synchronized emergence of branches in no preferential position along the filament. The partial loss of the E cell type in etl, resulting in a high abundance of R cells (90% compared with 50% in the wild type) can easily account for this phenotype, as in the wild type mainly R cells ensure branching (Le Bail et al., 2008a). This also accounts for the arrest in mitotic activity observed early in the apical parts of the etl filaments. Furthermore, etl cells displayed modifications of their structural features: in addition to slight modifications in their dimensions, etl cells showed a thicker ECM with a rougher surface, which was correlated with a higher activity of the Golgi apparatus. Moreover, several observations supported an alteration of positional information: (1) cell transition events were modified, and modeling showed that this could result from a partial loss in cell–cell communication, and (2) primarily Notch domain transmembrane proteins were underexpressed. Altogether, these results show that ETL regulates cell differentiation and developmental patterning in the brown alga E. siliculosus and point to a combined role of ECM and novel transmembrane proteins in cell–cell communication involved in these processes.
The understanding of the functional relationship between the ECM and ERG1-4 transmembrane proteins will require detailed analyses focused on these two cellular and molecular actors. Indeed, both E. siliculosus ECM and ERGs display peculiar features, which make interpretation of the data challenging. The brown alga ECM is composed mainly of alginate and sulfated fucan (fucoidan) matrix polysaccharides, which embrace a low proportion of rigid cellulose fibers (Kloareg and Quatrano, 1988). Interestingly, Michel et al. (2010) recently showed that the E. siliculosus ECM was synthesized by a patchwork of enzymes inherited from various ancestors through independent evolutionary events. While cellulose is synthesized by enzymes closely related to land plant cellulose synthases (CESAs), fucoidans are synthesized by an ancestral pathway shared with animals. In the latter organisms, this pathway is responsible for the biosynthesis of sulfated ECM polysaccharides (e.g., heparan sulfate). Even more surprisingly, a horizontal gene transfer from an Actinobacterium is thought to account for the acquisition of alginates as well as hemicellulose. Altogether, these polysaccharides provide brown algae with a unique ECM adapted to the marine environment (Michel et al., 2010).
The unique nature of the brown algal ECM sharing features with land plants and metazoans makes comparisons with these organisms both risky and tempting. In animals, numerous examples of the combined role of the ECM and transmembrane proteins in cell development have been reported (Friedl and Zallen, 2010). The ECM regulates cell polarization by activating mechanosensitive transmembrane proteins, so-called integrins, and by stimulating the assembly of focal adhesion complexes. Reciprocally, the transmembrane and intracellular Wnt signaling pathway impacts the organization of the ECM. The ECM also regulates cell differentiation. Sulfated polysaccharides present within the ECM play a determining role in cell fate by regulating ligand binding to transmembrane receptors. For example, heparan sulfate, a sulfated ECM glycosaminoglycan, regulates the diffusion of the ligand of the epidermal growth factor (EGF) receptor through the ECM by mediating its sequestration by proteoglycans (Prince et al., 2010). This sequestration engages the cell in growth inhibition instead of cell proliferation. Interestingly, brown algal fucoidans also inhibit the binding of various ligands to animal transmembrane receptors (Heinzelmann et al., 1998). Hence, in brown algae, fucoidans could similarly restrain ligand diffusion to the Notch domain receptors, a process that could be enhanced in etl because of its thicker ECM. The reported positive feedback loop exercised by Notch proteins on Notch genes (Liu et al., 2009) illustrates a mechanism of autoregulation that could also apply to ERG genes and could possibly account for the decrease in their transcript level observed in etl.
In land plants, the cell wall has long been viewed as a static compartment, through which signal molecules must diffuse (reviewed in Brownlee, 2002). However, it was recently recognized as a dynamic and responsive compartment with an important role in relaying information from external stimuli during development (Kohorn, 2000; Humphrey et al., 2007). Several cell wall proteins are thought to be involved in these processes. The family of receptor-like kinases, and more specifically the wall-associated kinases, are transmembrane proteins able to sense the cell wall integrity and to regulate machineries involved in cell wall modification. They are involved in several morphogenetic events (Kohorn, 2001). Wall-associated kinases contain two EGF-like repeats in their extracellular domain, and their interaction with the cell wall polysaccharide pectin as well as Gly-rich proteins (Steinwand and Kieber, 2010) regulates the expression of numerous downstream genes, such as transcription factors and phytohormone-responsive genes (Kohorn et al., 2009). Similarly, structural cell wall components, such as Leu-rich repeat extensin proteins, were shown to regulate proper cell wall organization, and the presence of a Leu-rich repeat domain in these proteins suggests that they act as regulator and/or receptor of extracellular signals (Baumberger et al., 2003).
Altogether, in both the green and the opisthokont lineages, the study of the functional interactions between the ECM and transmembrane receptors is still an emerging field, and the peculiar evolutionary history of the brown algal ECM emphasizes the interest in deciphering them in E. siliculosus using etl as a tool.
The putative receptors identified in this study illustrate once more the special features of brown algae. Most of the transmembrane proteins repressed in etl (ERG1-4) contain a Lin-Notch domain. On one hand, this domain is absent from the Plantae phylum, with the exception of one nontransmembrane protein (Q016E4_OSTTA) in the prasinophyte Ostreococcus tauri (Chlorophyta). On the other hand, the Notch domain is specifically found in metazoan transmembrane receptors called Notch receptors, where it regulates the interaction between the extracellular and the intracellular regions (Fiúza and Arias, 2007). These Notch receptors are fundamental for animal development, in which they control lateral inhibition, boundary formation, and cell fate choice upon direct interaction with another transmembrane protein expressed at the surface of the adjacent cells (Delta or Jagged/Serrate; Fortini, 2009). Deregulation of Notch proteins is responsible for numerous cancers, and their involvement as both an oncogene and a tumor suppressor has been demonstrated frequently (Bolós et al., 2007; Koch and Radtke, 2007; Dotto, 2008). ERG2-4 do not contain EGF-like repeats, which mediate the interaction of metazoan Notch receptors with their extracellular ligand (Artavanis-Tsakonas et al., 1995). The lack of these EGF-like domains in the E. siliculosus ERGs precludes solid presumptions about a putative conservation of function. However, the combination of the domains present in Notch receptors seems to result from a shuffling of ancestral domains specific to metazoans (Gazave et al., 2009). Hence, brown algae might have evolved alternative strategies to assemble domains at the cell surface. As a matter of fact, several predicted excreted or transmembrane EGF-like domain-containing proteins are encoded by the E. siliculosus genome (see Supplemental Data Set 4 online), and homologs were shown to be secreted from embryonic cells of the brown alga Fucus distichus (Belanger et al., 2003). Therefore, E. siliculosus could have evolved structurally different receptors involved in cell–cell signaling while conserving Notch domains. The chemical features of the brown algal ECM could easily support the hypothesis that these organisms have evolved different cell–cell communication strategies than metazoans. This hypothesis is compatible with the absence of homologs of the Notch ligands Delta/Serrate/Lag-2 in the genome of E. siliculosus (see Supplemental Data Set 5 online).
Many questions related to the etl mutant remain unanswered. In particular, whether the E. siliculosus ECM influences ERG signaling or vice versa is not clear, as several examples of reciprocal ECM-receptor interactions have been reported in animals and plants (Humphrey et al., 2007; Friedl and Zallen, 2010). Nevertheless, because of the unique features of their ECM, brown algae are interesting models to study ECM-transmembrane receptor interactions and their impact on cell differentiation. The presence of integrin genes and associated proteins (e.g., talin and α-actinin) recently reported in the genome of E. siliculosus illustrates the unexplored potential of brown algae as models to unravel novel ECM-cytoplasm communication systems (Cock et al., 2010). In this context, etl is a particularly relevant tool to start deciphering them. The identification of the potential ligands of the Lin-Notch-ERG transmembrane proteins is among the most challenging goals for future studies. The characterization of their diffusion through the ECM and their interaction with Notch domain proteins will enable a better understanding of the mechanisms accounting for the developmental pattern of E. siliculosus filaments. The other major challenge is the identification of the ETL locus, as we have shown it to be a major regulator of the developmental pattern in E. siliculosus, as a probable actor in the ECM receptor signaling processes. Positional cloning of ETL has recently been initiated, and the availability of other branching mutants in the laboratory might help to identify ETL.
In conclusion, this study complements the recent discovery on the role of auxin in the development of E. siliculosus (Le Bail et al., 2010) and illustrates that the development of brown algae is likely based on a combination of strategies with molecular actors conserved in either metazoans or land plants, although in each case, variations were found at the molecular level.
METHODS
Algal Strains, Culture Conditions, and Production of Mutants
The wild-type strain used was Ec32 (accession CCAP 1310/4; origin San Juan de Marcona, Peru). The etl mutant (CCAP 1310/337) was generated from unfertilized gametes of the Ec32 strain irradiated for 11 min (LD50) with UV-B (316 nm) and selected after 2 weeks of growth for its modified morphology compared with wild-type parthenogenetic sporophytes (parthenosporophytes). The Ec568 female strain (accession CCAP 1310/334; origin Arica, Chile) was used to perform crosses with the etl mutant. Thalli were grown in half-strength Provasoli-enriched (Starr and Zeikus, 1993), autoclaved natural seawater (pH 7.8) in Petri dishes located in a controlled environment cabinet at 13°C with a 14:10 light:dark cycle (light intensity 29 μmol photon·m−2·s−1).
Zygote Production
etl (Ec32 genetic background) was crossed with a wild-type sister strain (Ec568). Fertile gametophytes of each sex were placed in the dark in a small amount of medium for a few hours to induce synchronized gamete release after the addition of natural seawater. Five microliters of each gamete solution was then mixed in a single drop on a cover slip, which was inverted on a silicone joint fixed on a slide with Vaseline to isolate the drop. Zygotes, which can be distinguished from unfertilized gametes due to the presence of two stigmata, were localized in the drop and monitored for 2 weeks in the hanging drop with change of medium every 2 d. Thereafter, the cover slip with germlings was immersed in a Petri dish and further development was followed under an inverted microscope. Germlings from the germination of unfertilized gametes were removed. The sex of gametophytes was determined by microscopy observation of zygote formation in hanging-drop preparations in which the strains to be tested had been combined with fertile thalli of male and female reference strains. The heterozygous state of each zygote was confirmed by PCR amplification with markers specific to the two parents (see below). The 10 zygotes obtained from this cross all showed a wild-type phenotype, indicating that the etl mutation is recessive.
Cross-Verification
DNA was extracted from the diploid heterozygous sporophytes with the NucleoSpin Plant II kit (Macherey-Nagel). Nuclear DNA was amplified in a microsatellite region whose length differs by 40 bp between Ec32 (genetic background of the etl mutant) and Ec568 (Heesch et al., 2010; primer sequences are ESM27F, 5′-TGTAAAACGGCCAGTGCAACTGCCCTCGGAAAATAAG-3′, and ESM27R, 5′-GCGCGGTCGATGCTAATAATAACTA-3′). The amplification was performed in an Applied Biosystems thermocycler (35 cycles, 0.5 min 94°C, 0.5 min 65°C, 1 min 72°C) using GoTaq DNA polymerase (Promega).
Morphometric Analysis
Observations were performed using an Olympus IX 51 inverted microscope. Cell sizes were measured using ImageJ software (http://rsbweb.nih.gov/ij/).
Cells in a filament were numbered from the closest extremity. A cell within a filament of L cells in length was assigned the number c, where, for reasons of symmetry, c is an integer such that 0 ≤ c ≤ (L−1)/2. To compare positions between primary filaments of unequal length, the cell position was converted into a relative index using a procedure adapted from Le Bail et al. (2008a): a number of intervals, N, was chosen (in this study, we used n = 10). The whole filament was graduated into 2N intervals so that each extremity was marked as 0, and the center was marked N (note that N represents the limit between two cells if L is even, and the middle of the central cell if L is odd). If an event (such as an axial division or the emergence of a secondary filament) occurs at cell number c, then this event is shared between the intervals that overlap with this cell, each interval receiving an amount proportional to the extent of overlap. Mathematically, cell c overlaps with the nth interval (the interval between graduations n−1 and n) if and only if 2c/L ≤ n/N and (n−1)/n ≤ (2c+1)/L; then the nth interval undergoes an amount of event An = min (n/N,(2c+1)/L)−max((n-1)/N,2c/L). For each interval, the amounts computed in all individuals of the observed sample are added, giving a total amount of observed events for each interval, On. In parallel, for each observed individual, L expectations were computed by assigning the same probability (1/L) of event to each cell and shared between intervals using the same procedure. These expected amounts are also added over the whole population, giving a total amount of expected events for each interval: En. The value Sn = (On−En)/(On+En) represents the bias between observed and expected events in the nth interval.
TEM
Parthenosporophytes were fixed with 4% glutaraldehyde and 0.25 M sucrose at room temperature and washed with 0.2 M sodium cacodylate buffer containing graded concentrations of sucrose. The samples were postfixed in 1.5% osmium tetroxyde, dehydrated with a gradient of ethanol concentrations, and embedded in Epon. Polymerization was performed at 60°C for 24 h. Ultrathin serial sections were cut with a diamond knife on an ultramicrotome and were mounted on copper grids. After staining with 2% uranyl acetate for 10 min and 2% lead citrate for 3 min, the grids were examined with a Jeol 1400 transmission electron microscope.
Scanning Electron Microscopy
Twenty microliters of a gamete release (Le Bail et al., 2008a) was loaded and fixed on a polycarbonate filtration membrane (diameter 13 mm; Nuclepore, Whatman). After 2 weeks of growth, parthenosporophytes were fixed in artificial seawater (ASW) with 3% paraformaldehyde for 1 h and then washed for 10 min in ASW:water (3:2), ASW:water (2:3), and water. The parthenosporophytes were then dehydrated successively in a gradient of ethanol concentrations (30, 50, 70, and 90%, two times 95%, and three times 100%) and finally dried using a critical point dryer (Baltec CPD 030; Balzer). The membranes were then fixed on a glass plot and covered with a 25-nm-thick gold layer. Filaments were finally observed with a JEOL JSM 5200 scanning electron microscope.
Measurements of the cell wall thickness were performed using the ImageJ software (http://rsb.info.nih.gov/ij/). Because sections are not necessarily perpendicular to the cell wall, calculations of the mean were performed on both five random measurements per cell and a single value corresponding to the smallest thickness (n = 89 for wild-type and n = 82 for etl cells) and were statistically analyzed with the R software (http://www.r-project.org/; R Development Core Team, 2011). The two measurement protocols yielded similar results.
Modeling
The development of a wild-type primary filament of Ectocarpus siliculosus was initially modeled by the stochastic one-dimensional cell automaton ECTOMAT (Billoud et al., 2008). Filaments of etl mutants could not be simulated by the same model, and observations showed that in this mutant, new transition events had to be taken into account. As the initial configurations of these new rules were expressed by patterns of various lengths, we reimplemented ECTOMAT in the more general form of a stochastic L-system (for review, see Prusinkiewicz and Lindenmayer, 1996). The probabilities for the five or eight transitions were optimized using a random descent algorithm, in which the value to minimize was the sum of squared differences between the distributions of R cell number in simulated versus observed filaments at each stage, normalized by the total number of cells (=indicator of discrepancy).
Microarray Experiment
Microarray experiments were performed on four independent wild-type, etl, and mutant A cultures. Each of the 12 cultures was grown in a single Petri dish, in identical growth conditions (culture cabinet), and for different culture durations (between 20 and 40 d, until they reached similar developmental stages corresponding to immature parthenosporophytes). To avoid the putative effects of diurnal oscillations on gene expression, all replicates were harvested at the same hour of day (10:30 am). Inoculation and collection dates were different. These samples were frozen in liquid nitrogen and RNA extractions were performed as described by Le Bail et al. (2008b). cDNA synthesis and amplification were performed following the protocol outlined by Dittami et al. (2009). The array design contained probes for a total of 17,119 unigenes, each represented by four 60-mers, and was created by Roche NimbleGen, as described by Dittami et al. (2009). Roche NimbleGen performed cDNA labeling and hybridization (single-channel platform) as part of their Gene Expression Array Service. These experiments were based on amplified double-stranded cDNAs. Differentially expressed genes were identified in the data set with a significance analysis of microarrays test (Tusher et al., 2001) using TigrMEV 4.3 (Saeed et al., 2003; false discovery rate = 0.05). Normalized expression values were generated using quantile normalization (Bolstad et al., 2003) and the Robust Multichip Average algorithm (Irizarry et al., 2003a, 2003b) (see Supplemental Data Set 1 online). The data obtained for the etl mutant were uploaded to ArrayExpress under accession number E-TABM-799.
qRT-PCR
Validation of the microarray data was performed by qRT-PCR amplification (SYBR green) of the target genes as described by Le Bail et al. (2008b). Three biological replicates independent from the ones used for the microarray experiments were tested. Normalization of the transcript levels was performed with the three reference genes EsEF1α, EsR26S, and EsUBCE as described by Le Bail et al. (2008b). Oligonucleotides are indicated in Supplemental Table 1 online.
Sequence Analysis
The peptide sequences of the ERG proteins were initially obtained from the automated annotation of the E. siliculosus genome (Cock et al., 2010) and checked in the wild type by sequencing of PCR-amplified or cloned full-length cDNAs for ERG 1, 2, 3, and 5. Using a proofreading PCR-based approach, each of the five ERG coding sequences framed by ~1 kb upstream and downstream was obtained in the etl mutant. Conserved domains were identified using InterProScan (Zdobnov and Apweiler, 2001). BLASTP analyses were performed using the blast 2.2.21 package (Altschul et al., 1997).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression and DNA Copy Number of the Retrotransposon in Wild-Type and etl Genomes.
Supplemental Table 1. Oligonucleotides Used in the Quantitative RT-PCR Analysis of the ERG Genes.
Supplemental Data Set 1. Normalized Signals of the Hybridized Microarrays.
Supplemental Data Set 2. Statistical Analysis of the Normalized Microarray (SAM) Data.
Supplemental Data Set 3. E. siliculosus Proteins with a Notch Domain.
Supplemental Data Set 4. Presence of EGF Patterns in the Proteome of E. siliculosus (Results from Interproscan).
Supplemental Data Set 5. Best Matches for the Uniprot Entries Annotated Delta/Jagged/Serrate Proteins in the Proteome of E. siliculosus.
Acknowledgments
We thank Simon Dittami for help in the preparation and data analyses of the microarray experiment and for English corrections. We also thank Jean Sourimant for technical assistance with the scanning electron microscope and Béatrice Satiat-Jeunemaitre for her help with the interpretation of the Golgi apparatus structure. We thank Carole Maisonneuve for subculturing the etl mutant. A.L.B. was a fellow of the French Ministry of Higher Education and Research. The creation of the mutant library was financially supported by the Brittany Regional Council (Contract ACOMB ‘ECTOMUT-1790’).
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Matsuno K., Fortini M.E. (1995). Notch signaling. Science 268: 225–232 [DOI] [PubMed] [Google Scholar]
- Baldauf S.L. (2008). An overview of the phylogeny and diversity of eukaryotes. J. Syst. Evol. 46: 263–273 [Google Scholar]
- Baumberger N., Steiner M., Ryser U., Keller B., Ringli C. (2003). Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35: 71–81 [DOI] [PubMed] [Google Scholar]
- Belanger K.D., Wyman A.J., Sudol M.N., Singla-Pareek S.L., Quatrano R.S. (2003). A signal peptide secretion screen in Fucus distichus embryos reveals expression of glucanase, EGF domain-containing, and LRR receptor kinase-like polypeptides during asymmetric cell growth. Planta 217: 931–950 [DOI] [PubMed] [Google Scholar]
- Berger F., Taylor A., Brownlee C. (1994). Cell fate determination by the cell wall in early fucus development. Science 263: 1421–1423 [DOI] [PubMed] [Google Scholar]
- Billoud B., Le Bail A., Charrier B. (2008). A stochastic 1D nearest-neighbour automaton models early development of the brown alga Ectocarpus siliculosus. Funct. Plant Biol. 35: 1014–1024 [DOI] [PubMed] [Google Scholar]
- Bolós V., Grego-Bessa J., de la Pompa J.L. (2007). Notch signaling in development and cancer. Endocr. Rev. 28: 339–363 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bouget F.Y., Berger F., Brownlee C. (1998). Position dependent control of cell fate in the Fucus embryo: Role of intercellular communication. Development 125: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brownlee C. (2002). Role of the extracellular matrix in cell-cell signalling: Paracrine paradigms. Curr. Opin. Plant Biol. 5: 396–401 [DOI] [PubMed] [Google Scholar]
- Charrier B., Coelho S.M., Le Bail A., Tonon T., Michel G., Potin P., Kloareg B., Boyen C., Peters A.F., Cock J.M. (2008). Development and physiology of the brown alga Ectocarpus siliculosus: Two centuries of research. New Phytol. 177: 319–332 [DOI] [PubMed] [Google Scholar]
- Cock J.M., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621 [DOI] [PubMed] [Google Scholar]
- Dittami S.M., et al. (2009). Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol. 10: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G.P. (2008). Notch tumor suppressor function. Oncogene 27: 5115–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiúza U.M., Arias A.M. (2007). Cell and molecular biology of Notch. J. Endocrinol. 194: 459–474 [DOI] [PubMed] [Google Scholar]
- Fortini M.E. (2009). Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell 16: 633–647 [DOI] [PubMed] [Google Scholar]
- Fowler J.E., Vejlupkova Z., Goodner B.W., Lu G., Quatrano R.S. (2004). Localization to the rhizoid tip implicates a Fucus distichus Rho family GTPase in a conserved cell polarity pathway. Planta 219: 856–866 [DOI] [PubMed] [Google Scholar]
- Friedl P., Zallen J.A. (2010). Dynamics of cell-cell and cell-matrix interactions in morphogenesis, regeneration and cancer. Curr. Opin. Cell Biol. 22: 557–559 [DOI] [PubMed] [Google Scholar]
- Gazave E., Lapébie P., Richards G.S., Brunet F., Ereskovsky A.V., Degnan B.M., Borchiellini C., Vervoort M., Renard E. (2009). Origin and evolution of the Notch signalling pathway: An overview from eukaryotic genomes. BMC Evol. Biol. 9: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable W.E., Kropf D.L. (2005). The Arp2/3 complex nucleates actin arrays during zygote polarity establishment and growth. Cell Motil. Cytoskeleton 61: 9–20 [DOI] [PubMed] [Google Scholar]
- Heesch S., Cho G.Y., Peters A.F., Le Corguillé G., Falentin C., Boutet G., Coëdel S., Jubin C., Samson G., Corre E., Coelho S.M., Cock J.M. (2010). A sequence-tagged genetic map for the brown alga Ectocarpus siliculosus provides large-scale assembly of the genome sequence. New Phytol. 188: 42–51 [DOI] [PubMed] [Google Scholar]
- Heinzelmann M., Polk H.C., Jr, Miller F.N. (1998). Modulation of lipopolysaccharide-induced monocyte activation by heparin-binding protein and fucoidan. Infect. Immun. 66: 5842–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T.V., Bonetta D.T., Goring D.R. (2007). Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 176: 7–21 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003a). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003b). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kloareg B., Quatrano R.S. (1988). Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. 26: 259–315 [Google Scholar]
- Koch U., Radtke F. (2007). Notch and cancer: A double-edged sword. Cell. Mol. Life Sci. 64: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B.D. (2000). Plasma membrane-cell wall contacts. Plant Physiol. 124: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B.D. (2001). WAKs; cell wall associated kinases. Curr. Opin. Cell Biol. 13: 529–533 [DOI] [PubMed] [Google Scholar]
- Kohorn B.D., Johansen S., Shishido A., Todorova T., Martinez R., Defeo E., Obregon P. (2009). Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60: 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropf D.L. (1992). Establishment and expression of cellular polarity in fucoid zygotes. Microbiol. Rev. 56: 316–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bail A., Billoud B., Kowalczyk N., Kowalczyk M., Gicquel M., Le Panse S., Stewart S., Scornet D., Cock J.M., Ljung K., Charrier B. (2010). Auxin metabolism and function in the multicellular brown alga Ectocarpus siliculosus. Plant Physiol. 153: 128–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bail A., Billoud B., Maisonneuve C., Peters A.F., Cock J.M., Charrier B. (2008a). Early development of the brown alga Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) sporophyte. J. Phycol. 44: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Le Bail A., Dittami S.M., de Franco P.O., Rousvoal S., Cock M.J., Tonon T., Charrier B. (2008b). Normalisation genes for expression analyses in the brown alga model Ectocarpus siliculosus. BMC Mol. Biol. 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kennard S., Lilly B. (2009). NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ. Res. 104: 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel G., Tonon T., Scornet D., Cock J.M., Kloareg B. (2010). The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 188: 82–97 [DOI] [PubMed] [Google Scholar]
- Nagasato C., Motomura T. (2009). Effect of latrunculin B and brefeldin A on cytokinesis in the brown alga Scytosiphon lomentaria (Scytosiphonales, Phaeophyceae). J. Phycol. 45: 404–441 [DOI] [PubMed] [Google Scholar]
- Peters A.F., Marie D., Scornet D., Kloareg B., Cock J.M. (2004). Proposal of Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) as a model organism for brown algal genetics and genomics. J. Phycol. 40: 1079–1088 [Google Scholar]
- Peters A.F., Scornet D., Ratin M., Charrier B., Monnier A., Merrien Y., Corre E., Coelho S.M., Cock J.M. (2008). Life-cycle-generation-specific developmental processes are modified in the immediate upright mutant of the brown alga Ectocarpus siliculosus. Development 135: 1503–1512 [DOI] [PubMed] [Google Scholar]
- Prince R.N., Schreiter E.R., Zou P., Wiley H.S., Ting A.Y., Lee R.T., Lauffenburger D.A. (2010). The heparin-binding domain of HB-EGF mediates localization to sites of cell-cell contact and prevents HB-EGF proteolytic release. J. Cell Sci. 123: 2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P., Lindenmayer A. (1996). The Algorithmic Beauty of Plants. (Berlin: Springer-Verlag; ). [Google Scholar]
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Ritter A., Ubertini M., Romac S., Gaillard F., Delage L., Mann A., Cock J.M., Tonon T., Correa J.A., Potin P. (2010). Copper stress proteomics highlights local adaptation of two strains of the model brown alga Ectocarpus siliculosus. Proteomics 10: 2074–2088 [DOI] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Starr R.C., Zeikus J.A. (1993). Utex - The culture collection of algae at the University of Texas at Austin. J. Phycol. 29: 1–106 [Google Scholar]
- Steinwand B.J., Kieber J.J. (2010). The role of receptor-like kinases in regulating cell wall function. Plant Physiol. 153: 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Yoon H.S., Hackett J.D., Ciniglia C., Pinto G., Bhattacharya D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21: 809–818 [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. (2001). InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yu J., Li D., Zhang Z., Liu F., Zhou X., Wang T., Ling Y., Su Z. (2010). PMRD: Plant microRNA database. Nucleic Acids Res. 38 (Database issue): D806–D813 [DOI] [PMC free article] [PubMed] [Google Scholar]