Abstract
We describe an in situ technique for studying the chromatin binding of proteins in the fission yeast Schizosaccharomyces pombe. After tagging the protein of interest with green fluorescent protein (GFP), chromatin-associated protein is detected by GFP fluorescence following cell permeabilization and washing with a non-ionic detergent. Cell morphology and nuclear structure are preserved in this procedure, allowing structures such as the mitotic spindle to be detected by indirect immunofluorescence. Cell cycle changes in the chromatin association of proteins can therefore be determined from individual cells in asynchronous cultures. We have applied this method to the DNA replication factor mcm4/cdc21, and find that chromatin association occurs during anaphase B, significantly earlier than is the case in budding yeast. Binding of mcm4 to chromatin requires orc1 and cdc18 (homologous to Cdc6 in budding yeast). Release of mcm4 from chromatin occurs during S phase and requires DNA replication. Upon overexpressing cdc18, we show that mcm4 is required for re-replication of the genome in the absence of mitosis and is associated with chromatin in cells undergoing re-replication.
Keywords: cdc18/cell cycle/DNA replication/mcm proteins/ORC
Introduction
Many proteins involved in chromosome replication and segregation associate periodically with chromatin during the cell cycle. Regulated chromatin binding of such proteins is important for ensuring that the genome is replicated just once per cell cycle and that sister chromatids are segregated faithfully to daughter cells during mitosis. The multiple origins from which DNA replication initiates in eukaryotic chromosomes are important sites for the periodic binding of replication factors. In budding and fission yeasts, these origins are bound throughout the cell cycle by the origin recognition complex (ORC) (Diffley et al., 1994; Aparicio et al., 1997; Donovan et al., 1997; Tanaka et al., 1997; Lygerou and Nurse, 1999; Ogawa et al., 1999) but, early in the cell cycle, additional proteins bind at origins to form pre-replicative complexes (pre-RCs). Pre-RC formation establishes replication competence for the subsequent S phase, and this step in DNA replication has been particularly well characterized in Saccharomyces cerevisiae (for reviews, see Diffley, 1996; Stillman, 1996). The assembly of pre-RCs requires Cdc6 and involves the assembly of six minichromosome maintenance (MCM) proteins around origins (Cocker et al., 1996; Aparicio et al., 1997; Donovan et al., 1997; Tanaka et al., 1997). MCM proteins, which have been shown to have limited DNA helicase activity in vitro (Ishimi, 1997) and may move with replication forks (Aparicio et al., 1997), are displaced as S phase proceeds (for reviews, see Kearsey and Labib, 1998; Tye, 1999). The re-formation of pre-RCs is then blocked by cyclin-dependent kinase activity until late mitosis (Dahmann et al., 1995; Piatti et al., 1996; Tanaka et al., 1997).
The central features of pre-RC assembly in budding yeast are likely to be conserved in other eukaryotes, since studies in Xenopus have also shown that replication licensing involves both ORC and Cdc6-dependent loading of MCM proteins onto chromatin (Coleman et al., 1996; Romanowski et al., 1996; Rowles et al., 1996). In fission yeast, this process has not been examined in detail, although recently mcm6 was shown to associate with replication origins only during the G1 and S phases of the cell cycle (Ogawa et al., 1999). Schizosaccharomyces pombe shows certain differences compared with S.cerevisiae in terms of having a larger, more complex replication origin structure (Clyne and Kelly, 1995; Dubey et al., 1996; Okuno et al., 1999) and also in the specificity of ORC–DNA interactions (Chuang and Kelly, 1999; Moon et al., 1999). It will be interesting to determine whether these differences reflect general similarities between origin function in fission yeast and higher eukaryotes.
To examine steps leading to DNA replication initiation in S.pombe, we have developed a novel assay for the chromatin association of fission yeast proteins. Previously described methods involve: (i) partial purification of chromatin and analysis of associated proteins by immuno– blotting (Donovan et al., 1997; Liang and Stillman, 1997; Lygerou and Nurse, 1999; Ogawa et al., 1999); (ii) analysis by indirect immunofluorescence of chromatin-associated proteins in ‘chromosome spreads’ after cell lysis and loss of cell structure (Tanaka et al., 1997; Ogawa et al., 1999); and (iii) chromatin immunoprecipitation analysis of DNA sequences cross-linked to chromatin-bound proteins by formaldehyde (Aparicio et al., 1997; Tanaka et al., 1997; Ogawa et al., 1999). These methods require the use of synchronous cultures, and the degree of synchrony that can be achieved limits the precision with which changes in chromatin binding of proteins can be correlated with particular stages of the cell cycle. Also, experimental artefacts can be introduced by synchronization, depending upon the method used. The method described here preserves cell morphology, so that changes in the chromatin association of proteins can be correlated precisely with the stage of the cell cycle in individual cells from asynchronous cultures. This method is potentially useful for the analysis of any protein that is associated periodically with chromatin. We have used this approach to analyse the regulation of pre-RC formation in fission yeast by studying the mcm4 protein. We show that mcm4 binds to chromatin during anaphase B, and is displaced as DNA replication proceeds in the subsequent S phase. Chromatin association of mcm4 is dependent on both an ORC component and on cdc18. Our results imply that the mechanism of pre-RC formation in fission yeast is similar to that in other eukaryotes, although pre-RC assembly occurs significantly earlier in mitosis than in budding yeast and in mammalian cells.
Results
In situ chromatin binding assay
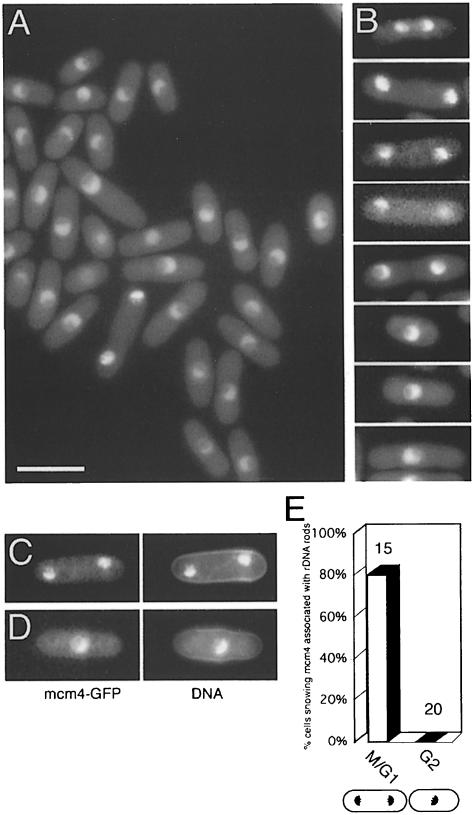
To study the controls regulating the initiation of DNA replication in fission yeast, we developed a simple cyto– logical assay to allow the chromatin binding of mcm4 to be monitored in individual cells. The assay is based on detection of mcm4–GFP fluorescence in permeabilized cells, after extraction with a non-ionic detergent. We first modified the cdc21+/mcm4+gene so that GFP is fused to the C–terminus of mcm4. mcm4–GFP is expressed from the native promoter as the only copy in the cell, and is functional at all temperatures normally permissive for fission yeast. mcm4 remains nuclear throughout the cell cycle (Figure 1A), confirming earlier results obtained using indirect immunofluorescence of fixed cells (Maiorano et al., 1996). Nuclear localization of mcm4 requires functional mcm2 and mcm6 (data not shown), and similar results, based on indirect immunofluorescence, have been reported recently (Pasion and Forsburg, 1999). These observations are consistent with data showing that MCM proteins exist as heterohexameric complexes (Adachi et al., 1997) and suggest that functional interactions between MCMs are needed for accumulation of mcm4 in the nucleus. Careful examination of live cells at different stages of the cell cycle shows subtle changes in the subnuclear localization of mcm4 (Figure 1B–D) that were not apparent in earlier studies of fixed cells (Maiorano et al., 1996). In binucleate unseptated (G1 phase) cells, the pattern of mcm4–GFP fluorescence is compact, and adopts the characteristic hemispherical (Martian) shape shown by 4′,6-diamidino-2-phenylindole (DAPI) staining of chromatin (Toda et al., 1981). This includes localization to the rDNA rods that protrude into the nucleolus (Uzawa and Yanagida, 1992) (Figure 1C and E), while such localization was not seen in uninucleate G2 phase cells (Figure 1D and E).
Fig. 1. mcm4 localization in live cells. (A) Mcm4–GFP in strain P560. Bar = 10 μm. (B) mcm4–GFP at different stages of the cell cycle (strain P560). (C and D) mcm4–GFP and DNA (DAPI) images of representative (C) binucleate unseptated (G1 phase) and (D) uninucleate (G2 phase) cells. (E) Percentage of cells showing mcm4 localization to rDNA nucleolar protrusions in binucleate unseptated and uninucleate cells (numbers of cells scored shown above the bars). Cells were only scored if rDNA nucleolar protrusions were visible by DAPI staining.
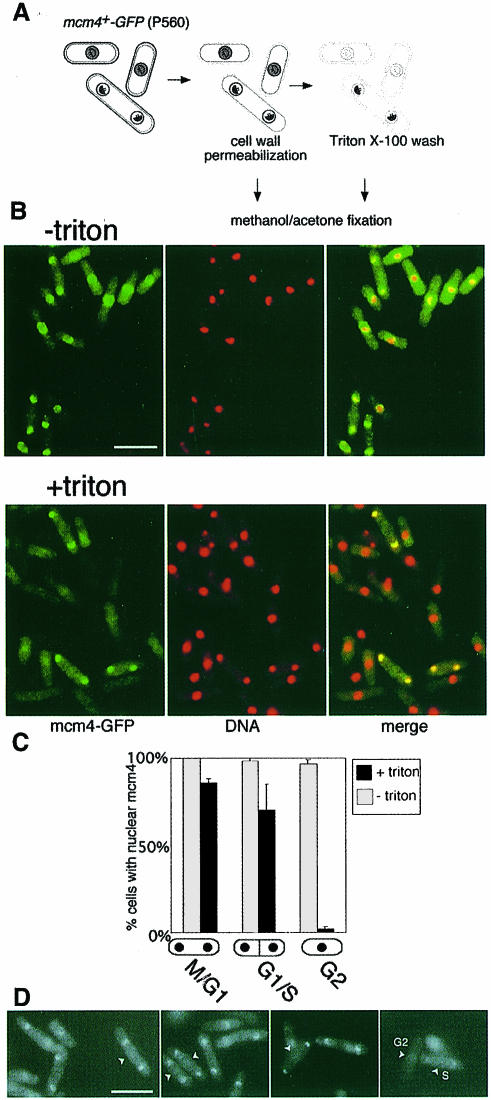
To determine whether these changes in the subnuclear distribution of mcm4 in wild-type cells reflect periodic changes in chromatin binding, we subjected cells to partial digestion of the cell wall, followed by washing with a Triton X-100-containing buffer before fixation (Figure 2A). We first examined an asynchronous population of wild-type cells. In permeabilized cells that have not been detergent washed, mcm4 is nuclear throughout the cell cycle (Figure 2B, –triton), as in live samples. Following detergent extraction, mcm4 nuclear localization is lost in uninucleate G2 cells, while binucleate (G1/S phase) cells predominantly retain nuclear mcm4 (Figure 2B, +triton). We note that in unseptated binucleate cells (predominantly in G1), there is no apparent reduction in mcm4–GFP fluorescence intensity after detergent extraction, indicating that most nuclear mcm4 is resistant to detergent extraction before S phase (Figure 2D).
Fig. 2. Chromatin association of mcm4 is periodic during the fission yeast cell cycle. (A) Assay procedure; for details see Materials and methods. Grey shading represents unbound, and black shading represents chromatin-bound mcm4. (B) mcm4–GFP localization (green) and DNA staining (DAPI, red) determined by fluorescence microscopy. In merged images, mcm4-positive nuclei appear yellow. Bar = 10 μm. (C) Proportion of binucleate unseptated, binucleate septated and uninucleate cells with mcm4-positive nuclei before and after extraction with a Triton X-100-containing buffer. (D) Triton-extracted and non-extracted cells were mixed, after labelling of the cell wall of the non-extracted cells with Texas red GS-1 lectin. In this way, the mcm4–GFP signal in both extracted and non-extracted cells can be compared under identical conditions in a single field of view, since the non-extracted cells can be identified by detection of Texas red fluorescence (these cells are marked by arrows). mcm4–GFP intensity, in binucleate unseptated cells, is similar before and after extraction, suggesting that the majority of nuclear mcm4 is chromatin bound in G1 phase. A proportion of binucleate septated cells are fainter than unextracted cells, e.g. cell ‘S’, presumably reflecting partial mcm4 chromatin displacement in mid-S phase; G2 cells are negative, as expected, e.g. cell ‘G2’.
The previous experiment suggests that mcm4 becomes sensitive to detergent extraction in permeabilized cells as cytokinesis is completed, which also corresponds to the time that S phase is executed. To test more directly whether DNA replication is required for mcm4 to become sensitive to detergent extraction, we arrested cells in early S phase with hydroxyurea, which inhibits ribonucleotide reductase (Figure 3). After 2 h in hydroxyurea, a high proportion of cells show a 1C DNA content, and >80% of uninucleate cells are positive for nuclear mcm4 after detergent extraction. As cells leak through the block (4 h, Figure 3B), the proportion of mcm4-positive nuclei drops and some nuclei show heterogeneous retention of mcm4. Digestion of hydroxyurea-arrested cells with DNase I before fixation eliminates mcm4 nuclear retention (Figure 3E and F). Taken together, these experiments show that mcm4 is associated periodically with chromatin during the fission yeast cell cycle, and replication of DNA is required for its release from chromatin.
Fig. 3. Displacement of mcm4 from chromatin requires progression through S phase or DNase I digestion. (A) Experimental procedure. (B) mcm4–GFP chromatin association and DNA staining (DAPI) were determined by fluorescence microscopy during a time course after addition of hydroxyurea. Bar = 10 μm. (C) Proportion of uninucleate cells with mcm4-positive nuclei before and after extraction with a Triton X-100-containing buffer. (D) Flow cytometric analysis of DNA contents of the cells shown in (B). (E) Cells from the ‘2h + hydroxyurea’ time point were digested with DNase I (for details, see Materials and methods). mcm4–GFP localization (left) and DNA staining (DAPI, right) were determined by fluorescence microscopy after Triton extraction. Bar = 10 μm. (F) Proportion of Triton-extracted cells with mcm4-positive nuclei with and without digestion with DNase I.
mcm4 binds to chromatin during anaphase B
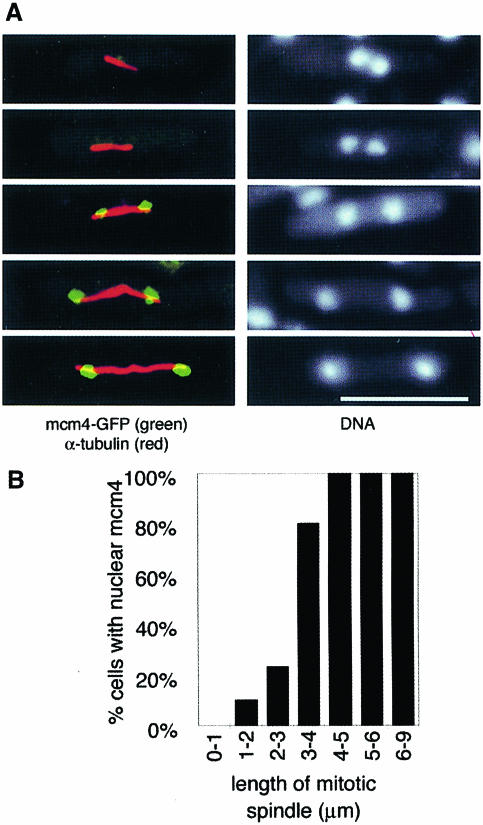
Studies from budding yeast and mammalian cells suggest that pre-RC formation occurs as cells complete mitosis, but in S.pombe this would only provide a brief interval for pre-RC formation, as the G1 phase is normally very short. Therefore, we considered the possibility that pre-RC formation may occur during mitosis. Using DAPI staining alone, as in Figure 2, it is difficult to distinguish cells in late mitosis from those in G1. Therefore, after the permeabilization and detergent extraction steps of the standard assay, cells were stained with an anti-tubulin antibody to reveal mitotic spindles (Figure 4A). Cells with short spindles (<3 μm) in metaphase, anaphase A or early stages of anaphase B are largely negative for mcm4, whereas cells in anaphase B with spindles longer than 3 μm are largely positive for mcm4 (Figure 4B). We did not observe cells where only one of the two segregating nuclei in an anaphase cell was positive for mcm4, suggesting that the association of mcm4 with chromatin is a synchronous event during nuclear division. Thus, the binding of mcm4 to chromatin occurs in mid-anaphase B. This suggests that pre-RC formation occurs significantly earlier during mitosis than is the case in budding yeast, where nuclear exclusion of MCM proteins is maintained until the end of anaphase, consequently delaying pre-RC formation until the end of mitosis (Hennessy et al., 1990; Yan et al., 1993; Labib et al., 1999).
Fig. 4. mcm4 binds to chromatin during anaphase B. Cells from an asynchronous culture (P560) in YE were processed using the in situ chromatin binding procedure, after which cells were stained with anti-α-tubulin antibody. (A) The left images indicate cells in different stages of anaphase B, showing mitotic spindles (red) and chromatin-bound mcm4 (green). Bar = 10 μm. The right images show the corresponding DNA staining (DAPI). (B) Proportion of mitotic cells with mcm4-positive nuclei shown according to mitotic spindle length. On average, 12 cells were scored for each length class (range 5–20 cells).
mcm4 chromatin binding requires ORC and cdc18
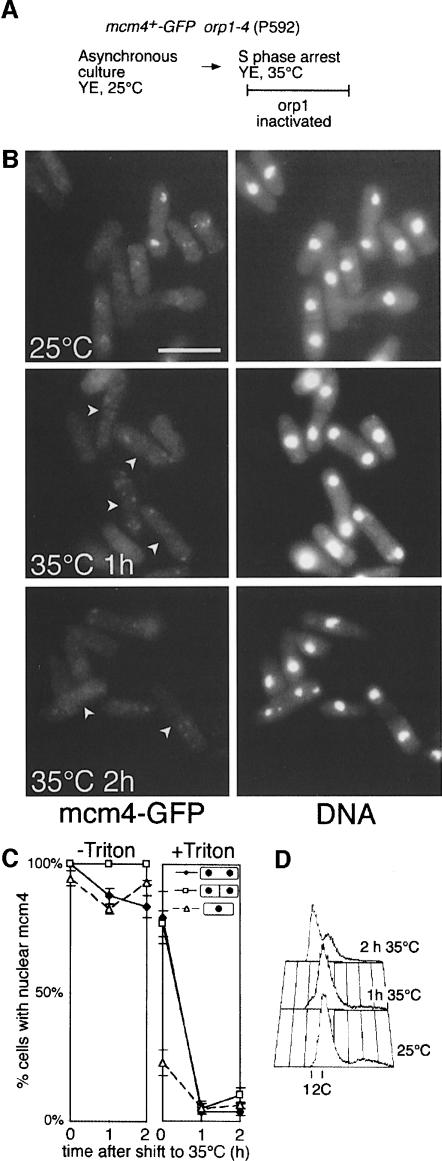
In S.cerevisiae and Xenopus, the ORC complex has been shown to be necessary for the Cdc6-mediated association of MCM proteins with chromatin (Rowles et al., 1996; Aparicio et al., 1997; Tanaka et al., 1997). Cdc6 binds to origins in an ORC-dependent manner and is required for MCM loading (Cocker et al., 1996; Coleman et al., 1996; Donovan et al., 1997). To determine whether binding of mcm4 to chromatin in fission yeast is also dependent on ORC and the cdc18 homologue of Cdc6, we used strains in which either ORC or cdc18 could be conditionally inactivated. We first examined mcm4 chromatin binding in a strain carrying a temperature-sensitive allele of the orc1 gene (Grallert and Nurse, 1996) (Figure 5A). Although shifting an asynchronous culture to the restrictive temperature had no effect on the nuclear localization of mcm4 in cells that had not been detergent extracted (Figure 5C, –Triton), there was a striking loss of chromatin binding in binucleate cells (Figure 5B and C, +Triton). This loss of mcm4 chromatin binding could be detected 1 h after shifting to 35°C, and therefore preceded the appearance of cells with a 1C DNA content, which were subsequently produced as cytokinesis was completed in the absence of DNA replication (Figure 5D). No effect on mcm4 chromatin binding was seen in wild-type cells after the same temperature shift (data not shown).
Fig. 5. mcm4 chromatin binding requires ORC function. (A) Experimental procedure. (B) mcm4–GFP chromatin association and DNA staining (DAPI) were determined by fluorescence microscopy during a time course after shifting the culture to the non-permissive temperature. Arrows on 35°C GFP panels indicate binucleate cells. Bar = 10 μm. (C) Proportion of binucleate unseptated, binucleate septated and uninucleate cells with mcm4-positive nuclei before and after extraction with a Triton X-100-containing buffer. (D) Flow cytometric analysis of DNA contents of the cells shown in (B).
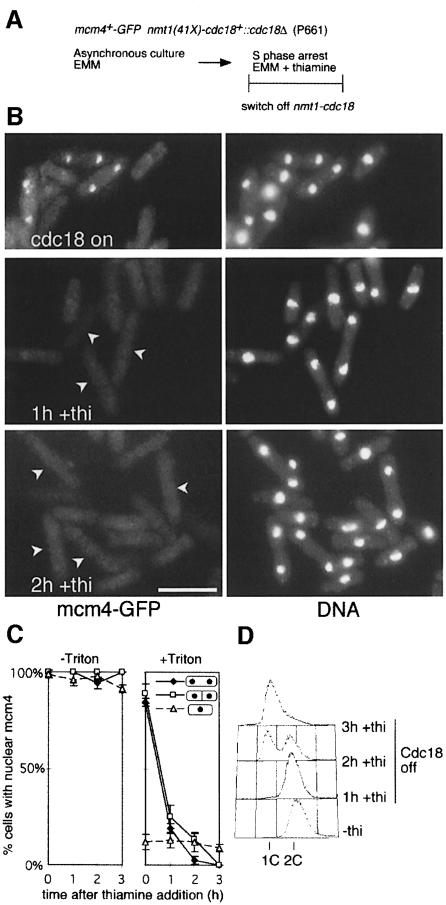
In a similar experiment, we examined the requirement for cdc18 in mcm4 chromatin binding, using a strain where cdc18 expression is regulated by a weak version of the thiamine-repressible nmt1 promoter (Muzi-Falconi et al., 1996). cdc18 expression was repressed in an asynchronous culture, and effects on mcm4 chromatin binding and DNA replication were followed over a 3 h time course (Figure 6A). As seen with inactivation of orc1, cdc18 shut-off had no effect on the nuclear localization of mcm4 in cells that had not been detergent extracted (Figure 6C, –Triton), but prevented chromatin binding during late mitosis (Figure 6B and C, +Triton). One hour after thiamine addition, binucleate cells were largely negative for mcm4 after detergent extraction, and again this change preceded the appearance of cells with a 1C DNA content. These experiments show that mcm4 chromatin association during anaphase is dependent on orc1 and cdc18, implying that pre-RC formation in fission yeast occurs by a mechanism similar to that operating in S.cerevisiae and Xenopus. In addition, these results indicate that the block to DNA replication in the absence of orc1 or cdc18 results from a failure of MCM proteins to associate with chromatin. These results are consistent with recent results from a conventional chromatin binding assay, where mcm6 chromatin binding was shown to be blocked in a cdc10 mutant at the restrictive temperature, presumably due to reduced expression of the cdc18+ gene (Ogawa et al., 1999).
Fig. 6. cdc18 is essential for chromatin association of mcm4. (A) Experimental procedure. (B) mcm4–GFP chromatin association and DNA staining (DAPI) were determined by fluorescence microscopy during a time course after addition of thiamine. Arrows on +thiamine GFP panels indicate binucleate cells. Bar = 10 μm. (C) Proportion of binucleate unseptated, binucleate septated and uninucleate cells with mcm4-positive nuclei before and after extraction with a Triton X-100-containing buffer. (D) Flow cytometric analysis of DNA contents of the cells shown in (B).
mcm4 is required for cdc18-induced re-replication and is associated with chromatin in re-replicating cells
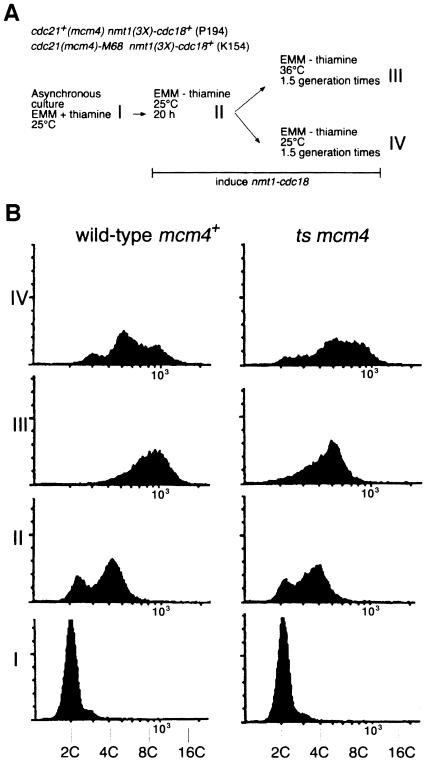
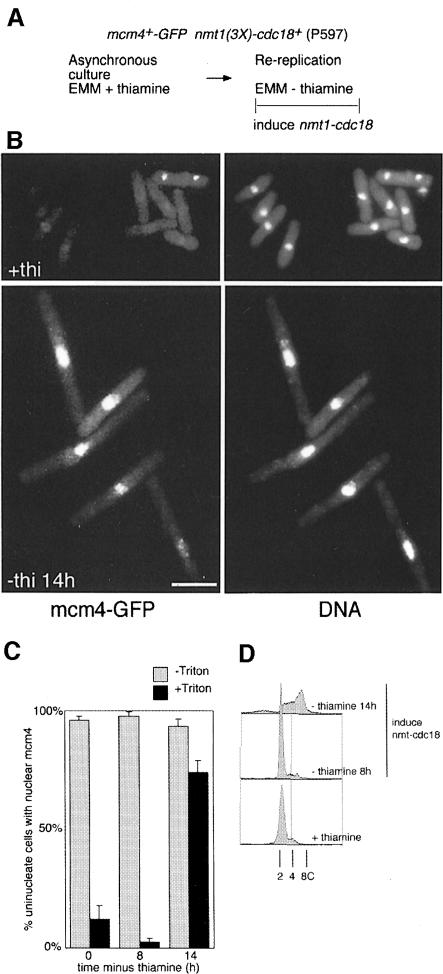
Overexpression of cdc18 in fission yeast results in dramatic re-replication of DNA, emphasizing the important role of this protein in replication control in this organism (Nishitani and Nurse, 1995; Muzi-Falconi et al., 1996). Since cdc18 is required for chromatin association of mcm4, we examined whether cdc18 overexpression causes chromatin association of mcm4 during re-replication. We first determined whether mcm4 is required for cdc18-induced re-replication (Figure 7). Strains carrying either the wild-type cdc21+/mcm4+ gene or the temperature-sensitive cdc21-M68 allele were grown at 25°C before induction of high levels of cdc18+ expression from the nmt1 promoter (Figure 7A). Once cells had started to re-replicate their DNA, the cultures were divided and one half was shifted to the non-permissive temperature, while the other was maintained at 25°C. In the strain with wild-type cdc21+/mcm4+, overexpression of cdc18+ at either temperature produced cells with a DNA content between 8C and 16C by the end of the experiment (Figure 7B, III and IV). In contrast, although re-replication at 25°C in the cdc21-M68 strain proceeded as in wild-type cells (Figure 7B, IV), inactivation of mcm4 at 36°C inhibited further re-replication (Figure 7B, III). Thus, mcm4 is required for cdc18-induced re-replication, which is consistent with MCMs acting after the function of cdc18 in the initiation of DNA replication. As shown in Figure 8, induction of nmt1-cdc18+ expression caused chromatin binding of mcm4 in uninucleate cells that are undergoing re-replication. It therefore appears that overexpression of cdc18+ is sufficient to effect chromatin association of mcm4 in uninucleate cells that consequently undergo re-replication, just as expression of cdc18+ at wild-type levels is necessary for the association of mcm4 with chromatin during anaphase.
Fig. 7. mcm4 is required for cdc18-induced re-replication. (A) Experimental procedure, see text for further details. (B) Flow cytometric analysis of DNA contents at different stages of the experiment shown in (A). DNA content is shown on a log scale.
Fig. 8. Overexpression of cdc18 causes mcm4 chromatin binding and re-replication. (A) Experimental procedure. (B) mcm4–GFP chromatin association and DNA staining (DAPI) were determined by fluorescence microscopy at t = 0 and t = 14 h after removal of thiamine. Bar = 10 μm. (C) Proportion of uninucleate cells with mcm4-positive nuclei before and after extraction with a Triton-X-100-containing buffer. (D) Flow cytometric analysis of DNA contents of the cells shown in (B). The DNA content is shown on a log scale.
Discussion
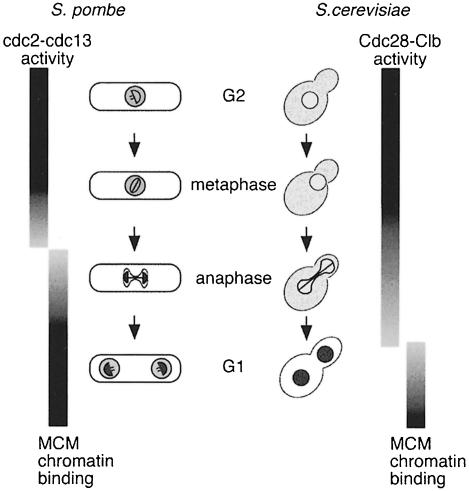
The cytological method described here allows a correlation between the chromatin binding of a specific protein and morphological criteria, such as spindle formation, in individual fission yeast cells. Using the method to analyse mcm4 chromatin binding, our results suggest that pre-RC assembly in fission yeast occurs by a similar mechanism to that operating in budding yeast and Xenopus. Association of mcm4 with chromatin requires a functional ORC complex as well as cdc18 (Figures 5 and 6). By correlating the binding of mcm4 in individual cells with the length of the mitotic spindle, we have shown that chromatin association of mcm4 occurs before the end of mitosis, and is prominent in mid-anaphase B (Figures 4 and 9). This contrasts with the budding yeast situation (Figure 9), where MCM proteins are excluded from the nucleus until the end of anaphase (Hennessy et al., 1990; Yan et al., 1993; Dalton and Whitbread, 1995; Labib et al., 1999), thereby delaying the point at which chromatin binding can occur. The situation in fission yeast may also be distinct from that in mammalian cells, where association of MCM proteins with chromatin appears to occur during telophase (Kubota et al., 1995; Tsuruga et al., 1997).
Fig. 9. Timing of association of MCM proteins with chromatin during the fission and budding yeast cell cycles. Unbound MCM protein is represented by light grey shading, bound by dark grey. For details see text.
In both budding and fission yeasts, it is likely that the timing of MCM chromatin association and pre-RC assembly is determined by the kinetics of CDK inactivation during mitosis (Figure 9). In budding yeast, Cdc28 kinase activity inhibits nuclear accumulation (Labib et al., 1999) and chromatin association of MCMs (Tanaka et al., 1997), together with the assembly of pre-RCs (Dahmann et al., 1995; Detweiler and Li, 1998). Cdc28 is associated with B-type (Clb) cyclin partners during mitosis, and B-cyclin degradation starts at the metaphase to anaphase transition, although persistence of Clb2–Cdc28 activity during anaphase is likely to inhibit pre-RC formation until the end of mitosis (Irniger et al., 1995; Visintin et al., 1997; reviewed by Zachariae and Nasmyth, 1999). In fission yeast, transcription of cdc18+ begins at metaphase (Baum et al., 1998), but cdc18 protein cannot accumulate at this point owing to its destabilization by cdc2 phosphorylation (Jallepalli et al., 1997; Baum et al., 1998; Lopez Girona et al., 1998). Cdc13, which is the major mitotic B-type cyclin partner of cdc2, is degraded during mid-anaphase (Booher et al., 1989; Moreno et al., 1989), and this is likely to trigger mcm4 chromatin binding at this point by allowing accumulation of cdc18 protein. It remains to be established whether other components of fission yeast pre-RCs associate with chromatin with similar kinetics to mcm4.
Our data suggest that pre-RC formation in fission yeast occurs earlier during mitosis than in budding yeast and mammalian cells. This is likely to be important, as the G1 phase of the S.pombe cell cycle is very short and the initiation of DNA replication occurs soon after mitotic exit. Mitotic pre-RC formation may also occur in other eukaryotic cell cycles where the G1 phase is very short or non-existent, such as in early Xenopus or Drosophila development, when embryonic cells cycle rapidly between S and M phases. Early in Xenopus development, individual chromosomes become surrounded by a membrane during anaphase, to form karyomeres. MCM proteins accumulate within such karyomeres (Lemaitre et al., 1998), and it is therefore possible that chromatin association may also occur during anaphase. In the plasmodial phase of Physarum, the absence of a G1 phase presumably also requires MCM chromatin association to occur in anaphase, to allow DNA replication to commence in telophase (Pierron and Bénard, 1996). The assembly of pre-RCs during anaphase implies that chromosome condensation during this phase of the cell cycle does not bar access of MCM and Cdc18 proteins to chromatin. In this regard, it is possible that mitotic chromosome organization may have an influence on replication origin distribution, and it will be interesting to determine whether mitotic pre-RC assembly requires additional factors not necessary for pre-RC formation in G1.
Many proteins, of which pre-RC components are just one class, show periodicity in chromatin binding, and the assay described here should be generally useful for their analysis. These include other replication components such as DNA polymerase α (Desdouets et al., 1998), and cohesins, which play a key role in the regulation of chromatin segregation during mitosis (Yanagida, 1998; Nasmyth, 1999).
Materials and methods
Fission yeast strains and methods
All strains used were constructed by standard genetic methods and are shown in Table I. Strains were grown in rich medium (YE) or minimal medium (EMM) as previously described (Moreno et al., 1991). Repression of transcription from the nmt1 promoter was achieved by addition of thiamine (5 μg/ml) to EMM.
Table I. Schizosaccharomyces pombe strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| K154 | cdc21-M68 nmt1(3X)-cdc18+::leu1+ leu1-32 | this work |
| P194 | nmt1(3X)-cdc18+::leu1+ leu1-32 | Nishitani and Nurse (1995) |
| P560 | cdc21+-GFP::ura4+ ura4-D18 leu-32 h+ | this work |
| P592 | cdc21+-GFP:: ura4+ orc1-4(orp1-4) leu1-32 | derived from Grallert and Nurse (1996) |
| P597 | cdc21+-GFP:: ura4+ nmt1(3X)-cdc18+-leu1+ leu1-32 ura4-D18 | derived from Nishitani and Nurse (1995) |
| P624 | cdc21+-GFP::ura4+ cdc19-P1 leu1-32 ura4 ade6 h– | derived from Nasmyth and Nurse (1981) |
| P661 | cdc21+-GFP:: ura4+ nmt1(41X)-cdc18+::cdc18Δ::leu1+ ura4-D18 ade6 leu1-32 | derived from YMF15 (Muzi-Falconi et al., 1996) |
| P669 | cdc21+-GFP::ura4+ mis5-268 leu1-32 ura4-D18 | derived from Takahashi et al. (1994) |
In situ chromatin binding assay
A 1/100 volume of 10% NaN3 was added to cultures typically containing 108 cells. Cells were washed in ZM buffer [50 mM sodium citrate pH 5.6, 1.2 M sorbitol, 0.5 mM MgAc, 10 mM dithiothreitol (DTT)], resuspended in ZM buffer containing 2 mg/ml zymolyase and incubated at 32°C until cells were >95% phase dark after lysis by SDS. Three volumes of cold STOP buffer (0.1 M MES pH 6.4, 1.2 M sorbitol, 1 mM EDTA, 0.5 mM MgAc) were added, and cells were washed twice in STOP buffer. Cells were extracted using conditions similar to those developed for use with the budding yeast chromatin extraction assay (Donovan et al., 1997). Cells were washed in EB (20 mM PIPES–KOH pH 6.8, 0.4 M sorbitol, 2 mM MgAc, 150 mM KAc) and resuspended in EB containing 1/1000 volume of protease inhibitor cocktail (Sigma P-8215). The suspension was split and 1/10 volume of EBT [EB containing 10% (w/v) Triton X-100] was added to half the culture. After incubation at 20°C for 7 min, cells were spun down and resuspended in methanol. Finally, cells were recentrifuged and resuspended in acetone. For fluorescence microscopy, cells in acetone were spread on polylysine-coated slides and mounted in 50% glycerol–phosphate-buffered saline (PBS) containing 0.4 μg/ml DAPI.
For DNase I digestion, following the STOP and EB washes, cells were resuspended in EB containing 5 mM MgAc, 1/1000 volume of protease inhibitor cocktail (Sigma P-8215) and 1% (w/v) Triton X-100. The suspension was split, and 1/10 volume of 1 mg/ml DNase I (Boehringer) was added to half the culture (Todorov et al., 1995). Following incubation at 0°C for 30 min, NaCl was added to 250 mM to both fractions, and the cells were spun down and fixed as above. The use of 250 mM NaCl was necessary in order to solubilize digested chromatin, and had no effect on mcm4 chromatin binding at the concentration used (data not shown; Donovan et al., 1997).
For anti-tubulin staining of extracted cells, cells in acetone were spread on polylysine-coated coverslips and incubated in PBSBAL (100 mM lysine hydrochloride, 10 mM sodium phosphate pH 6.9, 120 mM NaCl, 2.7 mM potassium chloride, 0.01% sodium azide, 1% bovine serum albumin) for 30 min. A 20 μl aliquot of primary anti-α-tubulin antibody in PBSBAL (TAT1; Woods et al., 1989) was added and coverslips were incubated under humid conditions for at least 1 h. Coverslips were washed in PBSBAL and incubated with secondary antibody (anti-mouse IgG, Texas red conjugated; Vector Labs) for at least 1 h. Finally, coverslips were washed in PBS and mounted in 50% glycerol–PBS containing 0.4 μg/ml DAPI.
For comparative fluorescence microscopy of Triton-extracted and non-extracted cells (Figure 2D), a log phase culture of P560 was split in two and one half was then processed as per the in situ chromatin binding assay, while the remaining cells were washed twice in 1 mM CaCl2/Tris-buffered saline (TBS), and incubated in 1 mM CaCl2/TBS containing 2 μg/ml Texas red-conjugated GS-1 lectin (EY Laboratories Inc., San Mateo, CA). Cells were washed with CaCl2/TBS, and fixed in methanol and acetone. Texas red-stained cells were mixed with an equal number of extracted cells in acetone and mounted. The mcm4–GFP signal of non-extracted cells could be compared with extracted cells in the same field, using Texas red fluorescence to identify non-extracted cells unambiguously.
For flow cytometry, methanol/acetone-fixed cells were rehydrated in 50 mM sodium citrate, 0.1 mg/ml RNase A, 2 μg/ml propidium iodide, and incubated at 37°C for 2 h. Cells were analysed using a Coulter Epics XL-MCL.
Construction of mcm4–GFP strains
A general purpose GFP-tagging vector, pSMUG (DDBJ/EMBL/GenBank accession No. AJ250107), was derived from pBluescript KS+ by inserting the ura4+ gene at the NgoMIV site (oligos used for ura4+ PCR were ATCGCCGGCTTAGCTACAAATCCCACTGGC and ATCGCCGGCTTGTGATATTGACGAAAC), and GFP5 (Siemering et al., 1996) into the XhoI and SacI sites (oligos used for GFP5 PCR were CGAACTCGAGAAGCTTTAATGAGTAAAGGAGAAGAACTTTTCAC and GGAGAGCTCAGGATCCGTCGACAAGCTCATCATGTT– TGTATAG). The C–terminal encoding region of the cdc21+ gene was inserted into the ApaI and XhoI sites of pSMUG to generate pSMUG-mcm4-GFP (oligos used for cdc21+ PCR were ATAGGGCCCATGCTACAGATATGGAGGTC and GCTCTCGAGCACCGGCACCATCAG– TCTGTGCAATTGAACG). In this construct, an eight amino acid linker region is generated between the C–terminus of mcm4 and GFP. GFP-tagged strains were generated by cleaving pSMUG-mcm4-GFP with EcoNI, and ura4– strains were transformed by electroporation. PCRs were carried out using Vent DNA polymerase (New England Biolabs). Ura+ transformants were checked for integration of pSMUG-mcm4-GFP at the cdc21 locus by colony PCR, and by Western blotting to confirm synthesis of an mcm4–GFP fusion protein (data not shown).
Fluorescence microscopy
To examine mcm4–GFP localization in live cells, cells grown in rich medium were washed twice in EMM, and mounted after mixing with an equal volume of 1.2% low melting temperature agarose in EMM at 37°C. For DAPI staining of live mcm4–GFP cells, the same procedure was used except that EMM was replaced with water and DAPI was included to give a final concentration of 10 μg/ml. All samples were examined using a Zeiss Axioskop microscope, and GFP fluorescence was detected as previously described (Labib et al., 1999). Phase, DAPI and GFP channels for each image were assembled into stacks using NIH Image 1.6 for data quantitation; at least 100 cells were counted for each data point (error bars show the range of at least two experiments). Final image assembly was carried out using Adobe Photoshop.
Protein extracts and Western blots
Protein extracts for Western blotting were made by trichloroacetic acid extraction, as described previously (Foiani et al., 1994). For Western blot analysis, the antibodies anti-mcm4 (Maiorano et al., 1996) and anti-GFP (Sawin et al., 1999) were used. The secondary antibodies were anti-rabbit or anti-mouse IgG–horseradish peroxidase conjugates, used at a dilution of 1/10 000. Detection was performed using the enhanced chemiluminescence procedure (Pierce Supersignal).
Acknowledgments
Acknowledgements
We thank the laboratories of Tom Kelly, Paul Nurse and Mitsuhiro Yanagida for strains, antibodies and plasmids. We are grateful to Jim Haseloff for GFP5, Keith Gull for TAT1 antibody and Tom Chappell for advice on lectin staining. We thank Sue Cotterill Haseloff Bass Hassan and Domenico Maiorano for comments on the manuscript. This work was supported by the EU TMR programme (contract ERB-MRX-CT970125), the Cancer Research Campaign and the Wellcome Trust.
References
- Adachi Y., Usukura, J. and Yanagida, M. (1997) A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells, 2, 467–479. [DOI] [PubMed] [Google Scholar]
- Aparicio O., Weinstein, D. and Bell, S. (1997) Components and dynamics of DNA replication complexes in S.cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Baum B., Nishitani, H., Yanow, S. and Nurse, P. (1998) Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J., 17, 5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R.N., Alfa, C.E., Hyams, J.S. and Beach, D.H. (1989) The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell, 58, 485–497. [DOI] [PubMed] [Google Scholar]
- Chuang R.Y. and Kelly, T.J. (1999) The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl Acad. Sci. USA, 96, 2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne R.K. and Kelly, T.J. (1995) Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J., 14, 6348–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker J.H., Piatti, S., Santocanale, C., Nasmyth, K. and Diffley, J.F. (1996) An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature, 379, 180–182. [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Carpenter, P.B. and Dunphy, W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley, J. and Nasmyth, K. (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol., 5, 1257–1269. [DOI] [PubMed] [Google Scholar]
- Dalton S. and Whitbread, L. (1995) Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl Acad. Sci. USA, 92, 2514–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouets C., Santocanale, C.S., Drury, L., Perkins, G., Foiani, M., Plevani, P. and Diffley, J.F.X. (1998) Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J., 17, 4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detweiler C. and Li, J. (1998) Ectopic induction of Clb2 in early G1 phase is sufficient to block prereplicative complex formation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 95, 2384–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. (1996) Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev., 10, 2819–2830. [DOI] [PubMed] [Google Scholar]
- Diffley J.F., Cocker, J.H., Dowell, S.J. and Rowley, A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood, J., Drury, L. and Diffley, J. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D.D., Kim, S.M., Todorov, I.T. and Huberman, J.A. (1996) Large, complex modular structure of a fission yeast DNA replication origin. Curr. Biol., 6, 467–473. [DOI] [PubMed] [Google Scholar]
- Foiani M., Marini, F., Gamba, D., Lucchini, G. and Plevani, P. (1994) The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol., 14, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert B. and Nurse, P. (1996) The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev., 10, 2644–2654. [DOI] [PubMed] [Google Scholar]
- Hennessy K., Clark, C. and Botstein, D. (1990) Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev., 4, 2252–2263. [DOI] [PubMed] [Google Scholar]
- Irniger S., Piatti, S., Michaelis, C. and Nasmyth, K. (1995) Genes involved in sister chomatid separation are needed for B-type cyclin proteolysis. Cell, 81, 269–288. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Jallepalli P.V., Brown, G.W., Muzi Falconi, M., Tien, D. and Kelly, T.J. (1997) Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev., 11, 2767–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S. and Labib, K. (1998) MCM proteins: evolution, properties and role in eukaryotic DNA replication. Biochim. Biophys. Acta, 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Mimura, S., Nishimoto, S., Takisawa, H. and Nojima, H. (1995) Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell, 81, 601–609. [DOI] [PubMed] [Google Scholar]
- Labib K., Diffley, J.F.X. and Kearsey, S.E. (1999) G1 and B-type cyclins exclude the DNA replication factor Mcm4 from the nucleus. Nature Cell Biol., 1, 415–422. [DOI] [PubMed] [Google Scholar]
- Lemaitre J.-M., Géraud, G., Méchali, M. (1998) Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. J. Cell Biol., 142, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. and Stillman, B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Girona A., Mondesert, O., Leatherwood, J. and Russell, P. (1998) Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol. Biol. Cell, 9, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z. and Nurse, P. (1999) The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci., 112, 3703–3712. [DOI] [PubMed] [Google Scholar]
- Maiorano D., Blom van Assendelft, G. and Kearsey, S.E. (1996) Fission yeast cdc21, a member of the MCM protein family, is required for onset of S phase and located in the nucleus throughout the cell cycle. EMBO J., 15, 861–872. [PMC free article] [PubMed] [Google Scholar]
- Moon K.Y., Kong, D., Lee, J.-K., Raychaudhuri, S. and Hurwitz, J. (1999) Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 96, 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Hayles, J. and Nurse, P. (1989) Regulation of p34cdc2 protein kinase during mitosis. Cell, 58, 361–372. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar, A. and Nurse, P. (1991) Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M., Brown, G.W. and Kelly, T.J. (1996) Cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 93, 1566–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. (1999) Separating sister chromatids. Trends Biochem. Sci., 24, 98–104.10203756 [Google Scholar]
- Nasmyth K.A. and Nurse, P. (1981) Cell division mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 182, 119–124. [DOI] [PubMed] [Google Scholar]
- Nishitani H. and Nurse, P. (1995) p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell, 83, 397–405. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Takahashi, T. and Masukata, H. (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol., 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Satoh, H., Sekiguchi, M. and Masukata, H. (1999) Clustered adenine/thymine stretches are essential for the functioning of a fission yeast replication origin. Mol. Cell. Biol., 19, 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasion S.G. and Forsburg, S.L. (1999) Nuclear localization of S.pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell, 10, 4043–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm, T., Cocker, J.H., Diffley, J.F. and Nasmyth, K. (1996) Activation of S-phase-promoting CDKs in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev., 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Pierron G. and Bénard,M. (1996) DNA replication in Physarum. In DePamphilis,M. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 933–946. [Google Scholar]
- Romanowski P., Madine, M.A., Rowles, A., Blow, J.J. and Laskey, R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Rowles A., Chong, J.P., Brown, L., Howell, M., Evan, G.I. and Blow, J.J. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- Sawin K., Hajibagheri, M.N.A. and Nurse, P. (1999) Misspecification of cortical identity in a fission yeast PAK mutant. Curr. Biol., 9, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Siemering K.R., Golbik, R., Sever, R. and Haseloff, J. (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol., 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Stillman B. (1996) Cell cycle control of DNA replication. Science, 274, 1659–1664. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamada, H. and Yanagida, M. (1994) Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell, 5, 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Knapp, D. and Nasmyth, K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Toda T., Yamamoto, M. and Yanagida, M. (1981) Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe: video fluorescence microscopy of synchronously growing wild-type and cold sensitive cdc mutants by using a DNA-binding fluorescent probe. J. Cell Sci., 52, 271–287. [DOI] [PubMed] [Google Scholar]
- Todorov I.T., Attaran, A. and Kearsey, S.E. (1995) BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J. Cell Biol., 129, 1433–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruga H., Yabuta, N., Hosoya, S., Tamura, K., Endo, Y. and Nojima, H. (1997) HsMCM6: a new member of the human MCM/P1 family encodes a protein homologous to fission yeast Mis5. Genes Cell, 2, 381–399. [DOI] [PubMed] [Google Scholar]
- Tye B. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- Uzawa S. and Yanagida, M. (1992) Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci., 101, 267–275. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz, S. and Amon, A. (1997) CDC20 and CDH1: a family of substrate-specific activators of APC–dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin, T., Sasse, R., McRae, T., Baines, A. and Gull, K. (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci., 93, 491–500. [DOI] [PubMed] [Google Scholar]
- Yan H., Merchant, A.M. and Tye, B.K. (1993) Cell cycle regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev., 7, 2149–2160. [DOI] [PubMed] [Google Scholar]
- Yanagida M. (1998) Fission yeast cut mutations revisited: the control of anaphase. Trends Cell Biol., 8, 144–149. [DOI] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth, K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]