Abstract
Acylphosphatase can be converted in vitro, by addition of trifluoroethanol (TFE), into amyloid fibrils of the type observed in a range of human diseases. The propensity to form fibrils has been investigated for a series of mutants of acylphosphatase by monitoring the range of TFE concentrations that result in aggregation. We have found that the tendency to aggregate correlates inversely with the conformational stability of the native state of the protein in the different mutants. In accord with this, the most strongly destabilized acylphosphatase variant forms amyloid fibrils in aqueous solution in the absence of TFE. These results show that the aggregation process that leads to amyloid deposition takes place from an ensemble of denatured conformations under conditions in which non-covalent interactions are still favoured. These results support the hypothesis that the stability of the native state of globular proteins is a major factor preventing the in vivo conversion of natural proteins into amyloid fibrils under non-pathological conditions. They also suggest that stabilizing the native states of amyloidogenic proteins could aid prevention of amyloidotic diseases.
Keywords: acylphosphatase/amyloid fibrils/conformational stability/protein engineering/protein misfolding
Introduction
A number of debilitating human diseases are associated with the deposition in tissue of protein aggregates known as amyloid fibrils (Tan and Pepys, 1994; Kelly, 1996; Dobson, 1999; Lansbury, 1999; Perutz, 1999). These include Alzheimer's disease, Huntington's disease, type II diabetes, primary and secondary systemic amyloidosis, Parkinson's disease and the spongiform encephalopathies. Under these pathological conditions, proteins or peptides that are normally soluble undergo aggregation, either in their intact forms or as fragments, and generate stable insoluble fibrils that accumulate in a variety of organs including the brain, liver and spleen. Despite the variation encountered in the amino acid sequences and native structures of proteins associated with these diseases, structural studies have revealed that amyloid fibrils from different sources share a common ultrastructure (Sunde and Blake, 1997). Electron microscopy has suggested that all fibrils are formed from unbranched protofilaments 2–5 nm wide that associate laterally or twist together to form fibrils of larger width (Glenner et al., 1971; Merz et al., 1983; Bauer et al., 1995; Serpell et al., 1995; Goldsbury et al., 1997; Conway et al., 1998; Jiminez et al., 1999). X-ray fibre diffraction studies indicate a characteristic structure in which the polypeptide chains form β–strands oriented perpendicularly to the long axis of the fibril, resulting in β–sheets propagating in the direction of the protofilaments (reviewed by Sunde and Blake, 1997). Although the cause and effect relationship between fibril deposition and the onset of disease has long been debated, evidence is mounting that amyloid plaques, or in some cases the precursor aggregates that lead to them, are the cause of the degenerative processes that occur in patients that suffer from amyloid-related diseases (Kelly, 1996; Selkoe, 1996; Lansbury, 1999; Perutz, 1999). A fundamental understanding of the mechanisms of amyloid formation is therefore central in the development of therapeutic strategies aimed at preventing the formation of amyloid and combating amyloidotic diseases.
An important recent development has been the discovery that amyloid formation is not restricted to the small number of protein sequences associated with diseases (Dobson, 1999). In vitro experiments show that amyloid formation is a property common to polypeptide chains under a variety of partially denaturing conditions. Amyloid fibrils structurally similar to those extracted from patients or reproduced in vitro from disease-related proteins have been characterized in vitro, under a variety of conditions, for the SH3 domain of phosphatidylinositol 3-kinase (Guijarro et al., 1998; Jimenez et al., 1999), a type III domain of fibronectin (Litvinovich et al., 1998), muscle acylphosphatase (AcP) (Chiti et al., 1999a), and peptides isolated from the sequence of the cold shock protein B (Gross et al., 1999). None of these proteins is linked to any human amyloidotic disease. This has led to the suggestion that the ability to adopt the amyloid form of protein molecules is a general property of polypeptide chains regardless of the amino acid sequence (Guijarro et al., 1998; Chiti et al., 1999a; Dobson, 1999). Formation of amyloid by natural proteins not related to disease is promoted in vitro by partially denaturing conditions such as low pH (Guijarro et al., 1998), high temperature (Litvinovich et al., 1998) or in the presence of moderate concentrations of alcohols (Chiti et al., 1999a). That partially denaturing conditions are required for the in vitro formation of amyloid can be attributed to the necessity of destabilizing the native fold of a protein under conditions in which non-covalent interactions involving the polypeptide chain remain favourable (Chiti et al., 1999a). It is the existence of such interactions that stabilize the intermolecular β–sheet structure that is the basis of the highly ordered structure of amyloid fibrils.
Here we investigate the propensity to form amyloid of a series of variants of AcP, a protein of 98 residues, each having a single point mutation. AcP has recently been shown to form amyloid fibrils in the presence of moderate concentrations of trifluoroethanol (TFE) (Chiti et al., 1999a). The protein in its native conformation has an ‘open sandwich’ structure with two α–helices packed against a five stranded β–sheet (Saudek et al., 1989; Pastore et al., 1992). We show in the present study that fibrils of the type associated with amyloid diseases can be formed in the absence of any denaturant for variants of this protein, provided that the destabilization caused by the mutation is large enough to generate a significant population of denatured conformations under such conditions. In addition, the relationship between conformational stability and the potential to generate amyloid fibrils for this model protein has been compared with similar correlations for disease-related proteins.
Results
Conformational stability of AcP mutants
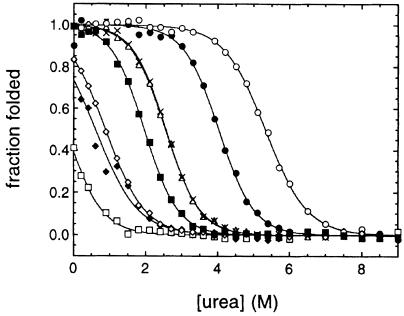
A series of mutants of AcP, involving single amino acid substitutions distributed throughout the sequence of the protein, was prepared previously to study the structure of the transition state for folding of this protein (Chiti et al., 1999b). Seven of these mutants (listed in Table I together with the location of the mutated residues in the sequence of the protein) were selected for their ability to display different conformational stabilities. The urea denaturation curves of wild-type and mutated forms of AcP are shown in Figure 1. Wild-type AcP folds in an apparent two-state fashion, that is intermediates are not significantly populated during the interconversion from the fully unfolded to the fully folded states (van Nuland et al., 1998). Urea denaturation of the various mutants analysed here showed in all cases the existence of a single transition between a fully folded and a fully unfolded state, demonstrating that a two-state approximation is also appropriate for these variants. Folding of AcP occurs cooperatively also in the presence of TFE, as a single transition has been observed upon TFE titration probed with either CD or intrinsic fluorescence (Chiti et al., 1999c). All mutants exhibit significant levels of enzymatic activity (Table I) suggesting that for these mutants the conformation in the absence of urea is globular with the topology characteristic of the wild-type protein. Analysis of the urea denaturation curves, performed as described by Santoro and Bolen (1988), allows the urea concentration at half-denaturation (i.e. the Cm value) and the free energy of unfolding in the absence of urea [i.e. the ΔG(H2O) value] to be determined for each variant (Table I). This analysis shows that the mutants studied here cover a wide range of conformational stabilities ranging from –1 to 23 kJ/mol. The Y11F mutant is more stable than the wild-type protein. The V51A, V47A and V13A mutants are slightly destabilized relative to the wild-type protein, and the M61A, F94L and L65V mutants are so substantially destabilized that they are partially denatured even in the absence of urea. The various mutants were tested for their propensity to aggregate in an in vitro model system based on TFE.
Table I. Conformational stabilities and enzymatic activities of AcP variants.
| AcP variant | Location of mutated residue in native structurea | Cm (M)b | ΔG(H2O) (kJ/mol)c | Specific activity (IU/mg)d |
|---|---|---|---|---|
| Wild-type | – | 4.02 ± 0.20 | 21.3 ± 1.2 | 4000 ± 400 |
| Y11F | β-strand 1 | 5.32 ± 0.20 | 23.1 ± 1.4 | 3700 ± 300 |
| V13A | β-strand 1 | 1.95 ± 0.20 | 10.3 ± 0.7 | 2500 ± 200 |
| V47A | β-strand 3 | 2.55 ± 0.20 | 13.5 ± 0.9 | 500 ± 50 |
| V51A | β-strand 3 | 2.54 ± 0.20 | 13.4 ± 0.8 | 3200 ± 300 |
| M61A | α-helix 2 | 0.88 ± 0.40 | 4.7 ± 1.1 | 2900 ± 300 |
| L65V | α-helix 2 | 0.21 ± 0.40 | –1.1 ± 1.0 | 3300 ± 300 |
| F94L | β-strand 5 | 0.61 ± 0.40 | 3.2 ± 1.1 | 2200 ± 200 |
aThe native structure of AcP has been determined by Saudek et al. (1989) and Pastore et al. (1992).
bUrea concentration at the mid-point of denaturation.
cFree energy of unfolding in the absence of denaturant.
dAcylphosphatase activity measured using benzoylphosphate as a substrate at 25°C at pH 5.5 and reported in international units per milligram of protein.
Fig. 1. Equilibrium denaturation curves for wild-type AcP and its mutational variants obtained by normalizing the raw data to the fraction of the protein molecules in the folded state, defined as (yobs – yu)/(yf – yu) where yf and yu are the intrinsic fluorescence signals of the native and denatured states, respectively, and yobs is the observed fluorescence intensity at the urea concentration under analysis. The AcP variants shown are Y11F (○), wild-type (•), V47A (×), V51A (▵), V13A (▪), M61A (⋄), F94L (♦) and L65V (□). The lines through the data represent the result of the fitting procedure used to estimate the parameters defining the conformational stability of each AcP variant (Santoro and Bolen, 1988).
Aggregation potential of the AcP mutants
It has recently been shown that wild-type AcP forms amyloid fibrils when incubated at a protein concentration of 0.375 mg/ml in the presence of 18–35% (vol/vol) TFE (Chiti et al., 1999a). Under these conditions amyloid formation is preceded by the formation of small protein aggregates within 2–3 h that develop very slowly to form, after a period of several days, amyloid protofilaments that associate further to form higher order structures (Chiti et al., 1999a). In order to investigate the propensity of the AcP mutants to aggregate, a number of samples containing different concentrations of TFE were prepared for each mutant at the same protein concentration. These were tested after an incubation period of 5 h for the presence of aggregated protein using far-UV circular dichroism spectroscopy (CD) and fluorescence in the presence of thioflavine T. After this time period, the aggregates are granular but their formation correlates well with the development of well defined amyloid fibrils at longer times (Chiti et al., 1999a). Moreover, these early aggregates possess an extensive β-sheet conformation and have the ability to bind specific dyes that are indicative of the presence of amyloid, suggesting that they possess even at this stage at least some elements of the ultrastructure characteristic of the fully developed fibrils (Chiti et al., 1999a). This procedure therefore enables the relatively rapid analysis of a large number of samples for the presence of aggregates when these are sufficiently small in size to detect using spectroscopic techniques.
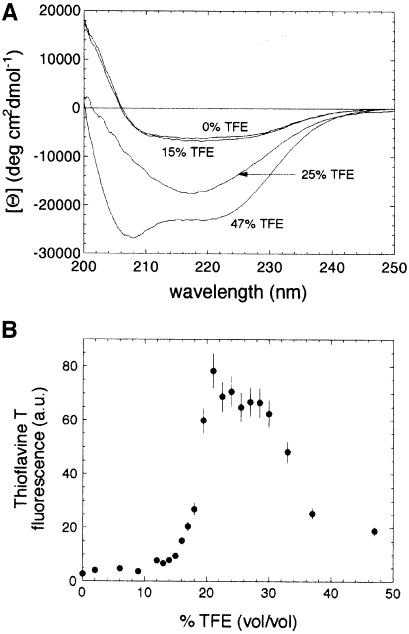
Figure 2A shows CD spectra acquired after 5 h for the wild-type protein at four representative TFE concentrations. The spectra of samples incubated at concentrations of TFE up to 19% (vol/vol) were virtually identical to each other and to that previously reported for the native protein in the absence of TFE (Chiti et al., 1998a, 1999c). At concentrations of TFE ranging from 19 to 35% (vol/vol) the CD spectra after this time period are typical of a largely β-sheet conformation as revealed by the single negative band at 215–220 nm. At concentrations of TFE higher than 35% (vol/vol) the CD spectrum exhibits two negative peaks at ∼208 and 222 nm, indicating that the protein has a largely α-helical conformation under these conditions. Only the samples incubated at TFE concentrations between 19 and 35% (vol/vol) produce the characteristic increase of thioflavine T fluorescence that is typical of ordered protein aggregates (Figure 2B). This therefore confirms that only within this range of TFE concentrations does the wild-type protein exist in a well-defined aggregated conformation after incubation for 5 h.
Fig. 2. (A) Far-UV CD spectra of wild-type AcP at various TFE concentrations. Representative spectra are shown for the native protein (obtained at TFE concentrations <19% vol/vol), aggregated protein (TFE concentrations of 19–35% vol/vol) and non-native protein in a monomeric conformation (TFE concentrations >35% vol/vol). In the latter case the high content of α–helical structure revealed by the CD spectrum is not that of a fully structured native conformation, but arises from the ability of TFE to generate this type of secondary structure in the denatured state. All spectra were acquired following 5 h of incubation at 25°C in 50 mM acetate buffer pH 5.5 at the TFE concentrations indicated. (B) 485 nm fluorescence (excitation 440 nm) of thioflavine T in the presence of wild-type AcP incubated for 5 h under the conditions described for (A). The changes in fluorescence with TFE concentration show that protein aggregates are present under these conditions at TFE concentrations of ∼19–35% (vol/vol).
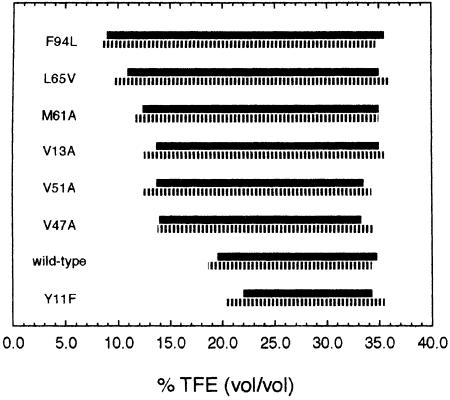
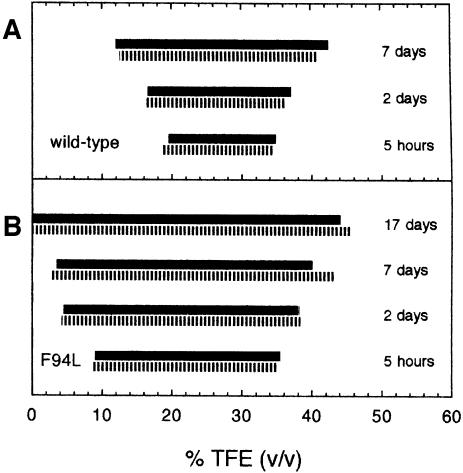
This analysis was repeated for each mutant of AcP and the ranges of TFE concentrations at which protein aggregates were observed in each case are shown in Figure 3. The boundaries of these intervals were defined by the TFE concentrations at which the protein produces ∼50% of the maximum observed increase in CD ellipticity at 215–220 nm and in thioflavine T fluorescence. As can be seen from Figure 3, the intervals determined using these two approaches are highly consistent, indicating that the conformational change observed by CD is attributable to formation of intermolecular β–sheet structures that are typical of amyloid fibrils or their precursors. This analysis shows that while the upper limit of the range of TFE concentrations at which the aggregates are present in the various mutants after 5 h is similar, within experimental error, to that for the wild-type protein, the lowest TFE concentration that is required for formation of protein aggregates is clearly different.
Fig. 3. Ranges of TFE concentration at which protein aggregates are present for each AcP variant after incubation at 25°C in 50 mM acetate buffer pH 5.5 and at protein concentrations of 0.375 mg/ml. Solid and hatched bars indicate results from far-UV CD spectroscopy and thioflavine T fluorescence, respectively. Experimental errors for both the lower and upper TFE concentrations at which aggregates occur are ∼1.5% (vol/vol).
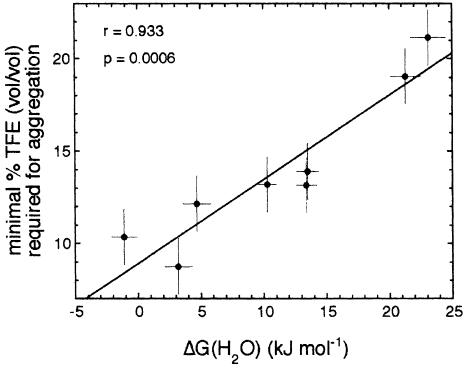
Figure 4 shows that there is a remarkable correlation between the minimum concentration of TFE that causes protein aggregation after 5 h and the free energy of unfolding. Values of 0.933 and 0.0006 are calculated for the linear correlation coefficient (r value) and the p value, respectively. This implies that the probability of obtaining an r value equal to or higher than that observed would be only 0.06% if the two plotted parameters were not correlated. The statistical analysis shows that the correlation remains significant when the observed values of TFE concentration and ΔG(H2O) are varied within their experimental errors. The fact that lower concentrations of TFE are required for the aggregation of destabilized AcP variants implies that less stable mutants form amyloid fibrils more readily than do the more stable ones.
Fig. 4. Correlation between the minimal TFE concentration required for producing aggregates after 5 h (taken as a measure of the amyloidogenic potential) and the ΔG(H2O) of unfolding (taken as an index of the conformational stability of the native protein). The high linear correlation coefficient (r value) of 0.933 and low p value of 0.0006 indicate that the correlation is highly significant. The best fitted equation has the form %TFE = (9 ± 1) + (0.46 ± 0.08) [ΔG(H2O)]. The values of TFE concentration on the y axis are an average of those determined with the thioflavine T and the CD data. The experimental errors are those indicated in the legend of Figure 3 for TFE concentration and in Table I for ΔG(H2O) values.
It is likely that high concentrations are not as effective as moderate concentrations of TFE in stimulating amyloid formation because the increasing content of α–helical structure within the denatured state at high TFE concentrations reduces the probability of the nucleation or growth of aggregates having largely β–sheet structure. It is also probable that high concentrations of TFE reduce the hydrophobic effect that is likely to be a significant factor in promoting the association of denatured molecules (Chiti et al., 1999a). As stated above, the upper limit of the range of TFE concentrations favouring aggregation is closely similar for the different mutants, i.e. it is independent of the conformational stability of the protein. This result can be rationalized since the protein is denatured under these conditions and the non-disruptive amino acid substitutions such as those investigated here are not expected to change substantially the overall solubility or helical propensity of the denatured state.
Figure 5 shows that the range of TFE concentrations at which aggregates are observed for AcP and its variance becomes larger with time. This indicates that the aggregation process occurs at TFE concentrations above and below those indicated in Figure 3, although at a slower rate. For wild-type AcP, for instance, TFE concentrations ranging from 19 to 35% (vol/vol) result in large quantities of aggregated protein after 5 h. At TFE concentrations of 12 and 41% the aggregation process is slower and only after ∼7 days can significant quantities of aggregates be detected. A similar situation holds for the F94L mutant of AcP that was examined as a representative case of a highly destabilized AcP variant (Figure 5B). Although small amounts of TFE facilitate rapid aggregation for this mutant, a very large quantity of aggregated protein develops after 17 days even in its absence.
Fig. 5. Change with time of the range of TFE concentrations within which aggregates are observed for the wild-type (A) and F94L variant (B) proteins. Solid and hatched bars indicate results from far-UV CD spectroscopy and thioflavine T fluorescence, respectively. Experimental conditions are as described in Figure 3. Experimental errors for both the lower and upper TFE concentrations at which aggregates occur are ∼1.5% (vol/vol).
Rate of aggregation
To assess whether the different propensities to aggregate for the various mutants of AcP arise from a specific effect of the mutated residues on the aggregation process, in addition to the effect on the stability of the native state, we studied the rate of aggregation in the presence of 25% TFE. At this TFE concentration AcP unfolds rapidly, on a time scale of seconds, and the aggregation process occurs subsequently and more slowly from monomeric conformations with extensive α–helical content (Chiti et al., 1999a). Starting from a denatured state of the protein, these kinetic experiments allow the process of aggregation to be studied independently of the contribution of conformational stability to the overall process.
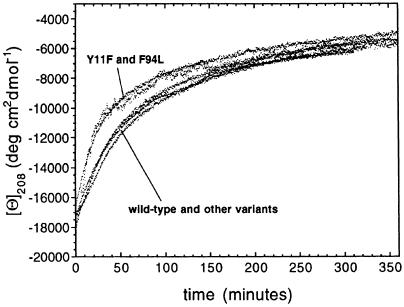
Figure 6 shows the change of the 208 nm mean residue ellipticity after mixing the various proteins in their native states with TFE to yield solutions of 25% (vol/vol) TFE. A rapid increase of the 208 nm CD signal resulting from the denaturation step occurs within the dead time of the manual mixing experiment and is not shown. This is followed by a slow decrease of the CD signal at this wavelength as a consequence of the ongoing aggregation. Seven kinetic traces obtained with the wild-type protein under identical conditions are largely superimposable, indicating that the results obtained from experiments of this type are fairly reproducible. It is evident that no significant relationship exists between the rate of aggregation and the amyloid-forming ability, monitored as described previously, within the set of AcP variants studied here. Indeed, mutants with very different propensities to aggregate appear to do so with very similar rates.
Fig. 6. Change of the mean residue ellipticity at 208 nm within the first 6 h of the aggregation process for AcP variants. The process shown in the figure is preceded by the conversion of the native protein into a denatured monomeric conformation with extensive α–helical structure (Chiti et al., 1999a). This occurs within the time required for manual mixing (∼10 s) and is not shown in the figure. The experiments were performed for all AcP variants at 25°C in 50 mM acetate buffer pH 5.5 and at protein and TFE concentrations of 0.375 mg/ml and 25% vol/vol, respectively.
The AcP variants have also been analysed following the change of thioflavin T fluorescence as the aggregates accumulate over time (data not shown). The kinetic traces obtained with this probe are also largely superimposable. No differences have been observed, in particular, between the Y11F and F94L mutants on the one hand and the wild-type protein and other AcP variants on the other hand. These results indicate that the small differences in the aggregation rate observed with CD spectroscopy for these two groups of AcP variants (Figure 6) are within experimental error and support further the idea that the substantive differences in the propensity for forming aggregates between these AcP variants have to be attributed to factors other than those associated with the intrinsic rate of aggregation from denatured conformations. We do not exclude, however, that specific mutations might exist in the AcP sequence that are able to alter the aggregation rate substantially. Indeed, it is reasonable to assume that there is a relatively small number of residues that play a critical role in the aggregation process of AcP.
Morphology of the protein aggregates by electron microscopy
It has been shown previously that aggregates formed from wild-type AcP in the presence of 25% (vol/vol) TFE are mixtures of amyloid fibrils and amorphous aggregates (Chiti et al., 1999a). Formation of ordered amyloid-like aggregates occurs preferentially at low protein concentrations; this can be attributed to the fact that at high concentrations of protein the aggregation process is too rapid to allow the development of highly ordered structures (Chiti et al., 1999a). The well defined fibrils formed in the TFE solutions appear to be largely isolated protofilaments or large bundles of protofilaments (Chiti et al., 1999a). The fact that the least stable mutants of AcP can aggregate in the absence of TFE provides an opportunity to examine the morphology of the AcP aggregates in aqueous solutions. Samples of the F94L mutant were therefore analysed using electron microscopy after incubation at room temperature for a period of 17 days.
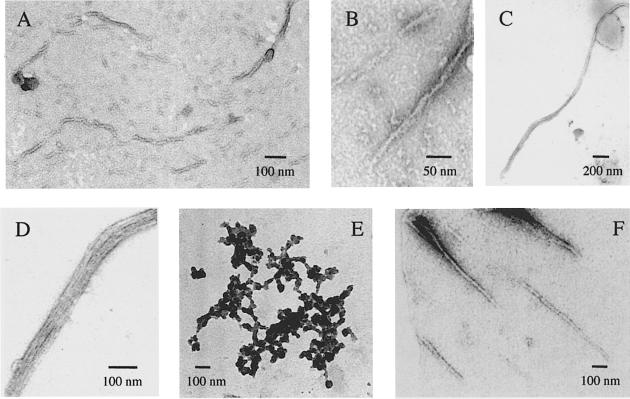
The aggregates obtained at protein concentrations >0.1 mg/ml appear predominantly amorphous in the electron micrographs. The preparations obtained with protein concentrations <0.1 mg/ml, however, can be seen to contain aggregates of a fibrillar nature (Figure 7A–D) in addition to amorphous aggregates (Figure 7E). Figure 7A and B shows fibrils with diameters of ∼13 nm formed from samples of F94L mutant incubated at 0.05 mg/ml in the absence of TFE. Although the ultrastructure of these fibrils is not very clear from these micrographs, some stretches of the fibrils can be seen to be composed of two or more protofilaments of smaller diameter, that associate laterally or twist together. Figure 7C shows a different morphology in which a bunch of protofilaments associates in a parallel manner to form rope-like structures with a diameter of ∼30–60 nm. High magnification images of this rope-like structure reveal that the protofilaments are straight and unbranched and possess widths of 2.5–4.0 nm (Figure 7D). The size and morphology of the protofilaments and fibrils observed here are typical of the amyloid fibrils extracted from patients suffering from amyloidotic diseases or reproduced in vitro with amyloid-related proteins (Shirahama and Cohen, 1967; Shirahama et al., 1973; Merz et al., 1983; Bauer et al., 1995; Serpell et al., 1995; Harper et al., 1997; Conway et al., 1998). Clusters of protofilaments of the type shown in Figure 7C and D can be generated in vitro with amyloid-related proteins such as calcitonin (Sletten et al., 1976).
Fig. 7. Electron micrographs showing the polymorphism of AcP aggregates formed at 25°C in 50 mM acetate buffer pH 5.5 at a protein concentration of 0.05 mg/ml and in the absence of TFE. (A and B) Aggregates formed from the highly destabilized F94L mutant of AcP showing fibrils ∼13 nm in width. (C and D) Group of protofilaments of the F94L mutant of AcP, ∼2.5–4.0 nm in width and associating laterally to form a 30–60 nm wide rope-like fibril. (E) Amorphous aggregates formed from the F94L mutant of AcP and present together with aggregates of fibrillar structure. (F) Aggregates formed from wild-type AcP at the relatively high concentration of 0.375 mg/ml (see text).
Samples of the wild-type protein, prepared as controls and incubated in the absence of TFE for 17 days in a manner similar to those of the F94L mutant, also showed the presence of a small number of amyloid fibrils (Figure 7F). However, fibrillar aggregates are apparent only in micrographs from samples containing the protein at concentrations >0.35 mg/ml. These samples retain ∼95% of the enzymatic activity of freshly prepared samples and their CD spectra are virtually identical to that of the native protein. These observations suggest that the fibrillar material revealed by electron microscopy represents the conversion into amyloid fibrils of a small amount of denatured protein present in the sample.
Discussion
Relationship between conformational stability and amyloidogenesis
The results presented here show that destabilization of the native conformation of AcP renders it more prone to amyloid formation. The ability of a highly destabilized mutant of AcP to assembly into amyloid fibrils in the absence of any denaturant, the necessity to use moderate concentrations of alcohol to generate amyloid fibrils from stable forms of AcP, and the significant correlation between the conformational stability of native AcP and the tendency to form aggregates, all indicate that amyloid fibrils originate from denatured conformations of the protein under conditions in which the formation of non-covalent interactions is still favourable.
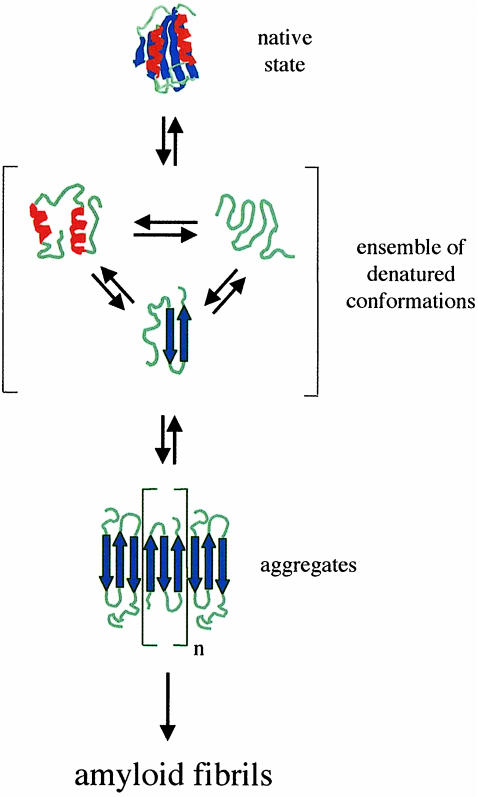
A simplified model that can be drawn to describe the relationship between conformational stability and amyloid formation for AcP is shown in Figure 8. The native conformation of AcP has little tendency to aggregate because interactions within the protein fold render the majority of hydrophobic side-chains, and the main-chain amide and carbonyl groups capable of forming strong hydrogen bonds, inaccessible to intermolecular interactions. The denatured state, by contrast, exposes such regions of the polypeptide chain providing an opportunity for intermolecular interactions to take place. The addition of moderate concentrations of TFE to highly stable variants of AcP, or the introduction of destabilizing mutations in the absence of denaturant, reduces the stability of the native fold without preventing the establishment of intermolecular contacts. In this respect, the effects exerted by a chemical denaturant such as TFE and by destabilizing mutations are similar. It is important to note, however, that the denatured monomeric species indicated in Figure 8 as a precursor of aggregated species is not necessarily a well defined conformation with β–sheet structure. Indeed, in the presence of TFE the denatured state of AcP from which the aggregates develop is an ensemble of conformations with a high helical content. A transient population with β–sheet structure will exist even in a denatured state where the ensemble is, on average, highly helical (Smith et al., 1996). Such a transient population of conformations in the denatured state ensemble can be sufficient to allow aggregation to take place (Fink, 1998).
Fig. 8. A simplified model that describes the conformations and equilibria involved in AcP amyloid formation in vitro. The native state is less prone to aggregation because intramolecular interactions within the protein fold render the majority of hydrophobic side-chains and the backbone amide and carbonyl groups unavailable for intermolecular interactions. In the ensemble of denatured conformations such regions of the polypeptide chain are exposed to the solvent enabling aggregation to take place. The highly ordered amyloid fibrils form following elongation and development of the initially formed aggregates. According to this model the addition of moderate concentrations of TFE or the introduction of destabilizing mutations shifts the equilibrium between the native state and the denatured ensemble towards the latter, thus favouring aggregation and amyloid formation.
Comparison with disease-related proteins
The influence of destabilizing mutations on the propensity to form amyloid fibrils has been investigated for several proteins associated with amyloidotic diseases. Clear relationships have been established between conformational stability, the tendency to form amyloid in vitro, and the age of onset of disease in patients, for a group of transthyretin variants associated with cases of familial amyloidotic polyneuropathy I (McCutchen et al., 1995). Mutations within the variable domain of the immuno– globulin light-chain, which are rare in healthy individuals but have been found to be responsible for the pathogenesis of light-chain amyloidosis, are also found to destabilize the native fold of the domain and to render it more prone to amyloid formation than the wild-type domain in in vitro model systems (Hurle et al., 1994). A correlation similar to that observed here for AcP between the free energy of unfolding and the tendency to form amyloid was also found with this protein for a number of naturally occurring amyloidogenic mutants (Hurle et al., 1994). Finally, the lysozyme variants isolated from the amyloid deposits of patients afflicted with hereditary non-neuropathic systemic amyloidosis have also been found to be unstable compared with wild-type lysozyme (Funahashi et al., 1996; Booth et al., 1997). In this case fibril formation occurs from an ensemble of denatured conformations whose average properties are reminiscent of the molten globule state (Booth et al., 1997). By contrast, in the case of transthyretin, fibril formation occurs under conditions in which a conformation with a substantial content of native-like structure, rather than an ensemble of conformations with a largely unstructured fold, is populated (Lai et al., 1996).
The observation that in all these cases the mutations have very little effect on the structure of the native conformation indicates that the enhanced in vitro and in vivo propensity to form amyloid of the protein variants is attributable to the destabilizing effect of the amino acid substitutions rather than to specific roles of the mutated residues in the aggregation process. The results described in this paper for a protein that is not linked to any of the known amyloidotic diseases, and involving the study not of known amyloidogenic mutants but of variants with mutations distributed through the sequence, provide strong support to the theory that conformational stability and propensity to form amyloid are highly correlated. In addition, they support the idea that the nature of the process by which a soluble protein converts into fibrillar aggregates, that has so far been investigated primarily with the small number of amino acid sequences associated with amyloid deposition in vivo, is likely to be common to natural proteins.
These arguments clearly do not apply to diseases where the amyloidogenic species is a peptide or protein that is unstructured under physiological conditions. In such cases alterations in the sequence of the polypeptide chain have effects on the propensity to aggregate for different reasons. Amyloidogenic mutations of α–synuclein, for instance, a protein linked to Parkinson's disease, do not change the conformational properties of the soluble protein significantly, which is highly unstructured under physiological conditions, but result in an increased rate of fibril growth (Conway et al., 1998). Similarly, variants of the activation domain of procarboxypeptidase A2, engineered in order to have a higher α–helical propensity, have been found to aggregate at a rate substantially lower than that of the wild-type protein under conditions where these variants are denatured (V.Villegas, J.Zurdo, V.V.Filimonov, F.X.Aviles, C.M.Dobson and L.Serrano, unpublished data). These observations suggest that in addition to altering the conformational stability of the native state, mutations can in some cases significantly influence the process of amyloid formation by changing the rate of aggregation of molecules in the denatured state. It is interesting in this regard that pathological mutations in the prion protein include a number that do not significantly destabilize the native fold (Swietnicki et al., 1998; Liemann and Glockshuber, 1999).
Implications for understanding and combating amyloidogenesis in vivo
A protein with a destabilized globular structure, the F94L mutant of AcP, has been converted into amyloid fibrils in the absence of denaturant and under conditions of pH that were previously found to be optimal for the stability and enzymatic activity of the wild-type protein (Chiti et al., 1998b). The amyloid fibrils formed under these conditions are similar to those formed by the wild-type protein under denaturing conditions and indeed to those formed by proteins associated with human amyloidotic diseases. This indicates that the effects of highly destabilizing mutations on conformational stability produce similar effects in vitro to destabilizing non-physiological conditions such as extremes of pH, high temperature or addition of alcohols. This provides clear evidence that destabilization of the native fold is a key feature of the ability to form amyloid, rather than the need to generate a specific alternative folded conformation.
One source of inspiration for the search for strategies aimed at combating amyloidosis in vivo is Nature itself. A protein such as the F94L mutant of AcP has a free energy of unfolding in water of ∼3.2 kJ/mol. Seventy per cent of the protein molecules are therefore in their native state under physiological conditions and the enzyme possesses considerable catalytic activity. A protein of this type would therefore probably be adequate for the implementation of the cellular functions associated with AcP. Nevertheless, a protein of such low stability would have a high tendency to aggregate as a consequence of the significant population of the protein in at least a partially unfolded state, although a more complex situation must exist in vivo due to the presence of chaperones that can bind specifically to such denatured conformations. This is likely to be a significant factor in the evolution of natural proteins whose conformational stabilities are generally higher than 10–15 kJ/mol. The mutations associated with hereditary amyloidosis are therefore likely to be predominantly linked with destabilization of the native state, at least in those cases where amyloid formation occurs from proteins that normally adopt a globular conformation. By inference, therefore, it is the stability and cooperativity of native proteins under physiological conditions that enable the avoidance of amyloid formation by natural proteins.
A number of approaches are being examined for the prevention of amyloid diseases (Wetzel, 1997; Baures et al., 1999; Perutz, 1999). These include strategies designed to inhibit formation of fibril nuclei, to inhibit fibril growth or to stimulate fibril degradation. Of particular interest, however, are those strategies directed at stabilizing the native state of the protein involved, as these have the potential to inhibit the formation of the precursor of aggregated species and hence to avoid the onset of disease. One system where this approach is particularly promising involves familial amyloid polyneuropathy I and senile systemic amyloidosis, through stabilization of the native tetrameric state of transthyretin by binding of analogues of the thyroid hormone (Miroy et al., 1996; Baures et al., 1999). Interestingly, similar strategies have also been proposed in the fight against diseases other than those involving deposition of amyloid. It has been suggested, for instance, that stabilization of p53, the protein regarded as the ‘guardian of genome’, can be an effective strategy against cancer (Bullock et al., 1997). The results presented here with a model protein provide support for approaches of this type and suggest that they could have widespread applicability in the prevention of amyloid formation and indeed of the aggregation or degradation of proteins more generally.
Materials and methods
Mutant production and purification
Site-directed mutagenesis experiments and purification of mutants of AcP were performed according to the procedures previously described (Taddei et al., 1996). DNA sequencing and electrospray mass spectrometry were used to ensure the presence of the desired mutation. Protein concentration was measured by UV absorption using an ɛ280 value of 1.42 ml⋅mg/cm. All proteins used in this study have cysteine at position 21 replaced by a serine residue in order to avoid complexities arising from a free cysteine residue (van Nuland et al., 1998). The C21S mutation does not alter significantly the stability or the folding properties of AcP (Modesti et al., 1996). For simplicity, the protein carrying only the C21S mutation is referred to as wild-type AcP and the mutants presented in this study, all carrying the C21S substitution beside the mutation of interest, are referred to as single point mutants.
Equilibrium denaturation experiments
Equilibrium urea denaturation curves for wild-type AcP and its mutants were obtained by measuring the intrinsic fluorescence of 25–30 equilibrated samples containing various concentrations of urea in 50 mM acetate buffer pH 5.5 at 28°C. A Perkin Elmer LS 50 B spectrofluorimeter with excitation and emission wavelengths of 280 and 335 nm, respectively, was used for these measurements. The data were analysed according to the method of Santoro and Bolen (1988) to yield the free energy of unfolding in the absence of denaturant [ΔG(H2O)], the dependence of ΔG on urea concentration (m value) and the urea concentration of half denaturation (Cm). For the mutants that were found to be partially denatured in the absence of urea (M61A, L65V and F94L) the fluorescence properties of the native protein and their dependence on urea concentration were assumed to be identical to those of the wild-type protein and were constrained in the fitting procedure.
Circular dichroism
Far-UV CD spectra were acquired at 25°C using a Jasco J-720 spectropolarimeter and cuvettes of 1 mm path length. For wild-type AcP and each mutant, 20–25 samples of protein at a concentration of 0.375 mg/ml (34 μM) were incubated at different TFE concentrations ranging from 0 to 45% (vol/vol) in acetate buffer pH 5.5 at 25°C. The far-UV CD spectrum of each sample was acquired after 5 h of incubation. For the wild-type protein and the F94L mutant the CD analysis was repeated after longer periods of incubation (2, 7 and 17 days). All mother solutions containing the protein mutants were centrifuged and their protein concentrations measured immediately before preparation of the samples. CD was also used to follow the time-course of the aggregation process at the early stages for all mutants. In this case the CD signal at 208 nm was followed after mixing the native protein with a TFE/acetate buffer mixture to yield a solution of 0.375 mg/ml protein in 25% (vol/vol) TFE, 50 mM acetate buffer pH 5.5 at 25°C.
Thioflavine T dye binding
Aliquots of the samples prepared as described in the previous paragraph for the CD measurements were also used to test for thioflavine T binding (LeVine III, 1995). A 133 μl sample was mixed with 867 μl of 25 μM thioflavine T in 25 mM phosphate buffer pH 6.0. The fluorescence was measured immediately after mixing using excitation and emission wavelengths of 440 and 485 nm, respectively.
Electron microscopy
Electron micrographs were acquired using a JEM 1010 transmission electron microscope at 80 kV excitation voltage. Samples of wild-type or F94L mutated AcP were incubated for 3 weeks at room temperature in 50 mM acetate buffer pH 5.5 at protein concentrations ranging from 0.05 to 5 mg/ml. A 3 μl sample of protein solution was placed on a formvar and carbon-coated grid. The sample was then negatively stained with 30 μl of 1% uranyl acetate and observed at a magnification of 12–30 000×.
Acknowledgments
Acknowledgements
We are grateful to Francesca Magherini, Cristina Capanni and Jesus Zurdo for their assistance with this work. We are also very grateful to Massimo Stefani for continuous encouragement and for useful discussions. We are very grateful for support from the Accademia Nazionale dei Lincei and the Consorzio Interuniversitario di Biotecnologie (F.C.), the European Community (G.R. and C.M.D.), the Howard Hughes Medical Institute (C.M.D.) and the Wellcome Trust (C.M.D.). The Oxford Centre for Molecular Sciences is supported by the Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council and the Medical Research Council. The Dipartimento di Scienze Biochimiche in Florence is supported by the Consiglio Nazionale delle Ricerche and the Ministero dell'Università e della Ricerca Scientifica e Tecnologica (target project: folding and misfolding delle proteine, fondi ex 40%).
References
- Bauer H.H., Aebi, U., Haner, M., Hermann, R., Muller, M., Arvinte, T. and Merkle, H.P. (1995) Architecture and polymorphism of fibrillar supramolecular assemblies produced by in vitro aggregation of human calcitonin. J. Struct. Biol., 115, 1–15. [DOI] [PubMed] [Google Scholar]
- Baures P.W., Oza, V.B., Peterson, S.A. and Kelly, J.W. (1999) Synthesis and evaluation of inhibitors of transthyretin amyloid formation based on the non-steroidal anti-inflammatory drug flufenamic acid. Bioorg. Med. Chem., 7, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Booth D.R. et al. (1997)Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature, 385, 787–793. [DOI] [PubMed] [Google Scholar]
- Bullock A.N., Henckel, J., DeDecker, B.S., Johnson, C.M., Nikolova, P.V., Proctor, M.R., Lane, D.P. and Fersht, A.R. (1997) Thermodynamic stability of wild-type and mutant p53 core domain. Proc. Natl Acad. Sci. USA, 94, 14338–14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Taddei, N., van Nuland, N.A.J., Magherini, F., Stefani, M., Ramponi, G. and Dobson, C.M. (1998a) Structural characterisation of the transition state for folding of muscle acylphosphatase. J. Mol. Biol., 283, 893–903. [DOI] [PubMed] [Google Scholar]
- Chiti F., van Nuland, N.A.J., Taddei, N., Magherini, F., Stefani, M., Ramponi, G. and Dobson, C.M. (1998b) Conformational stability of muscle acylphosphatase. The role of temperature, denaturant concentration and pH. Biochemistry, 37, 1447–1455. [DOI] [PubMed] [Google Scholar]
- Chiti F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G. and Dobson, C.M. (1999a) Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl Acad. Sci. USA, 96, 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Taddei, N., White, P.W., Bucciantini, M., Magherini, F., Stefani, M. and Dobson, C.M. (1999b) Mutational analysis of AcP suggests the importance of topology and contact order in protein folding. Nature Struct. Biol., 6, 1005–1010. [DOI] [PubMed] [Google Scholar]
- Chiti F., Taddei, N., Webster, P., Hamada, D., Fiaschi, T., Ramponi, G. and Dobson, C.M. (1999c) Acceleration of the folding of acylphosphatase by stabilisation of local secondary structure. Nature Struct. Biol., 6, 380–387. [DOI] [PubMed] [Google Scholar]
- Conway K.A., Harper, J.D. and Lansbury, P.T. (1998) Accelerated in vitro fibril formation by a mutant α–synuclein linked to early-onset Parkinson disease. Nature Med., 11, 1318–1320. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. (1999) Protein misfolding, evolution and disease. Trends Biochem. Sci., 9, 329–332. [DOI] [PubMed] [Google Scholar]
- Fink A.L. (1998) Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des., 3, R9–R23. [DOI] [PubMed] [Google Scholar]
- Funahashi J., Takano, K., Ogasahara, K., Yamagata, Y. and Yutani, K. (1996) The structure, stability and folding process of amyloidogenic mutant human lysozyme. J. Biochem., 120, 1216–1223. [DOI] [PubMed] [Google Scholar]
- Glenner G.G., Ein, D., Eanes, E.D., Bladen, H.A., Terry, W. and Page, D.L. (1971) Creation of ‘amyloid’ fibrils from Bence Jones proteins in vitro. Science, 174, 712–714. [DOI] [PubMed] [Google Scholar]
- Goldsbury C.S. et al. (1997)Polymorphic fibrillar assembly of human amylin. J. Struct. Biol., 119, 17–27. [DOI] [PubMed] [Google Scholar]
- Gross M., Wilkins, D.K., Pitkeathly, M.C., Chung, E.W., Higham, C., Clark, A. and Dobson, C.M. (1999) Formation of amyloid fibrils by peptides derived from the bacterial cold shock protein CspB. Protein Sci., 8, 1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro J.I., Sunde, M., Jones, J.A., Campbell, I.D. and Dobson, C.M. (1998) Amyloid fibril formation by an SH3 domain. Proc. Natl Acad. Sci. USA, 95, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.D., Lieber, C.M. and Lansbury, P.T. (1997) Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem. Biol., 4, 951–959. [DOI] [PubMed] [Google Scholar]
- Hurle M.R., Helms, L.R., Li, L., Chan, W. and Wetzel, R. (1994) A role for destabilising amino acid replacements in light chain amyloidosis. Proc. Natl Acad. Sci. USA, 91, 5446–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J.L., Guijarro, J.L., Orlova, E., Zurdo, J., Dobson, C.M., Sunde, M. and Saibil, H.R. (1999) Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J., 18, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.W. (1996) Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol., 6, 11–17. [DOI] [PubMed] [Google Scholar]
- Lai Z., Colon, W. and Kelly, J.W. (1996) The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry, 35, 6470–6482. [DOI] [PubMed] [Google Scholar]
- Lansbury P.T. (1999) Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc. Natl Acad. Sci. USA, 96, 3342–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine H. III (1995) Thioflavine T interaction with amyloid β-sheet structures. Amyloid: Int. J. Exp. Clin. Invest., 2, 1–6. [Google Scholar]
- Liemann S. and Glockshuber, R. (1999) Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry, 38, 3258–3267. [DOI] [PubMed] [Google Scholar]
- Litvinovich S.V., Brew, S.A., Aota, S., Akiyama, S.K., Haudenschild, C. and Ingham, K.C. (1998) Formation of amyloid-like fibrils by self-association of a partially unfolded fibronectin type III module. J. Mol. Biol., 280, 245–258. [DOI] [PubMed] [Google Scholar]
- McCutchen S.L., Lai, Z.H., Miroy, G.J., Kelly, J.W. and Colon, W. (1995) Comparison of lethal and nonlethal transthyrethin variants and their relationship to amyloid disease. Biochemistry, 34, 13527–13536. [DOI] [PubMed] [Google Scholar]
- Merz P.A., Wisniewski, H.M., Somerville, R.A., Bobin, S.A., Masters, C.L. and Iqbal, K. (1983) Ultrastructural morphology of amyloid fibrils from neuritic and amyloid plaques. Acta Neuropathol., 60, 113–124. [DOI] [PubMed] [Google Scholar]
- Miroy G.J., Lai, Z.H., Lashuel, H.A., Peterson, S.A., Strang, C. and Kelly, J.W. (1996) Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc. Natl Acad. Sci. USA, 93, 15051–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti A., Taddei, N., Chiti, F., Bucciantini, M., Magherini, F., Rigacci, S., Stefani, M., Raugei, G. and Ramponi, G. (1996) Properties of Cys21-mutated muscle acylphosphatases. J. Protein Chem., 15, 27–34. [DOI] [PubMed] [Google Scholar]
- Pastore A., Saudek, V., Ramponi, G. and Williams, R.J.P. (1992) Three-dimensional structure of acylphosphatase. Refinement and structure analysis. J. Mol. Biol., 224, 427–440. [DOI] [PubMed] [Google Scholar]
- Perutz M.F. (1999) Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem. Sci., 24, 58–63. [DOI] [PubMed] [Google Scholar]
- Santoro M.M. and Bolen, D.W. (1988) Unfolding free-energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry, 27, 8063–8068. [DOI] [PubMed] [Google Scholar]
- Saudek V., Boyd, J., Williams, R.J.P., Stefani, M. and Ramponi, G. (1989) The sequence-specific assignment of the 1H NMR spectrum of an enzyme, horse muscle acylphosphatase. Eur. J. Biochem., 182, 85–93. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J. (1996) Amyloid β-protein and the genetics of Alzheimer's disease. J. Biol. Chem., 271, 18295–18298. [DOI] [PubMed] [Google Scholar]
- Serpell L.C., Sunde, M., Fraser, P.E., Luther, P.K., Morris, E.P., Sangren, O., Lundgren, E. and Blake, C.C.F. (1995) Examination of the structure of the transthyretin amyloid fibril by image reconstruction from electron micrographs. J. Mol. Biol., 254, 113–118. [DOI] [PubMed] [Google Scholar]
- Shirahama T. and Cohen, A.S. (1967) High resolution electron microscopic analysis of the amyloid fibril. J. Cell Biol., 33, 679–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama T., Benson, M.D., Cohen, A.S. and Tanaka, A. (1973) Fibrillar assemblage of variable segments of immunoglobulin light chains: an electron microscopy study. J. Immunol., 110, 21–30. [PubMed] [Google Scholar]
- Sletten K., Westermark, P. and Natvig, J.B. (1976) Characterisation of amyloid fibril proteins from medullary carcinoma of the thyroid. J. Exp. Med., 143, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.J., Fiebig, K.M., Schwalbe, H. and Dobson, C.M. (1996) The concept of a random coil—Residual structure in peptides and denatured proteins. Fold. Des., 1, R95–R106. [DOI] [PubMed] [Google Scholar]
- Sunde M. and Blake, C. (1997) The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Protein Chem., 50, 123–159. [DOI] [PubMed] [Google Scholar]
- Swietnicki W., Petersen, R.B., Gambetti, P. and Surewicz, W.K. (1998) Familial mutations and the thermodynamic stability of the recombinant human prion protein. J. Biol. Chem., 273, 31048–31052. [DOI] [PubMed] [Google Scholar]
- Taddei N., Stefani, M., Magherini, F., Chiti, F., Modesti, A., Raugei, G. and Ramponi, G. (1996) Looking for residues involved in muscle acylphosphatase catalytic mechanism and structural stabilization: the role of Asn41, Thr42 and Thr46. Biochemistry, 35, 7077–7083. [DOI] [PubMed] [Google Scholar]
- Tan S.Y. and Pepys, M.B. (1994) Amyloidosis. Histopathology, 25, 403–414. [DOI] [PubMed] [Google Scholar]
- van Nuland N.A.J., Chiti, F., Taddei, N., Raugei, G., Ramponi, G. and Dobson, C.M. (1998) Slow folding of muscle acylphosphatase in the absence of intermediates. J. Mol. Biol., 283, 883–891. [DOI] [PubMed] [Google Scholar]
- Wetzel R. (1997) Inhibition of amyloid assembly. J. Neurochem., 69, S104. [Google Scholar]